Genetic Diversity and Selection of MHC I-UAA in Clariid Catfish from Thailand: Implications for Breeding and Conservation
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimen Collection and DNA Extraction
2.2. PCR Amplification and IlluminaTM Short-Read Sequencing
2.3. Sequence Quality Control and Read Processing
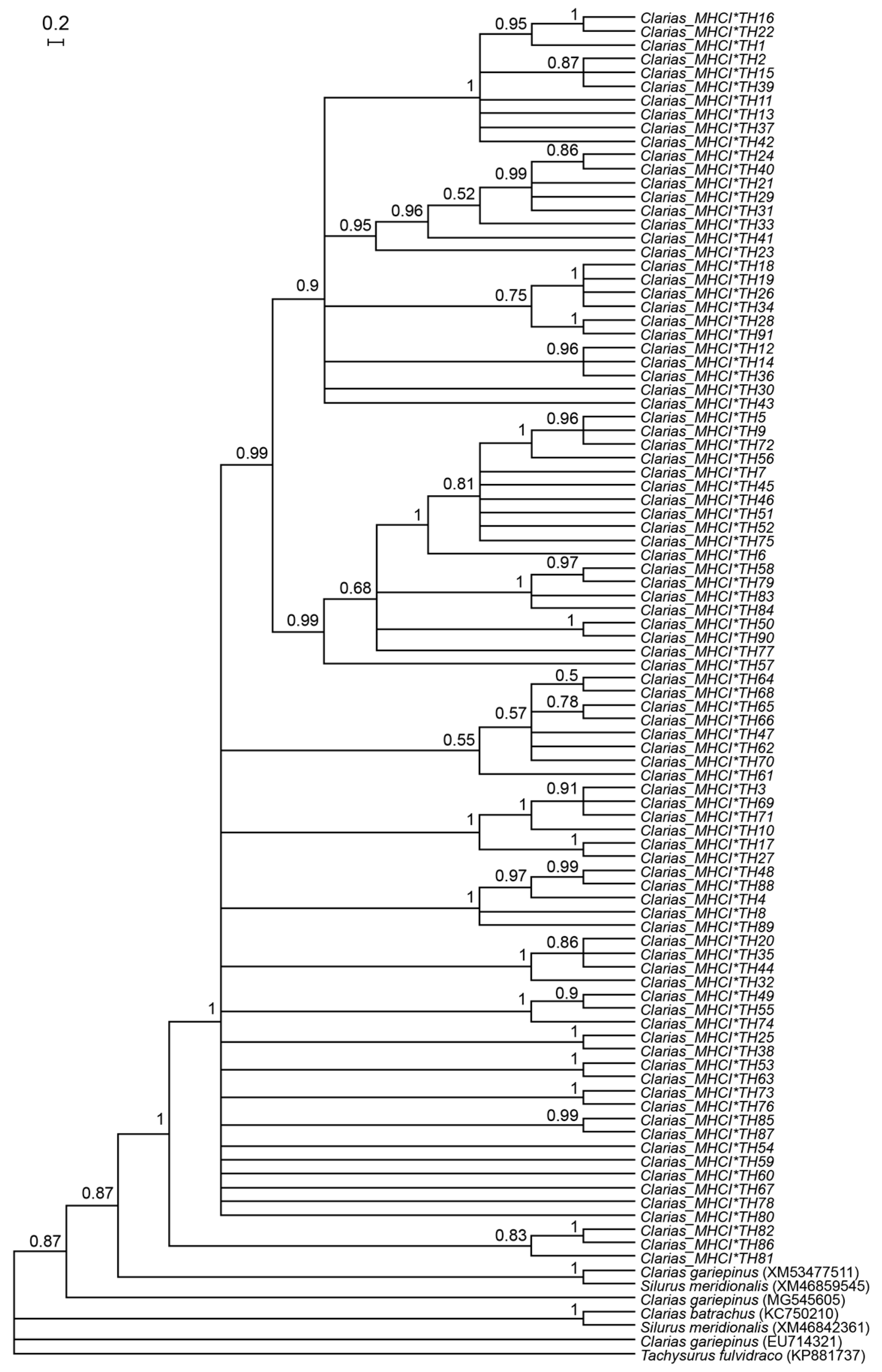
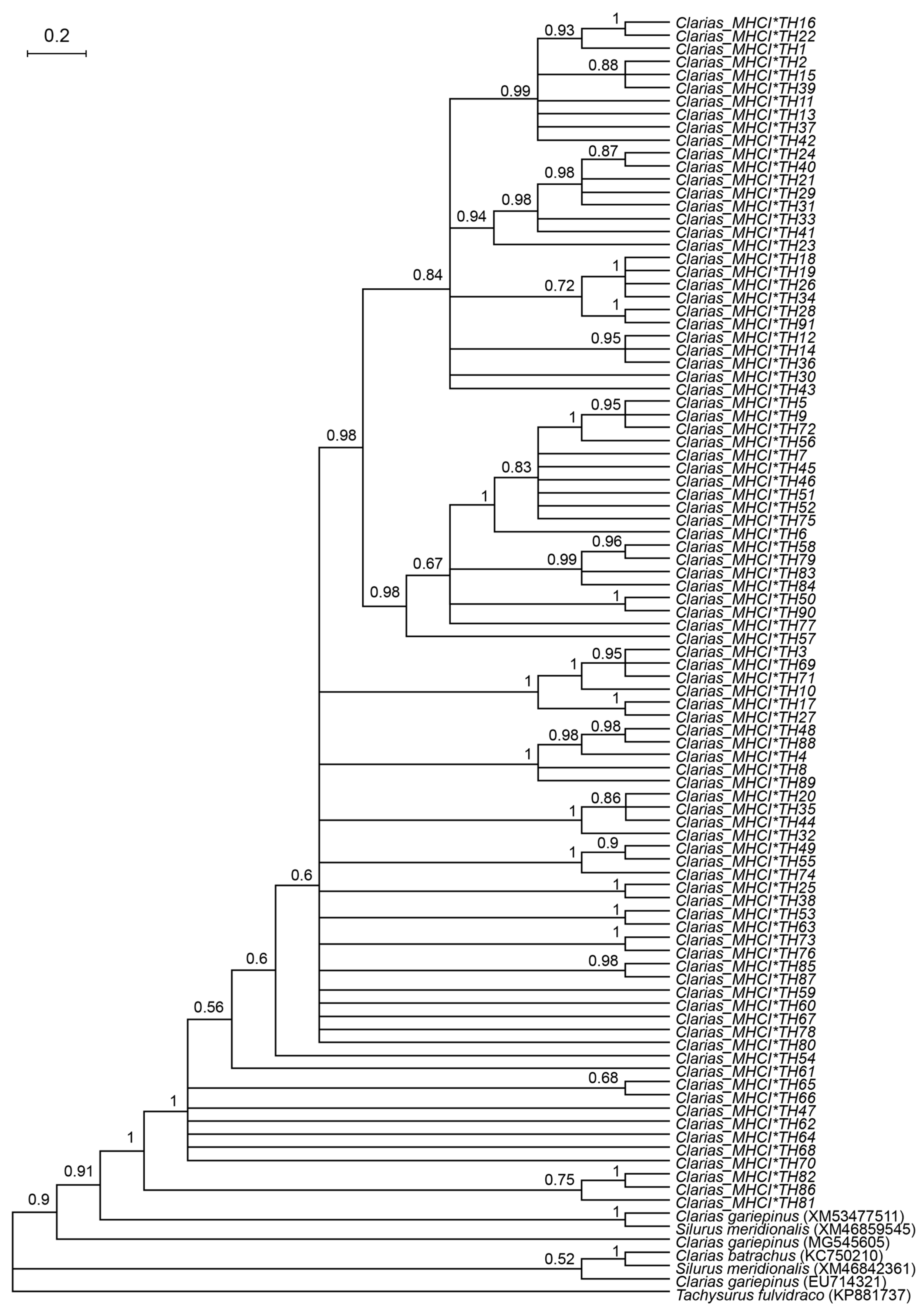
2.4. Genetic Diversity and Phylogenetic Analysis
2.5. Selection Analysis
2.6. Multiple Sequence Alignment of MHC I Amino Acid Residues
3. Results
3.1. Genetic Diversity of Catfish Based on MHC I Gene
3.2. Selection and Selective-Sweep Analyses of Clariid Catfish
3.3. Multiple Sequence Alignment of MHC I Amino Acid Residues and Prediction
4. Discussion
4.1. MHC I-UAA Diversity in Clariid Catfish
4.2. Evidence of Selection at the MHC I-UAA Locus
4.3. Advances and Future Directions in MHC Genotyping
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMOVA | Analysis of Molecular Variance |
| BIC | Bayesian Information Criterion |
| ESU | Evolutionarily Significant Unit |
| FEL | Fixed Effects Likelihood |
| FUBAR | Fast Unconstrained Bayesian Approximation |
| MCMC | Markov Chain Monte Carlo |
| MHC I | Major Histocompatibility Complex class I |
| NCBI | National Center for Biotechnology Information |
| PCR | Polymerase Chain Reaction |
References
- FAO. Top 10 Species Groups in Global Aquaculture; Food and Agriculture Organization of The United Nations: Rome, Italy, 2023. [Google Scholar]
- Garcia de Leaniz, C.; Fleming, I.A.; Einum, S.; Verspoor, E.; Jordan, W.C.; Consuegra, S.; Aubin-Horth, N.; Lajus, D.; Letcher, B.H.; Youngson, A.F.; et al. A critical review of adaptive genetic variation in Atlantic salmon: Implications for conservation. Biol. Rev. 2007, 82, 173–211. [Google Scholar] [CrossRef]
- Neff, B.D.; Garner, S.R.; Pitcher, T.E. Conservation and enhancement of wild fish populations: Preserving genetic quality versus genetic diversity. Can. J. Fish. Aquat. Sci. 2011, 68, 1139–1154. [Google Scholar] [CrossRef]
- Wachirachaikarn, A.; Rungsin, W.; Srisapoome, P.; Na-Nakorn, U. Crossing of African catfish, Clarias gariepinus (Burchell, 1822), strains based on strain selection using genetic diversity data. Aquaculture 2009, 290, 53–60. [Google Scholar] [CrossRef]
- Gjedrem, T.; Robinson, N.; Rye, M. The importance of selective breeding in aquaculture to meet future demands for animal protein: A review. Aquaculture 2012, 350, 117–129. [Google Scholar] [CrossRef]
- Sonesson, A.K.; Hallerman, E.; Humphries, F.; Hilsdorf, A.W.S.; Leskien, D.; Rosendal, K.; Bartley, D.; Hu, X.; Gomez, R.G.; Mair, G.C. Sustainable management and improvement of genetic resources for aquaculture. J. World Aquac. Soc. 2023, 54, 364–396. [Google Scholar] [CrossRef]
- Bijlsma, R.; Loeschcke, V. Environmental stress, adaptation and evolution: An overview. J. Evol. Biol. 2005, 18, 744–749. [Google Scholar] [CrossRef]
- Hoffmann, A.A.; Sgrò, C.M. Climate change and evolutionary adaptation. Nature 2011, 470, 479–485. [Google Scholar] [CrossRef]
- Patta, C.; Sriphairoj, K.; Budi, T.; Quanoo, D.K.; Nguyen, T.H.D.; Jaito, W.; Chalermwong, P.; Pongsanarm, T.; Thatukan, C.; Wongloet, W.; et al. Genetic Structural Changes While Maintaining Effective Population Size of Bighead Catfish in Nong Han Lake: Implications of Metapopulation Dynamics or Release Activities. Fish. Manag. Ecol. 2024, 32, e12778. [Google Scholar] [CrossRef]
- Department of Fisheries. Marine Fisheries Management Plan of Thailand 2020–2022; Ministry of Agriculture and Cooperatives, Thailand: Bangkok, Thailand, 2023.
- Srikulnath, K.; Budi, T.; Panthum, T.; Sinthuvanich, C.; Haileselasie, T.H.; Kuldilok, K.; Photchanaprasert, N.; Sayruamyat, S.; Muangmai, N.; Duengkae, P.; et al. Sustainable Aquaculture in Thailand: Balancing Economic Growth and Ecological Integrity with North African Catfish Integration. Aquac. Res. 2025, in press. [CrossRef]
- Sciuto, S.; Colli, L.; Fabris, A.; Pastorino, P.; Stoppani, N.; Esposito, G.; Prearo, M.; Esposito, G.; Ajmone-Marsan, P.; Acutis, P.L.; et al. What can genetics do for the control of infectious diseases in aquaculture? Animals 2022, 12, 2176. [Google Scholar] [CrossRef]
- Wang, B.; Du, H.-h.; Huang, H.-q.; Xian, J.-a.; Xia, Z.-h.; Hu, Y.-h. Major histocompatibility complex class I (MHC Iα) of Japanese flounder (Paralichthys olivaceus) plays a critical role in defense against intracellular pathogen infection. Fish Shellfish. Immunol. 2019, 94, 122–131. [Google Scholar] [CrossRef]
- Olson, E.; Raghavan, M. Major histocompatibility complex class I assembly within endolysosomal pathways. Curr. Opin. Immunol. 2023, 84, 102356. [Google Scholar] [CrossRef]
- Janeway, C.A.; Travers, P.; Walport, M.; Shlomchik, M.J. The major histocompatibility complex and its functions. In Immunobiology: The Immune System in Health and Disease, 5th ed.; Garland Science: New York, NY, USA, 2001. [Google Scholar]
- Grimholt, U. MHC and Evolution in Teleosts. Biology 2016, 5, 6. [Google Scholar] [CrossRef]
- Piertney, S.B.; Oliver, M.K. The evolutionary ecology of the major histocompatibility complex. Heredity 2006, 96, 7–21. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Dijkstra, J.M. Major histocompatibility complex (MHC) genes and disease resistance in fish. Cells 2019, 8, 378. [Google Scholar] [CrossRef] [PubMed]
- Spurgin, L.G.; Richardson, D.S. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc. R. Soc. B Biol. Sci. 2010, 277, 979–988. [Google Scholar] [CrossRef] [PubMed]
- Azis; Alimuddin; Sukenda; Zairin, M. MHC I molecular marker inheritance and first generation catfish (Clarias sp.) resistance against Aeromonas hydrophila infection. Pak. J. Biotechnol. 2015, 12, 131–137. [Google Scholar]
- Oyebola, O.O.; Babaoye, E.O.; Anifowose, O.R. Growth performance, survival rate and expression of MHC I gene in Clarias gariepinus at post Vagococcus carniphilus challenge period. Ecol. Genet. Genom. 2025, 34, 100319. [Google Scholar] [CrossRef]
- Castro-Prieto, A.; Wachter, B.; Sommer, S. Cheetah paradigm revisited: MHC diversity in the world’s largest free-ranging population. Mol. Biol. Evol. 2011, 28, 1455–1468. [Google Scholar] [CrossRef]
- Smallbone, W.; Ellison, A.; Poulton, S.; van Oosterhout, C.; Cable, J. Depletion of MHC supertype during domestication can compromise immunocompetence. Mol. Ecol. 2021, 30, 736–746. [Google Scholar] [CrossRef]
- Sommer, S. The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Front. Zool. 2005, 2, 16. [Google Scholar] [CrossRef] [PubMed]
- O’Farrell, B.; Benzie, J.A.H.; McGinnity, P.; de Eyto, E.; Dillane, E.; Coughlan, J.; Cross, T.F.; Tramontano, A. Selection and phylogenetics of salmonid MHC class I: Wild brown trout (Salmo trutta) differ from a non-native introduced strain. PLoS ONE 2013, 8, e63035. [Google Scholar] [CrossRef] [PubMed]
- Schenekar, T.; Weiss, S. Selection and genetic drift in captive versus wild populations: An assessment of neutral and adaptive (MHC-linked) genetic variation in wild and hatchery brown trout (Salmo trutta) populations. Conserv. Genet. 2017, 18, 1011–1022. [Google Scholar] [CrossRef]
- Stet, R.M.; van Erp, S.H.; Hermsen, T.; Sültmann, H.A.; Egberts, E. Polymorphism and estimation of the number of MhcCyca class I and class I genes in laboratory strains of the common carp (Cyprinus carpio L.). Dev. Comp. Immunol. 1993, 17, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Yáñez, J.M.; Houston, R.D.; Newman, S. Genetics and genomics of disease resistance in salmonid species. Front. Genet. 2014, 5, 415. [Google Scholar] [CrossRef]
- Liu, W.; Liu, Y.; Liu, P.; Zhao, W. Genetic diversity of major histocompatibility complex class I genes in Zootoca vivipara. Biosci. Rep. 2020, 40, BSR20193809. [Google Scholar] [CrossRef]
- Supikamolseni, A.; Ngaoburanawit, N.; Sumontha, M.; Chanhome, L.; Suntrarachun, S.; Peyachoknagul, S.; Srikulnath, K. Molecular barcoding of venomous snakes and species-specific multiplex PCR assay to identify snake groups for which antivenom is available in Thailand. Genet. Mol. Res. 2015, 14, 13981–13997. [Google Scholar] [CrossRef]
- Imron, I.; Marnis, H.; Iswanto, B.; Suprapto, R. Development of a PCR marker for the identification of resistance to Motile Aeromonad Septicemia disease in African catfish (Clarias gariepinus). Aquac. Aquar. Conserv. Legis. 2020, 13, 1255–1267. [Google Scholar]
- Simon, A. FastQC: A Quality Control Tool for High Throughput Sequence Data, Version 0.10; Babraham Bioinformatics: Cambridge, UK, 2010.
- Sebastian, A.; Herdegen, M.; Migalska, M.; Radwan, J. amplisas: A web server for multilocus genotyping using next-generation amplicon sequencing data. Mol. Ecol. Resour. 2016, 16, 498–510. [Google Scholar] [CrossRef]
- Lighten, J.; Van Oosterhout, C.; Bentzen, P. Critical review of NGS analyses for de novo genotyping multigene families. Mol. Ecol. 2014, 23, 3957–3972. [Google Scholar] [CrossRef]
- Antao, A.B.; Chinchar, V.G.; McConnell, T.J.; Miller, N.W.; Clem, L.W.; Wilson, M.R. MHC class I genes of the channel catfish: Sequence analysis and expression. Immunogenetics 1999, 49, 303–311. [Google Scholar] [CrossRef]
- FBN. BioProject: Clarias Gariepinus RefSeq Genome Sequencing and Assembly; National Center for Biotechnology Information: Bethesda, MD, USA, 2022. [Google Scholar]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023; Available online: https://www.R-project.org/ (accessed on 5 May 2025).
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2014, e281. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Murrell, B.; O Wertheim, J.; Moola, S.; Weighill, T.; Scheffler, K.; Pond, S.L.K. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012, 8, e1002764. [Google Scholar] [CrossRef]
- Kosakovsky Pond, S.L.; Frost, S.D. Not so different after all: A comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 2005, 22, 1208–1222. [Google Scholar] [CrossRef] [PubMed]
- Murrell, B.; Moola, S.; Mabona, A.; Weighill, T.; Sheward, D.; Pond, S.L.K.; Scheffler, K. FUBAR: A fast, unconstrained bayesian approximation for inferring selection. Mol. Biol. Evol. 2013, 30, 1196–1205. [Google Scholar] [CrossRef]
- Pearson, W.R. An introduction to sequence similarity (“homology”) searching. Curr. Protoc. Bioinform. 2013, 42, 3.1.1–3.1.8. [Google Scholar] [CrossRef] [PubMed]
- Meiklejohn, K.A.; Damaso, N.; Robertson, J.M.; Fugmann, S.D. Assessment of BOLD and GenBank–Their accuracy and reliability for the identification of biological materials. PLoS ONE 2019, 14, e0217084. [Google Scholar] [CrossRef]
- Sato, A.; Dongak, R.; Hao, L.; Takezaki, N.; Shintani, S.; Aoki, T.; Klein, J. MHC class I genes of the cichlid fish Oreochromis niloticus. Immunogenetics 2006, 58, 917–928. [Google Scholar] [CrossRef]
- Zhou, T.; Yuan, Z.; Tan, S.; Jin, Y.; Yang, Y.; Shi, H.; Wang, W.; Niu, D.; Gao, L.; Jiang, W.; et al. A review of molecular responses of catfish to bacterial diseases and abiotic stresses. Front. Physiol. 2018, 9, 1113. [Google Scholar] [CrossRef] [PubMed]
- Segaran, T.C.; Azra, M.N.; Piah, R.M.; Lananan, F.; Téllez-Isaías, G.; Gao, H.; Torsabo, D.; Kari, Z.A.; Noordin, N.M. Catfishes: A global review of the literature. Heliyon 2023, 9, e20081. [Google Scholar] [CrossRef]
- Zhou, X.; Tang, J.; Wang, S.; Zhang, Y.; Ye, H.; Zhang, Q.; Xiang, W.; Cai, T.; Zeng, C. Whole genome sequencing and transcriptomics-based characterization of a novel β-cypermethrin-degrading Gordonia alkanivorans GH-1 isolated from fermented foods. Chemosphere 2023, 320, 138017. [Google Scholar] [CrossRef]
- De Alwis, P.S.; Kundu, S.; Gietbong, F.Z.; Amin, M.H.F.; Lee, S.-R.; Kim, H.-W.; Kim, A.R. Mitochondriomics of Clarias fishes (Siluriformes: Clariidae) with a new assembly of Clarias camerunensis: Insights into the genetic characterization and diversification. Life 2023, 13, 482. [Google Scholar] [CrossRef]
- Klein, J.; Sato, A.; Nagl, S.; O’hUigín, C. Molecular trans-species polymorphism. Annu. Rev. Ecol. Syst. 1998, 29, 1–21. [Google Scholar] [CrossRef]
- Hedrick, P.W. Adaptive introgression in animals: Examples and comparison to new mutation and standing variation as sources of adaptive variation. Mol. Ecol. 2013, 22, 4606–4618. [Google Scholar] [CrossRef] [PubMed]
- Gómez, D.; Conejeros, P.; Consuegra, S.; Marshall, S.H. MHC mediated resistance to Piscirickettsia salmonis in salmonids farmed in Chile. Aquaculture 2011, 318, 15–19. [Google Scholar] [CrossRef]
- Million, K.M.; Lively, C.M. Trans-specific polymorphism and the convergent evolution of supertypes in major histocompatibility complex class II genes in darters (Etheostoma). Ecol. Evol. 2022, 12, e8485. [Google Scholar] [CrossRef]
- Charlesworth, B.; Jensen, J.D. Effects of selection at linked sites on patterns of genetic variability. Annu. Rev. Ecol. Evol. Syst. 2021, 52, 177–197. [Google Scholar] [CrossRef]
- Scheiblhofer, S.; Laimer, J.; Machado, Y.; Weiss, R.; Thalhamer, J. Influence of protein fold stability on immunogenicity and its implications for vaccine design. Expert Rev. Vaccines 2017, 16, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Domnick, A.; Winter, C.; Sušac, L.; Hennecke, L.; Hensen, M.; Zitzmann, N.; Trowitzsch, S.; Thomas, C.; Tampé, R. Molecular basis of MHC I quality control in the peptide loading complex. Nat. Commun. 2022, 13, 4701. [Google Scholar] [CrossRef] [PubMed]
- Alimuddin, A.; Putri, F.M.; Wahjuningrum, D.; Hardiantho, D.; Sunarma, A.; Nuryati, S. Resistance against Aeromonas hydrophila infection and growth of second generation (F2) African catfish (Clarias gariepinus) using selected molecular markers. Biotropia 2018, 25, 95–102. [Google Scholar] [CrossRef]
- Lenz, T.L. Adaptive value of novel MHC immune gene variants. Proc. Natl. Acad. Sci. USA 2018, 115, 1414–1416. [Google Scholar] [CrossRef]
- López, M.E.; Benestan, L.; Moore, J.; Perrier, C.; Gilbey, J.; Di Genova, A.; Maass, A.; Diaz, D.; Lhorente, J.; Correa, K.; et al. Comparing genomic signatures of domestication in two Atlantic salmon (Salmo salar L.) populations with different geographical origins. Evol. Appl. 2019, 12, 137–156. [Google Scholar] [CrossRef] [PubMed]
- Dionne, M.; Miller, K.M.; Dodson, J.J.; Bernatchez, L. MHC standing genetic variation and pathogen resistance in wild Atlantic salmon. Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 1555–1565. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sun, Y.; Xu, T. Identification of 48 full-length MHC-DAB functional alleles in miiuy croaker and evidence for positive selection. Fish Shellfish. Immunol. 2016, 54, 544–550. [Google Scholar] [CrossRef]
- Wegner, K. Historical and contemporary selection of teleost MHC genes: Did we leave the past behind? J. Fish Biol. 2008, 73, 2110–2132. [Google Scholar] [CrossRef]
- Loh, Z.; Huan, X.; Awate, S.; Schrittwieser, M.; Renia, L.; Ren, E.C. Molecular characterization of MHC class I alpha 1 and 2 domains in Asian seabass (Lates calcarifer). Int. J. Mol. Sci. 2022, 23, 10688. [Google Scholar] [CrossRef] [PubMed]
- Larson, W.A.; Seeb, J.E.; Dann, T.H.; Schindler, D.E.; Seeb, L.W. Signals of heterogeneous selection at an MHC locus in geographically proximate ecotypes of sockeye salmon. Mol. Ecol. 2014, 23, 5448–5461. [Google Scholar] [CrossRef] [PubMed]
- D’ambrosio, J.; Phocas, F.; Haffray, P.; Bestin, A.; Brard-Fudulea, S.; Poncet, C.; Quillet, E.; Dechamp, N.; Fraslin, C.; Charles, M.; et al. Genome-wide estimates of genetic diversity, inbreeding and effective size of experimental and commercial rainbow trout lines undergoing selective breeding. Genet. Sel. Evol. 2019, 51, 26. [Google Scholar] [CrossRef]
- Hedrick, P.W. Pathogen resistance and genetic variation at MHC loci. Evolution 2002, 56, 1902–1908. [Google Scholar] [CrossRef]
- Evans, K.M.; Wortley, A.H.; Mann, D.G. An assessment of potential diatom “barcode” genes (cox1, rbcL, 18S and ITS rDNA) and their effectiveness in determining relationships in Sellaphora (Bacillariophyta). Protist 2007, 158, 349–364. [Google Scholar] [CrossRef]
| Species | Population | Code | N | Number of Allele per Population | Nucleotide Diversity |
|---|---|---|---|---|---|
| C. gariepinus | Nakhon Nayok | NYK-CG-C | 5 | 3 | 0.074 |
| Kalasin 1 | KSN1-CG-C | 94 | 20 | 0.111 | |
| Kalasin 2 | KSN2-CG-C | 134 | 37 | 0.082 | |
| Ubon Ratchathani | UBR-CG-C | 6 | 3 | 0.091 | |
| Sing Buri | SB-CG-C | 7 | 7 | 0.118 | |
| C. gariepinus | - | 246 | 41 | 0.095 | |
| SD | - | - | - | 0.017 | |
| C. macrocephalus | Sing Buri | SB-CM-C | 4 | 4 | 0.240 |
| Sakon Nakhon 1 | SNK1-CM-W | 182 | 38 | 0.133 | |
| Sakon Nakhon 2 | SNK2-CM-W | 74 | 34 | 0.121 | |
| Sakon Nakhon 3 | SNK3-CM-W | 82 | 16 | 0.104 | |
| Sakon Nakhon 4 | SNK4-CM-C | 14 | 5 | 0.132 | |
| Suphan Buri 1 | SPB1-CM-W | 6 | 11 | 0.123 | |
| Suphan Buri 2 | SPB2-CM-W | 3 | 10 | 0.127 | |
| Nakhon Pathom 1 | NPT1-CM-W | 2 | 8 | 0.131 | |
| Nakhon Pathom 2 | NPT2-CM-W | 2 | 7 | 0.144 | |
| Nakhon Si Thammarat 1 | NST1-CM-W | 3 | 6 | 0.145 | |
| Nakhon Si Thammarat 2 | NST2-CM-C | 10 | 13 | 0.136 | |
| Surat Thani | STN-CM-C | 25 | 26 | 0.114 | |
| Nakhon Phanom | NKPN-CM-C | 7 | 4 | 0.084 | |
| Ubon Ratchathani | UBR-CM-C | 6 | 10 | 0.128 | |
| C. macrocephalus | - | 420 | 59 | 0.133 | |
| SD | - | - | - | 0.033 | |
| C. batrachus | Ubon Ratchathani | UBR-CB-C | 8 | 6 | 0.087 |
| Overall mean value | - | 674 | 91 | 0.121 | |
| SD | - | - | - | 0.034 |
| Species | Population | Code | Tajima’s D | Fu and Li’s D | Fu and Li’s F |
|---|---|---|---|---|---|
| C. gariepinus | Nakhon Nayok | NYK-CG-C | 0.604 ns | 0.750 ns | 0.792 ns |
| Kalasin 1 | KSN1-CG-C | 0.228 ns | 1.356 ns | 1.001 ns | |
| Kalasin 2 | KSN2-CG-C | −0.699 ns | 0.045 ns | −0.388 ns | |
| Ubon Ratchathani | UBR-CG-C | −0.507 ns | −0.683 ns | −0.716 ns | |
| Sing Buri | SB-CG-C | −0.869 ns | −0.957 ns | −1.064 ns | |
| Mean | - | −0.249 | 0.102 | −0.075 | |
| C. macrocephalus | Sing Buri | SB-CM-C | −1.292 ns | −1.188 ns | −1.330 ns |
| Sakon Nakhon 1 | SNK1-CM-W | −0.936 ns | −3.082 * | −2.392 * | |
| Sakon Nakhon 2 | SNK2-CM-W | −1.142 ns | −2.370 * | −2.114 ns | |
| Sakon Nakhon 3 | SNK3-CM-W | −0.032 ns | 0.336 ns | 0.206 ns | |
| Sakon Nakhon 4 | SNK4-CM-C | −1.700 ns | −2.704 * | −2.800 * | |
| Suphan Buri 1 | SPB1-CM-W | −0.721 ns | 0.012 ns | −0.247 ns | |
| Suphan Buri 2 | SPB2-CM-W | −0.521 ns | 0.607 ns | 0.326 ns | |
| Nakhon Pathom 1 | NPT1-CM-W | −0.614 ns | 0.112 ns | −0.085 ns | |
| Nakhon Pathom 2 | NPT2-CM-W | −0.800 ns | −0.347 ns | −0.505 ns | |
| Nakhon Si Thammarat 1 | NST1-CM-W | −0.085 ns | 0.613 ns | 0.495 ns | |
| Nakhon Si Thammarat 2 | NST2-CM-C | −0.542 ns | −0.727 ns | −0.786 ns | |
| Surat Thani | STN-CM-C | −0.935 ns | 1.288 ns | 0.404 ns | |
| Nakhon Phanom | NKPN-CM-C | −0.879 ns | −0.906 ns | −1.010 ns | |
| Ubon Ratchathani | UBR-CM-C | −1.478 ns | −1.495 ns | −1.713 ns | |
| Mean | - | −0.834 | −0.704 | −0.825 | |
| C. batrachus | Ubon Ratchathani | UBR-CB-C | −0.670 ns | −1.453 ns | −1.422 ns |
| Overall mean value | - | −0.679 | −0.540 | −0.667 |
| Species | Population | Code | dN | dS | ω |
|---|---|---|---|---|---|
| C. gariepinus | Nakhon Nayok | NYK-CG-C | 0.049 ± 0.018 | 0.023 ± 0.016 | 2.130 |
| Kalasin 1 | KSN1-CG-C | 0.094 ± 0.020 | 0.037 ± 0.017 | 2.541 | |
| Kalasin 2 | KSN2-CG-C | 0.064 ± 0.012 | 0.020 ± 0.008 | 3.200 | |
| Ubon Ratchathani | UBR-CG-C | 0.035 ± 0.013 | 0.011 ± 0.008 | 3.182 | |
| Sing Buri | SB-CG-C | 0.077 ± 0.021 | 0.044 ± 0.019 | 1.750 | |
| Mean | - | 0.064 ± 0.017 | 0.027 ± 0.014 | 2.561 | |
| C. macrocephalus | Sing Buri | SB-CM-C | 0.502 ± 0.130 | 0.141 ± 0.049 | 3.560 |
| Sakon Nakhon 1 | SNK1-CM-W | 0.120 ± 0.023 | 0.090 ± 0.033 | 1.333 | |
| Sakon Nakhon 2 | SNK2-CM-W | 0.120 ± 0.021 | 0.012 ± 0.037 | 10.000 | |
| Sakon Nakhon 3 | SNK3-CM-W | 0.102 ± 0.023 | 0.084 ± 0.037 | 1.214 | |
| Sakon Nakhon 4 | SNK4-CM-C | 0.200 ± 0.043 | 0.063 ± 0.021 | 3.175 | |
| Suphan Buri 1 | SPB1-CM-W | 0.088 ± 0.019 | 0.097 ± 0.040 | 0.907 | |
| Suphan Buri 2 | SPB2-CM-W | 0.102 ± 0.021 | 0.098 ± 0.043 | 1.041 | |
| Nakhon Pathom 1 | NPT1-CM-W | 0.050 ± 0.012 | 0.034 ± 0.015 | 1.471 | |
| Nakhon Pathom 2 | NPT2-CM-W | 0.066 ± 0.018 | 0.093 ± 0.046 | 0.710 | |
| Nakhon Si Thammarat 1 | NST1-CM-W | 0.059 ± 0.018 | 0.087 ± 0.041 | 0.678 | |
| Nakhon Si Thammarat 2 | NST2-CM-C | 0.070 ± 0.016 | 0.080 ± 0.037 | 0.875 | |
| Surat Thani | STN-CM-C | 0.122 ± 0.022 | 0.117 ± 0.043 | 1.043 | |
| Nakhon Phanom | NKPN-CM-C | 0.062 ± 0.017 | 0.052 ± 0.027 | 1.192 | |
| Ubon Ratchathani | UBR-CM-C | 0.139 ± 0.022 | 0.132 ± 0.049 | 1.053 | |
| Mean | - | 0.129 ± 0.029 | 0.084 ± 0.037 | 2.018 | |
| C. batrachus | Ubon Ratchathani | UBR-CB-C | 0.107 ± 0.029 | 0.097 ± 0.052 | 1.103 |
| Overall mean value | - | 0.111 ± 0.097 | 0.071 ± 0.039 | 1.563 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, T.H.D.; Chalermwong, P.; Patta, C.; Jaito, W.; Singchat, W.; Panthum, T.; Budi, T.; Sriphairoj, K.; Hatachote, S.; Srisapoome, P.; et al. Genetic Diversity and Selection of MHC I-UAA in Clariid Catfish from Thailand: Implications for Breeding and Conservation. Genes 2025, 16, 1106. https://doi.org/10.3390/genes16091106
Nguyen THD, Chalermwong P, Patta C, Jaito W, Singchat W, Panthum T, Budi T, Sriphairoj K, Hatachote S, Srisapoome P, et al. Genetic Diversity and Selection of MHC I-UAA in Clariid Catfish from Thailand: Implications for Breeding and Conservation. Genes. 2025; 16(9):1106. https://doi.org/10.3390/genes16091106
Chicago/Turabian StyleNguyen, Ton Huu Duc, Piangjai Chalermwong, Chananya Patta, Wattanawan Jaito, Worapong Singchat, Thitipong Panthum, Trifan Budi, Kednapat Sriphairoj, Sittichai Hatachote, Prapansak Srisapoome, and et al. 2025. "Genetic Diversity and Selection of MHC I-UAA in Clariid Catfish from Thailand: Implications for Breeding and Conservation" Genes 16, no. 9: 1106. https://doi.org/10.3390/genes16091106
APA StyleNguyen, T. H. D., Chalermwong, P., Patta, C., Jaito, W., Singchat, W., Panthum, T., Budi, T., Sriphairoj, K., Hatachote, S., Srisapoome, P., Muangmai, N., Griffin, D. K., Antunes, A., Duengkae, P., & Srikulnath, K. (2025). Genetic Diversity and Selection of MHC I-UAA in Clariid Catfish from Thailand: Implications for Breeding and Conservation. Genes, 16(9), 1106. https://doi.org/10.3390/genes16091106












