Hi-C Technology Reveals Actionable Gene Fusions and Rearrangements in Diffuse Large B-Cell Lymphoma Unidentified by Conventional FISH
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Selection
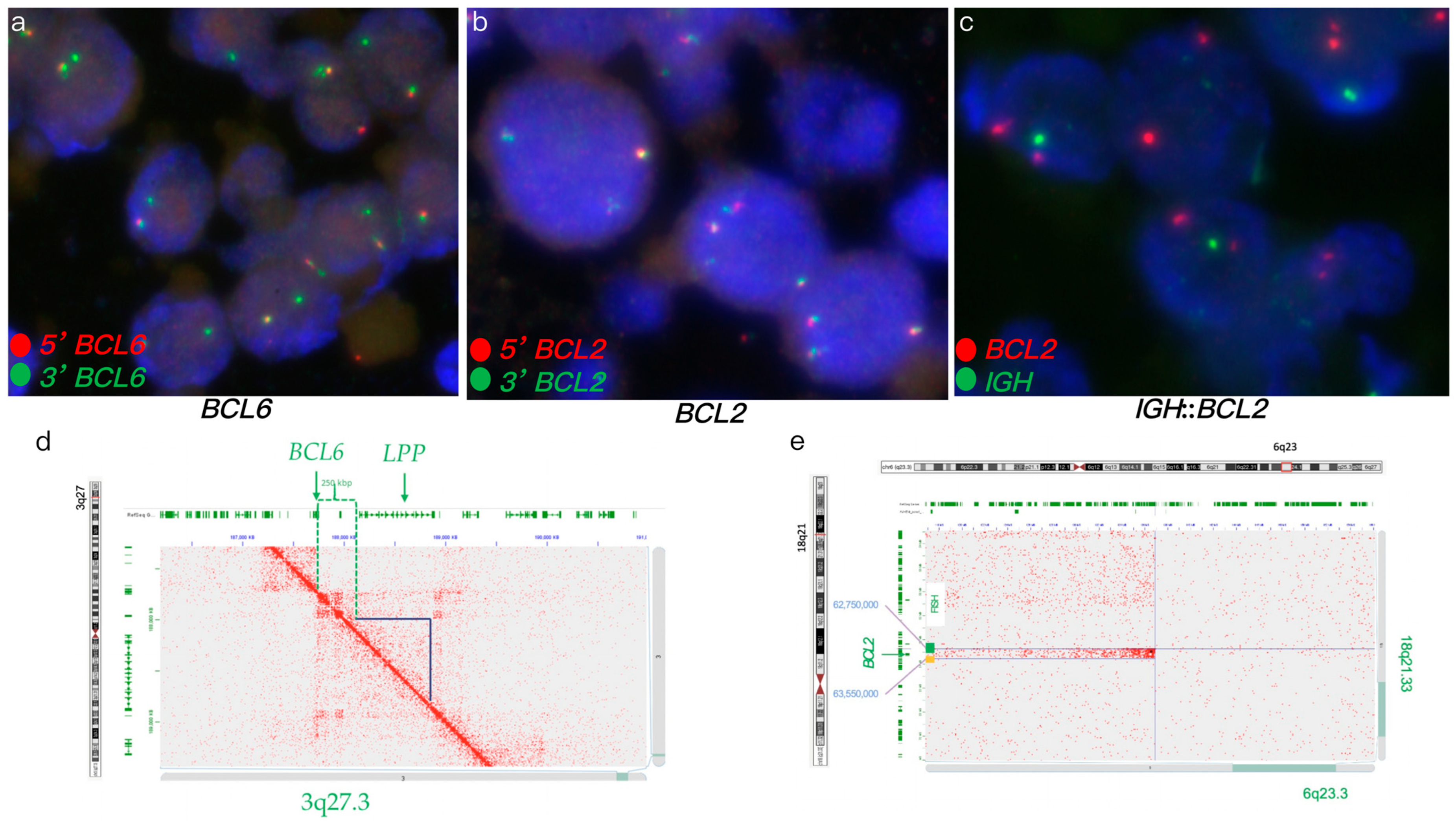
2.2. FISH Analysis
2.3. Hi-C-Based Lymphoma Assay
2.4. Data Analysis
2.5. Immunohistochemistry Assay
3. Results
3.1. Case-Specific Findings
3.1.1. Case #1
3.1.2. Case #2
3.1.3. Case #3
3.1.4. Case #4
3.1.5. Case #5
3.2. Comparative Performance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DLBCL | Diffuse large B-cell lymphoma |
| FISH | Fluorescence in situ hybridization |
| FFPE | Formalin-fixed paraffin-embedded |
| HGBCL | High grade B-cell lymphoma |
| NHL | Non-Hodgkin lymphoma |
| WGS | Whole genome sequencing |
References
- Sehn, L.H.; Salles, G. Diffuse Large B-Cell Lymphoma. N. Engl. J. Med. 2021, 384, 842–858. [Google Scholar] [CrossRef]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef] [PubMed]
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; Araujo, I.B.d.O.; Berti, E.; Bhagat, G.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 2022, 36, 1720–1748. [Google Scholar] [CrossRef]
- Scott, D.W.; King, R.L.; Staiger, A.M.; Ben-Neriah, S.; Jiang, A.; Horn, H.; Mottok, A.; Farinha, P.; Slack, G.W.; Ennishi, D.; et al. High-grade B-cell lymphoma with MYC and BCL2 and/or BCL6 rearrangements with diffuse large B-cell lymphoma morphology. Blood 2018, 131, 2060–2064. [Google Scholar] [CrossRef]
- Petrich, A.M.; Gandhi, M.; Jovanovic, B.; Castillo, J.J.; Rajguru, S.; Yang, D.T.; Shah, K.A.; Whyman, J.D.; Lansigan, F.; Hernandez-Ilizaliturri, F.J.; et al. Impact of induction regimen and stem cell transplantation on outcomes in double-hit lymphoma: A multicenter retrospective analysis. Blood 2014, 124, 2354–2361. [Google Scholar] [CrossRef]
- Johnson, N.A.; Slack, G.W.; Savage, K.J.; Connors, J.M.; Ben-Neriah, S.; Rogic, S.; Scott, D.W.; Tan, K.L.; Steidl, C.; Sehn, L.H.; et al. Concurrent expression of MYC and BCL2 in diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J. Clin. Oncol. 2012, 30, 3452–3459. [Google Scholar] [CrossRef]
- Van Imhoff, G.W.; Boerma, E.J.G.; Van Der Holt, B.; Schuuring, E.; Verdonck, L.F.; Kluin-Nelemans, H.C.; Kluin, P.M. Prognostic impact of germinal center-associated proteins and chromosomal break-points in poor-risk diffuse large B-cell lymphoma. J. Clin. Oncol. 2006, 24, 4135–4142. [Google Scholar] [CrossRef] [PubMed]
- Chapuy, B.; Stewart, C.; Dunford, A.J.; Kim, J.; Kamburov, A.; Redd, R.A.; Lawrence, M.S.; Roemer, M.G.M.; Li, A.J.; Ziepert, M.; et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat. Med. 2018, 24, 679–690. [Google Scholar] [CrossRef]
- Basso, K.; Dalla-Favera, R. Germinal centres and B cell lymphomagenesis. Nat. Rev. Immunol. 2015, 15, 172–184. [Google Scholar] [CrossRef] [PubMed]
- Kuppers, R.; Dalla-Favera, R. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene 2001, 20, 5580–5594. [Google Scholar] [CrossRef]
- Bertrand, P.; Bastard, C.; Maingonnat, C.; Jardin, F.; Maisonneuve, C.; Courel, M.-N.; Ruminy, P.; Picquenot, J.-M.; Tilly, H. Mapping of MYC breakpoints in 8q24 rearrangements involving non-immunoglobulin partners in B-cell lymphomas. Leukemia 2007, 21, 515–523. [Google Scholar] [CrossRef]
- Czabotar, P.E.; Lessene, G.; Strasser, A.; Adams, J.M. Control of apoptosis by the BCL-2 protein family: Implications for physiology and therapy. Nat. Rev. Mol. Cell Biol. 2014, 15, 49–63. [Google Scholar] [CrossRef]
- Aukema, S.M.; Siebert, R.; Schuuring, E.; van Imhoff, G.W.; Kluin-Nelemans, H.C.; Boerma, E.-J.; Kluin, P.M. Double-hit B-cell lymphomas. Blood 2011, 117, 2319–2331. [Google Scholar] [CrossRef]
- King, R.L.; Hsi, E.D.; Chan, W.C.; Piris, M.A.; Cook, J.R.; Scott, D.W.; Swerdlow, S.H. Diagnostic approaches and future directions in Burkitt lymphoma and high-grade B-cell lymphoma. Virchows Arch. 2023, 482, 193–205. [Google Scholar] [CrossRef]
- Muñoz-Marmol, A.M.; Sanz, C.; Tapia, G.; Marginet, R.; Ariza, A.; Mate, J.L. MYC status determination in aggressive B-cell lymphoma: The impact of FISH probe selection. Histopathology 2013, 63, 418–424. [Google Scholar] [CrossRef] [PubMed]
- King, R.L.; McPhail, E.D.; Meyer, R.G.; Vasmatzis, G.; Pearce, K.; Smadbeck, J.B.; Baughn, L.B. False-negative rates for MYC fluorescence in situ hybridization probes in B-cell neoplasms. Haematologica 2019, 104, e248–e251. [Google Scholar] [CrossRef]
- Hilton, L.K.; Tang, J.; Ben-Neriah, S.; Alcaide, M.; Jiang, A.; Grande, B.M.; Rushton, C.K.; Boyle, M.; Meissner, B.; Scott, D.W.; et al. The double-hit signature identifies double-hit diffuse large B-cell lymphoma with genetic events cryptic to FISH. Blood 2019, 134, 1528–1532. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, A.; Ay, F. Identification of copy number variations and translocations in cancer cells from Hi-C data. Bioinformatics 2018, 34, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Durand, N.C.; Shamim, M.S.; Machol, I.; Rao, S.S.P.; Huntley, M.H.; Lander, E.S.; Aiden, E.L. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 2016, 3, 95–98. [Google Scholar] [CrossRef]
- Kim, K.; Kim, M.; Kim, Y.; Lee, D.; Jung, I. Hi-C as a molecular rangefinder to examine genomic rearrangements. Semin. Cell Dev. Biol. 2022, 121, 161–170. [Google Scholar] [CrossRef]
- Song, F.; Xu, J.; Dixon, J.; Yue, F. Analysis of Hi-C data for discovery of structural variations in cancer. Methods Mol. Biol. 2022, 2301, 143–161. [Google Scholar]
- Okabe, A.; Kaneda, A. Hi-C analysis to identify genome-wide chromatin structural aberration in cancer. Methods Mol. Biol. 2023, 2519, 127–140. [Google Scholar]
- Prior, D.; Schmitt, A.D.; Louissaint, A.; Mata, D.A.; Massaro, S.; Nardi, V.; Xu, M.L. Large B-cell lymphoma with mystery rearrangement: Applying Hi-C to the detection of clinically relevant structural abnormalities. Br. J. Haematol. 2024, 205, 1225–1229. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, A.D.; Sikkink, K.; Ahmed, A.A.; Melnyk, S.; Reid, D.; Van Meter, L.; Farooqi, M.S. Evaluation of Hi-C sequencing for detection of gene fusions in hematologic and solid tumor pediatric cancer samples. Cancers 2024, 16, 17. [Google Scholar] [CrossRef] [PubMed]
- Galbraith, K.; Wu, J.; Sikkink, K.; Mohamed, H.; Reid, D.; Perez-Arreola, M.; Snuderl, M. Detection of gene fusions and rearrangements in formalin-fixed, paraffin-embedded solid tumor specimens using high-throughput chromosome conformation capture. J. Mol. Diagn. 2025, 27, 346–359. [Google Scholar] [CrossRef]
- Dixon, J.R.; Xu, J.; Dileep, V.; Zhan, Y.; Song, F.; Le, V.T.; Yardımcı, G.G.; Chakraborty, A.; Bann, D.V.; Wang, Y.; et al. Integrative detection and analysis of structural variation in cancer genomes. Nat. Genet. 2018, 50, 1388–1398. [Google Scholar] [CrossRef]
- Wu, J.; Chu, S.C.A.; Cho, J.; Movahed-Ezazi, M.; Galbraith, K.; Fang, C.S.; Ryan, R.J. Hi-C for genome-wide detection of enhancer-hijacking rearrangements in routine lymphoid cancer biopsies. bioRxiv 2025. [Google Scholar] [CrossRef]
- Haley, L.; Parimi, V.; Jiang, L.; Pallavajjala, A.; Hardy, M.; Yonescu, R.; Gocke, C.D. Diagnostic utility of gene fusion panel to detect gene fusions in fresh and formalin-fixed, paraffin-embedded cancer specimens. J. Mol. Diagn. 2021, 23, 1343–1358. [Google Scholar] [CrossRef] [PubMed]
- Wingett, S.W.; Ewels, P.; Furlan-Magaril, M.; Nagano, T.; Schoenfelder, S.; Fraser, P.; Andrews, S. HiCUP: Pipeline for mapping and processing Hi-C data. F1000Research 2015, 4, 1310. [Google Scholar]
- Wang, X.; Luan, Y.; Yue, F. EagleC: A deep-learning framework for detecting a full range of structural variations from bulk and single-cell contact maps. Sci. Adv. 2022, 8, eabn9215. [Google Scholar] [CrossRef]
- Wang, X.; Xu, J.; Zhang, B.; Hou, Y.; Song, F.; Lyu, H.; Yue, F. Genome-wide detection of enhancer-hijacking events from chromatin interaction data in rearranged genomes. Nat. Methods 2021, 18, 661–668. [Google Scholar] [CrossRef]
- Schieppati, F.; Balzarini, P.; Fisogni, S.; Re, A.; Pagani, C.; Bianchetti, N.; Micheli, L.; Passi, A.; Ferrari, S.; Maifredi, A.; et al. An increase in MYC copy number has a progressive negative prognostic impact in patients with diffuse large B-cell and high-grade lymphoma, who may benefit from intensified treatment regimens. Haematologica 2020, 105, 1369–1378. [Google Scholar] [PubMed]
- Quesada, A.E.; Medeiros, L.J.; Desai, P.A.; Lin, P.; Westin, J.R.; Hawsawi, H.M.; Wei, P.; Tang, G.; Seegmiller, A.C.; Reddy, N.M.; et al. Increased Myc copy number is an independent prognostic factor in patients with diffuse large B-cell lymphoma. Mod. Pathol. 2017, 30, 1688–1697. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.-X.; Fan, L.; Wang, L.; Wu, J.-Z.; Miao, K.-R.; Liang, J.-H.; Gong, Q.-X.; Wang, Z.; Young, K.H.; Xu, W.; et al. Myc or Bcl2 copy number aberration is a strong predictor of outcome in patients with diffuse large B-cell lymphoma. Oncotarget 2015, 6, 18374–18388. [Google Scholar] [CrossRef]
- Pophali, P.A.; Marinelli, L.M.; Ketterling, R.P.; Meyer, R.G.; McPhail, E.D.; Kurtin, P.J.; Mwangi, R.; Maurer, M.J.; Habermann, T.; King, R.L. High level MYC amplification in B-cell lymphomas: Is it a marker of aggressive disease? Blood Cancer J. 2020, 10, 5. [Google Scholar] [CrossRef]
- Collinge, B.; Ben-Neriah, S.; Chong, L.; Boyle, M.; Jiang, A.; Miyata-Takata, T.; Farinha, P.; Craig, J.W.; Slack, G.W.; Ennishi, D.; et al. The impact of MYC and BCL2 structural variants in tumors of DLBCL morphology and mechanisms of false-negative MYC IHC. Blood 2021, 137, 2196–2208. [Google Scholar] [CrossRef] [PubMed]
- Copie-Bergman, C.; Cuillière-Dartigues, P.; Baia, M.; Briere, J.; Delarue, R.; Canioni, D.; Salles, G.; Parrens, M.; Belhadj, K.; Fabiani, B.; et al. MYC-IG rearrangements are negative predictors of survival in DLBCL patients treated with immunochemotherapy: A GELA/LYSA study. Blood 2015, 126, 2466–2474. [Google Scholar] [CrossRef]
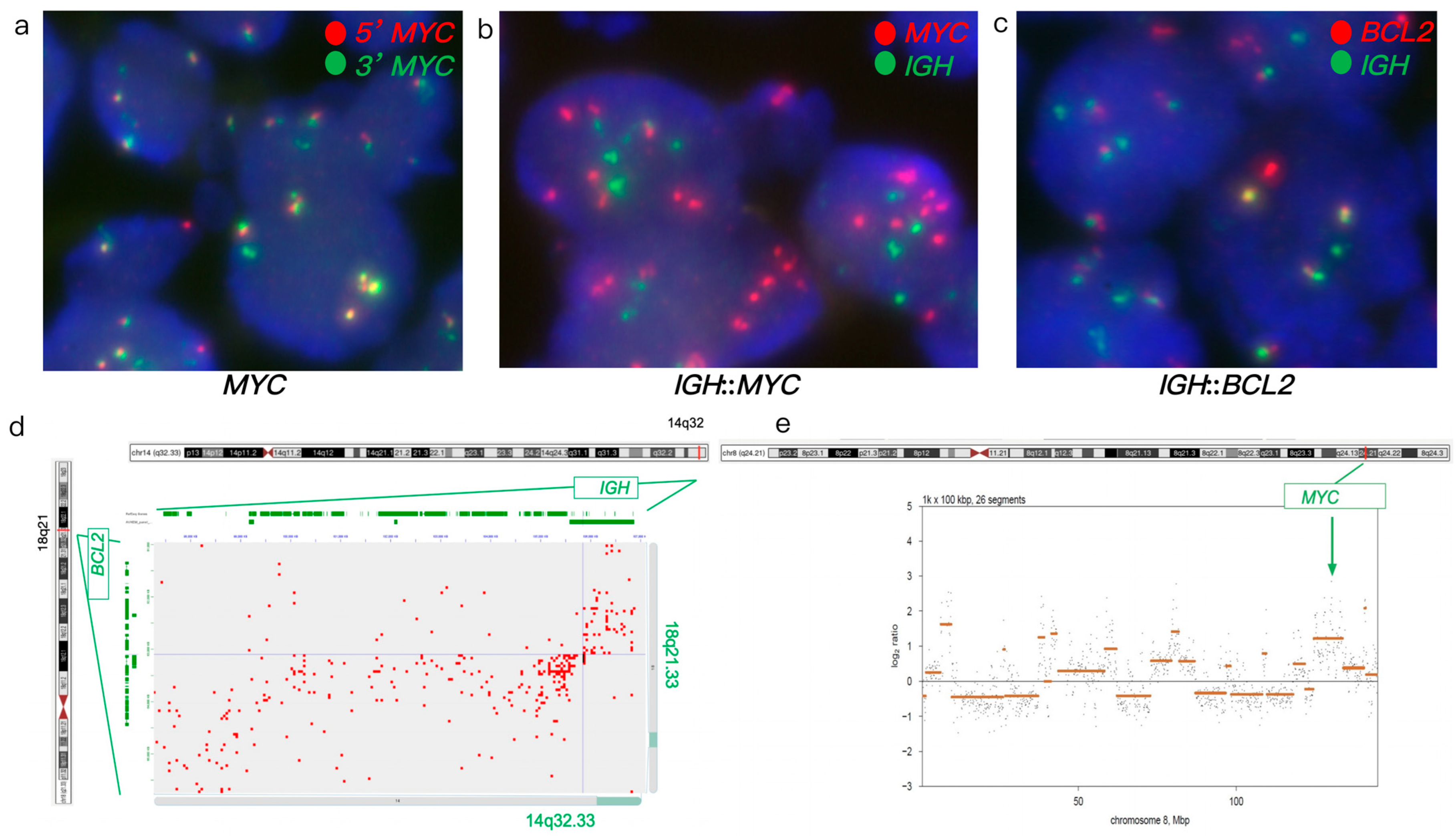
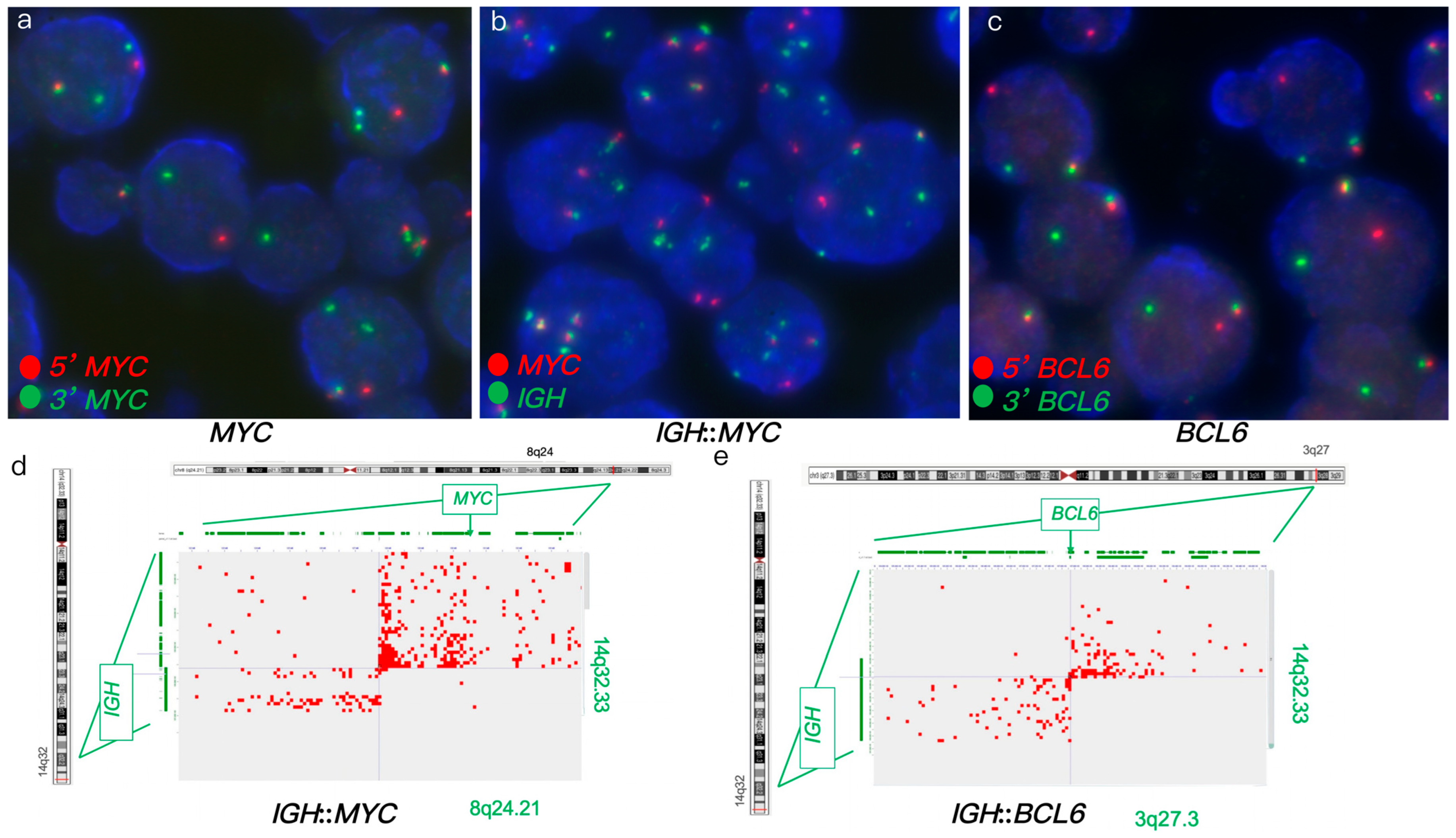
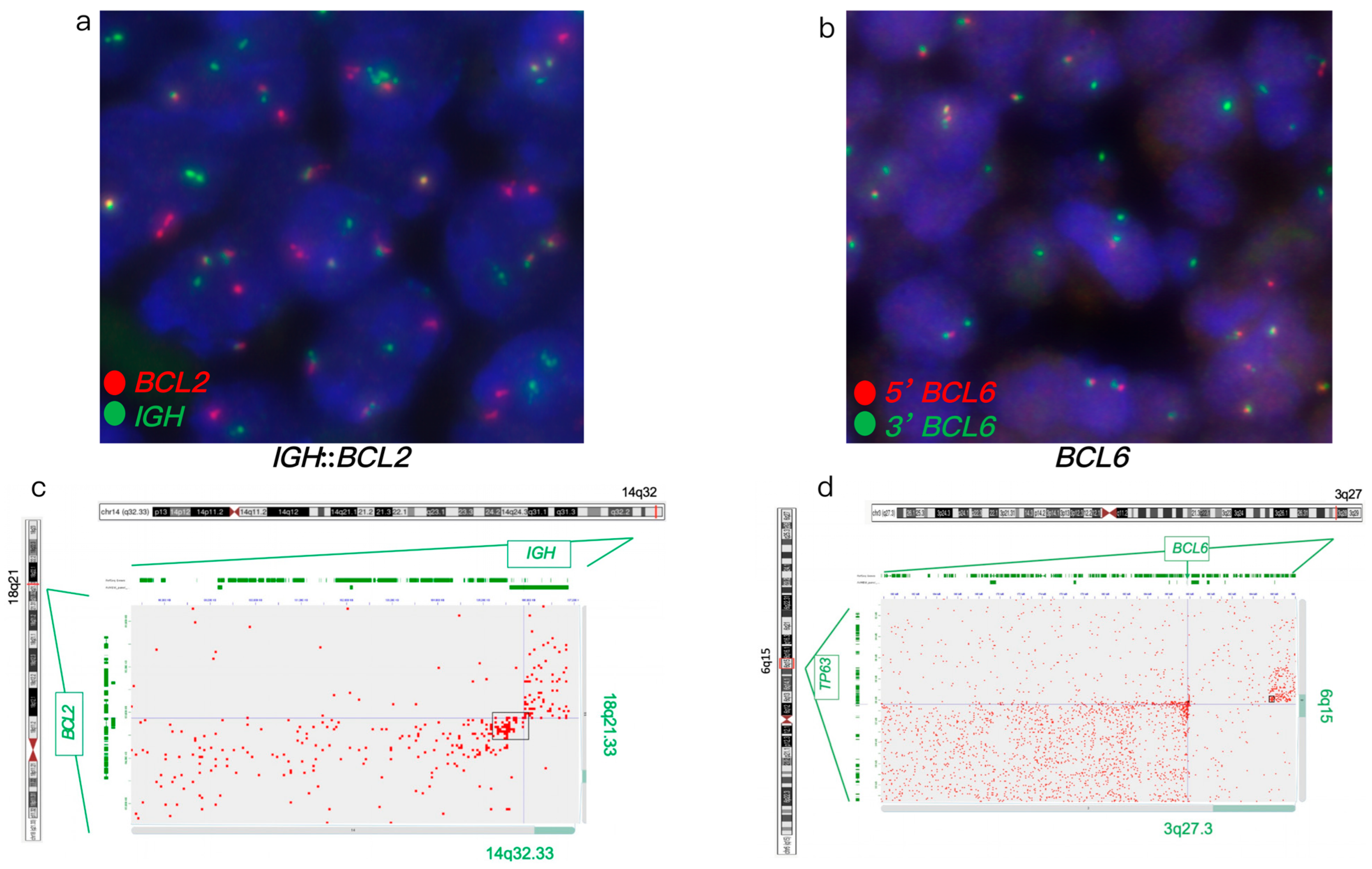
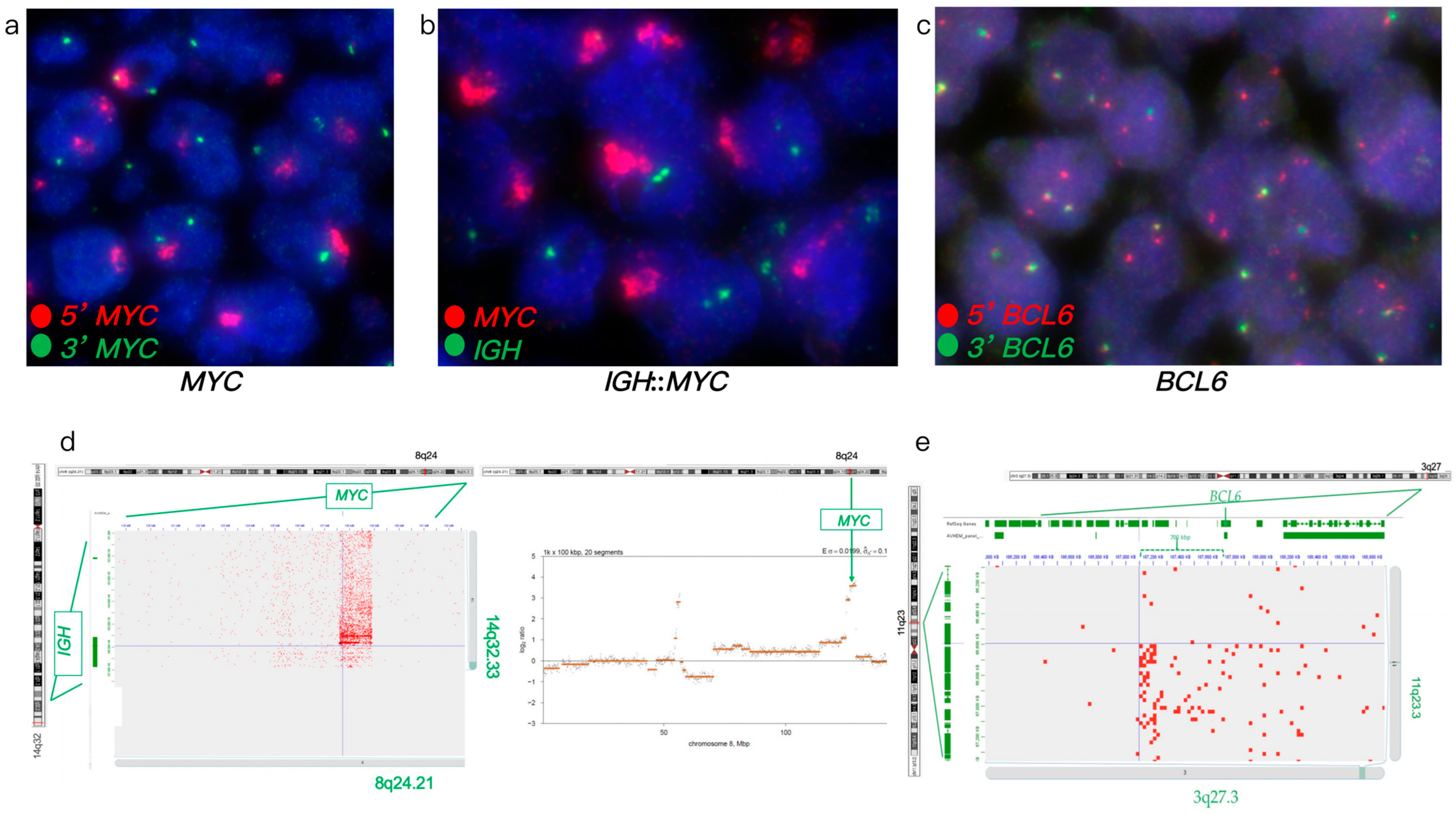
| Case # | FISH Results | Hi-C Lymphoma Assay Results | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MYC-R. | IGH::MYC Fusion | BCL2-R. | IGH::BCL2 Fusion | BCL6-R. | MYC-R. | IGH::MYC Fusion | BCL2-R. | IGH::BCL2 Fusion | BCL6-R. | Additional Gene Rearrangement/Fusions | Additional Copy Number Variants | |
| 1 | Atypical with gain/amp. (ampF1-2G) | - | + | + | - | Gain/amp. | - | + | + | - | KDSR, TNFRSF11A, BCR, LYN, PLAG1, and DENND3 | |
| 2 | Atypical (1R2G1F) | Atypical (1R2G1-3F, 2R2G2F) | - | - | + | + | + | - | - | IGH::BCL6 | POU2AF1, E2F2, ID3, and MDS2 | |
| 3 | - | - | + | + | Atypical (1G1F) | - | - | + | + | Atypical with a 7 Mb deletion 5′ of BCL6 | TNFRSF11A, KDSR, CLTC, HLF, LYN, TFRC, KAT6A, PLAG1, and NDRG1 | Loss of RAB29 and MSI2 |
| 4 | Atypical with amp. (ampR1-3G) | - | - | - | Atypical (1-3R2-3F) | Atypical with amp. (7–9 copies) | + | - | - | Atypical t(3;11) ~700 Kb from breakpoint | ERBB2, FOXO1, S1PR2, TYK2, NF1, PYCR1, FSTL3, LPP, PRDM16, KMT2A, LYN, PLAG1, IGH::MIR4507 fusion | Loss of TP53, amp. of KMT2A and PLAG1, gain of LYN, complex rearrangement of Chromosome 3 |
| 5 | - | - | - | - | Atypical (1G1F) | - | - | +: t(6;18) insertion and t(4;18) | - | Atypical with deletion of LPP | FGFR1, FGFR2, TP63, KDSR, TET1, LYN, and NSD3 | losses of CDNK2A/2B, TNFAIP3, BCL7A, and PCM1, gain of 18q |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, S.; Ament, C.; Klausner, M.; Stinnett, V.; Morsberger, L.; Ghabrial, J.; Middlezong, W.; Schmitt, A.D.; Hastie, A.R.; Zou, Y.S. Hi-C Technology Reveals Actionable Gene Fusions and Rearrangements in Diffuse Large B-Cell Lymphoma Unidentified by Conventional FISH. Genes 2025, 16, 1093. https://doi.org/10.3390/genes16091093
Liang S, Ament C, Klausner M, Stinnett V, Morsberger L, Ghabrial J, Middlezong W, Schmitt AD, Hastie AR, Zou YS. Hi-C Technology Reveals Actionable Gene Fusions and Rearrangements in Diffuse Large B-Cell Lymphoma Unidentified by Conventional FISH. Genes. 2025; 16(9):1093. https://doi.org/10.3390/genes16091093
Chicago/Turabian StyleLiang, Sichen, Candice Ament, Melanie Klausner, Victoria Stinnett, Laura Morsberger, Jen Ghabrial, William Middlezong, Anthony D. Schmitt, Alex R. Hastie, and Ying S. Zou. 2025. "Hi-C Technology Reveals Actionable Gene Fusions and Rearrangements in Diffuse Large B-Cell Lymphoma Unidentified by Conventional FISH" Genes 16, no. 9: 1093. https://doi.org/10.3390/genes16091093
APA StyleLiang, S., Ament, C., Klausner, M., Stinnett, V., Morsberger, L., Ghabrial, J., Middlezong, W., Schmitt, A. D., Hastie, A. R., & Zou, Y. S. (2025). Hi-C Technology Reveals Actionable Gene Fusions and Rearrangements in Diffuse Large B-Cell Lymphoma Unidentified by Conventional FISH. Genes, 16(9), 1093. https://doi.org/10.3390/genes16091093






