Updated Sequence and Annotation of the Broad Host Range Rhizobial Symbiont Sinorhizobium fredii HH103 Genome
Abstract
1. Introduction
2. Materials and Methods
3. Results and Discussion
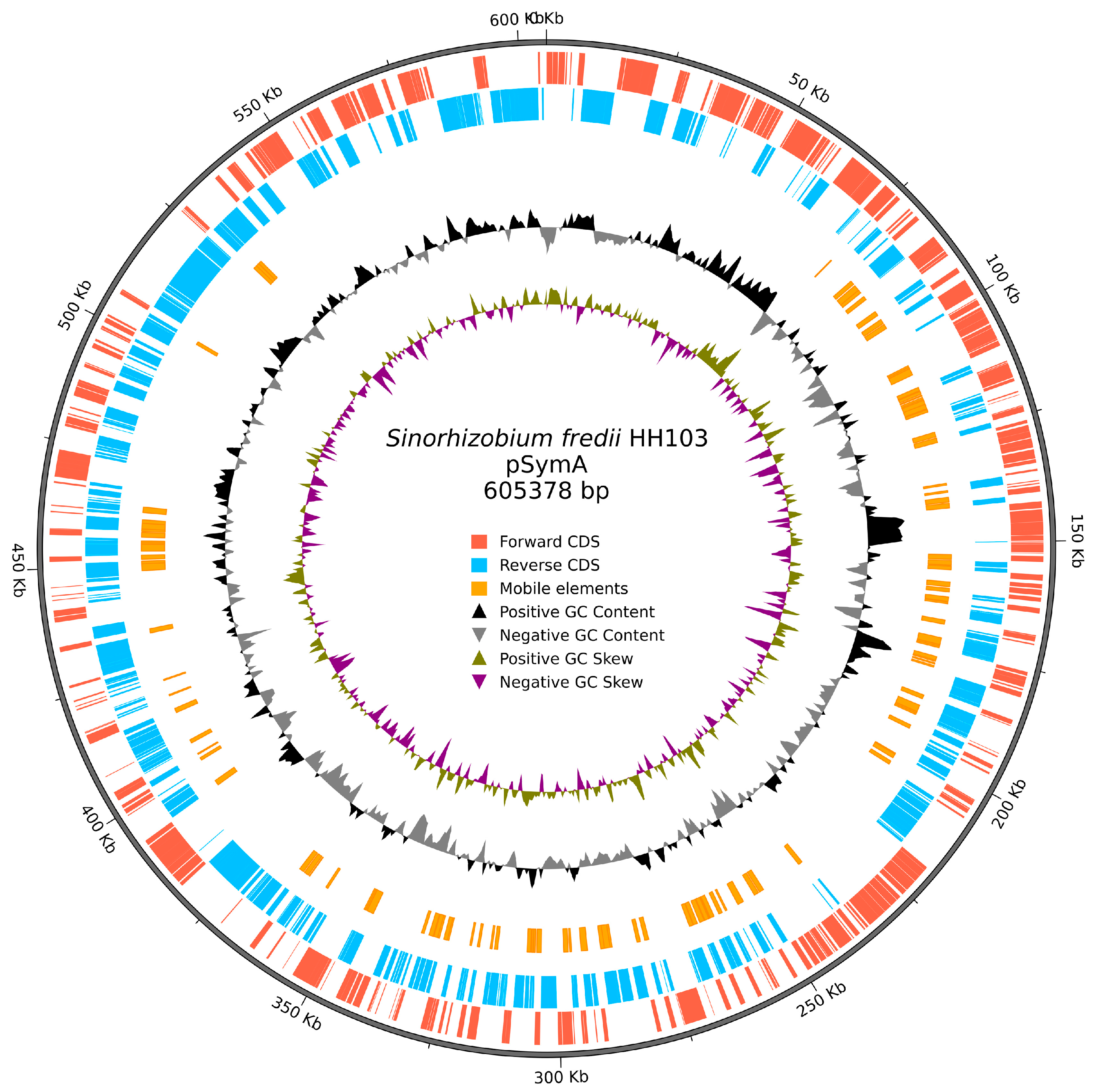
3.1. Characteristics of the Updated Version of the S. fredii HH103 Genome
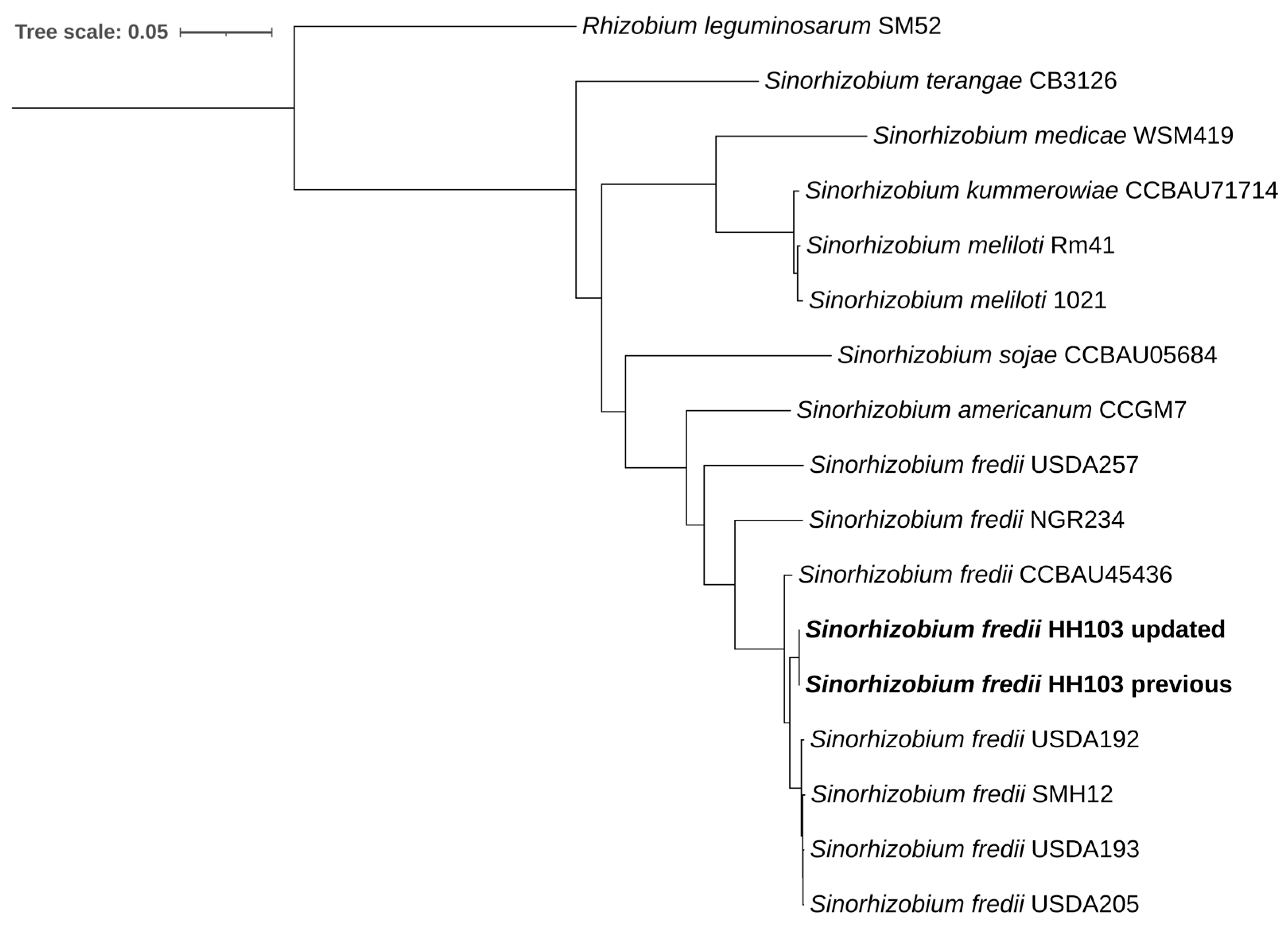
3.2. Comparison of the Genome of HH103 with Other Rhizobia
3.3. The HH103 Genome Is the Most Complex Among the Different S. fredii Strains Characterized So Far
3.4. S. fredii Strains Harbor Higher Numbers of Mobile Elements than Other Sinorhizobium Species
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pueppke, S.G.; Broughton, W.J. Rhizobium sp. strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host ranges. Mol. Plant-Microbe Interact. 1999, 12, 293–318. [Google Scholar] [CrossRef]
- López-Baena, F.J.; Ruiz-Sainz, J.E.; Rodríguez-Carvajal, M.A.; Vinardell, J.M. Bacterial molecular signals in the Sinorhizobium fredii-soybean symbiosis. Int. J. Mol. Sci. 2016, 17, 755. [Google Scholar] [CrossRef] [PubMed]
- El Idrissi, M.M.; Kaddouri, K.; Bouhnik, O.; Lamrabet, M.; Alami, S.; Abdelmoumen, H. Conventional and unconventional symbiotic nitrogen fixing bacteria associated with legumes. In Developments in Applied Microbiology and Biotechnology, Microbial Symbionts; Dharumadurai, D., Ed.; Academic Press: Cambridge, MA, USA, 2023; pp. 75–109. ISBN 9780323993340. [Google Scholar] [CrossRef]
- Kawaka, F. Characterization of symbiotic and nitrogen fixing bacteria. AMB Express 2022, 12, 99. [Google Scholar] [CrossRef]
- Margaret, I.; Becker, A.; Blom, J.; Bonilla, I.; Goesmann, A.; Göttfert, M.; Lloret, J.; Mittard-Runte, V.; Rückert, C.; Ruiz-Sainz, J.E.; et al. Symbiotic properties and first analyses of the genomic sequence of the fast-growing model strain Sinorhizobium fredii HH103 nodulating soybean. J. Biotechnol. 2011, 155, 11–19. [Google Scholar] [CrossRef]
- Cleyet-Marel, J.C. Dynamique des Populations de Rhizobium et de Bradyrhizobium dans le Sol et la Rhizosphere. Ph.D. Thesis, University Claude Bernard, Lyon, France, 1987. [Google Scholar]
- Rodríguez-Navarro, D.N.; Bellogín, R.; Camacho, M.; Daza, A.; Medina, C.; Ollero, F.J.; Santamaría, C.; Ruíz-Saínz, J.E.; Vinardell, J.M.; Temprano, F. Field assessment and genetic stability of Sinorhizobium fredii strain SMH12 for commercial soybean inoculants. Eur. J. Agron. 2003, 19, 299–309. [Google Scholar] [CrossRef]
- Temprano-Vera, F.; Rodríguez-Navarro, D.N.; Acosta-Jurado, S.; Perret, X.; Fossou, R.K.; Navarro-Gómez, P.; Zhen, T.; Yu, D.; An, Q.; Buendía-Clavería, A.M.; et al. Sinorhizobium fredii strains HH103 and NGR234 form nitrogen fixing nodules with diverse wild soybeans (Glycine soja) from Central China but are ineffective on Northern China accessions. Front. Microbiol. 2018, 9, 2843. [Google Scholar] [CrossRef]
- Videira, L.B.; Pastorino, G.N.; Balatti, P.A. Incompatibility may not be the rule in the Sinorhizobium fredii–soybean interaction. Soil Biol. Biochem. 2001, 33, 837–840. [Google Scholar] [CrossRef]
- Jiménez-Guerrero, I.; Medina, C.; Vinardell, J.M.; Ollero, F.J.; López-Baena, F.J. The rhizobial type 3 secretion system: The Dr. Jekyll and Mr. Hyde in the rhizobium–legume symbiosis. Int. J. Mol. Sci. 2022, 23, 11089. [Google Scholar] [CrossRef]
- Weidner, S.; Becker, A.; Bonilla, I.; Jaenicke, S.; Lloret, J.; Margaret, I.; Pühler, A.; Ruiz-Sainz, J.E.; Schneiker-Bekel, S.; Szczepanowski, R.; et al. Genome sequence of the soybean symbiont Sinorhizobium fredii HH103. J. Bacteriol. 2012, 194, 1617–1618. [Google Scholar] [CrossRef]
- Vinardell, J.M.; Acosta-Jurado, S.; Zehner, S.; Göttfert, M.; Becker, A.; Baena, I.; Blom, J.; Crespo-Rivas, J.C.; Goesmann, A.; Jaenicke, S.; et al. The Sinorhizobium fredii HH103 genome: A comparative analysis with S. fredii strains differing in their symbiotic behavior with soybean. Mol. Plant Microbe Interact. 2015, 28, 811–824. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.F.; Zhou, Y.J.; Zhang, Y.M.; Li, Q.Q.; Zhang, Y.Z.; Li, D.F.; Wang, S.; Wang, J.; Gilbert, L.B.; Li, Y.R.; et al. Comparative genomics of rhizobia nodulating soybean suggests extensive recruitment of lineage-specific genes in adaptations. Proc. Natl. Acad. Sci. USA 2012, 109, 8629–8634. [Google Scholar] [CrossRef]
- Beringer, J.E. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 1974, 84, 188–198. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Hunt, M.; Silva, N.D.; Otto, T.D.; Parkhill, J.; Keane, J.A.; Harris, S.R. Circlator: Automated circularization of genome assemblies using long sequencing reads. Genome Biol. 2015, 16, 294. [Google Scholar] [CrossRef]
- Bedtools: A Powerful Toolset for Genome Arithmetic. Available online: https://bedtools.readthedocs.io/en/latest/ (accessed on 15 March 2025).
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genom. 2021, 7, 000685. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Espinosa-Sáiz, D.; Velázquez, E.; Menéndez, E.; diCenzo, G.C. Complete genome sequences of the species type strains Sinorhizobium garamanticum LMG 24692 and Sinorhizobium numidicum LMG 27395 and CIP 109850. Microbiol. Resour. Announc. 2023, 12, e0025123. [Google Scholar] [CrossRef] [PubMed]
- Galibert, F.; Finan, T.M.; Long, S.R.; Puhler, A.; Abola, P.; Ampe, F.; Barloy-Hubler, F.; Barnett, M.J.; Becker, A.; Boistard, P.; et al. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 2001, 293, 668–672. [Google Scholar] [CrossRef]
- Schmeisser, C.; Liesegang, H.; Krysciak, D.; Bakkou, N.; Le Quéré, A.; Wollherr, A.; Heinemeyer, I.; Morgenstern, B.; Pommerening-Röser, A.; Flores, M.; et al. Rhizobium sp. strain NGR234 possesses a remarkable number of secretion systems. Appl. Environm. Microbiol. 2009, 75, 4035–4045. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Jurado, S.; Fuentes-Romero, F.; Ruiz-Sainz, J.E.; Janczarek, M.; Vinardell, J.M. Rhizobial exopolysaccharides: Genetic regulation of their synthesis and relevance in symbiosis with legumes. Int. J. Mol. Sci. 2021, 22, 6233. [Google Scholar] [CrossRef] [PubMed]
- Ausubel, F.M. Molecular genetics of symbiotic nitrogen fixation. Cell 1982, 29, 1–2. [Google Scholar] [CrossRef]
- Rodrigues, J.A.; López-Baena, F.J.; Ollero, F.J.; Vinardell, J.M.; Espuny, R.; Bellogín, R.A.; Ruiz-Sainz, J.E.; Thomasm, J.R.; Sumpton, D.; Ault, J.; et al. NopM and NopD are rhizobial nodulation outer proteins: Identification using LC-MALDI and LC-ESI with amonolithic capillary column. J. Proteome Res. 2007, 6, 1029–1037. [Google Scholar] [CrossRef]
- Toledo, I.; Lloret, L.; Martínez-Romero, E. Sinorhizobium americanus sp. nov., a new Sinorhizobium species nodulating native Acacia spp. in Mexico. Syst. Appl. Microbiol. 2003, 26, 54–64. [Google Scholar] [CrossRef]
- Mnasri, B.; Saïdi, S.; Chihaoui, S.A.; Mhamdi, R. Sinorhizobium americanum symbiovar mediterranense is a predominant symbiont that nodulates and fixes nitrogen with common bean (Phaseolus vulgaris L.) in a Northern Tunisian field. Syst. Appl. Microbiol. 2012, 35, 263–269. [Google Scholar] [CrossRef]
- Li, Q.Q.; Wang, E.T.; Chang, Y.L.; Zhang, Y.Z.; Zhang, Y.M.; Sui, X.H.; Chen, W.F.; Chen, W.X. Ensifer sojae sp. nov., isolated from root nodules of Glycine max grown in saline-alkaline soils. Int J. Syst. Evol. Microbiol. 2011, 61, 1981–1988. [Google Scholar] [CrossRef]
- Kuzmanović, N.; Fagorzi, C.; Mengoni, A.; Lassalle, F.; diCenzo, G.C. Taxonomy of Rhizobiaceae revisited: Proposal of a new framework for genus delimitation. Int. J. Syst. Evol. Microbiol. 2022, 72, 005243. [Google Scholar] [CrossRef] [PubMed]
- Schuldes, J.; Rodriguez Orbegoso, M.; Schmeisser, C.; Krishnan, H.B.; Daniel, R.; Streit, W.R. Complete genome sequence of the broad-host-range strain Sinorhizobium fredii USDA257. J. Bacteriol. 2012, 194, 4483. [Google Scholar] [CrossRef]
- Cutiño, A.M.; Del Carmen Sánchez-Aguilar, M.; Ruiz-Sáinz, J.E.; Del Rosario Espuny, M.; Ollero, F.J.; Medina, C. A novel system to selective tagging of Sinorhizobium fredii symbiotic plasmids. Meth. Mol. Biol. 2024, 2751, 247–259. [Google Scholar] [CrossRef]
- Siguier, P.; Gourbeyre, E.; Chandler, M. Bacterial insertion sequences: Their genomic impact and diversity. FEMS Microbiol. Rev. 2014, 38, 865–891. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Liu, L.X.; Zhang, Y.Z.; Jiao, J.; Cui, W.J.; Zhang, B.; Wang, X.L.; Li, M.L.; Chen, Y.; Xiong, Z.Q.; et al. Adaptive evolution of rhizobial symbiotic compatibility mediated by co-evolved insertion sequences. ISME J. 2018, 12, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Jiao, J.; Tian, C.-F. Adaptive evolution of rhizobial symbiosis beyond horizontal gene transfer: From genome innovation to regulation reconstruction. Genes 2023, 14, 274. [Google Scholar] [CrossRef] [PubMed]
- Riaz, M.R.; Sosa Marquez, I.; Lindgren, H.; Levin, G.; Doyle, R.; Romero, M.C.; Paoli, J.C.; Drnevich, J.; Fields, C.J.; Geddes, B.A.; et al. Mobile gene clusters and coexpressed plant-rhizobium pathways drive partner quality variation in symbiosis. Proc. Natl. Acad. Sci. USA 2025, 122, e2411831122. [Google Scholar] [CrossRef]

| BioProject accession no. | PRJNA1233244 |
| BioSample accession no. | SAMN47263981 |
| GenBank Assembly accession no. | GCF_048585425.1 |
| GenBank accession no. | CP183939, CP183940, CP183941, CP183942, CP183943, CP183944, CP183945 |
| SRA accession no. | - |
| PacBio reads | SRR34701253 |
| Illumina reads | SRR34710307 |
| Total PacBio read length (nt) | 1,170,389,454 |
| No. of PacBio reads | 98,229 |
| PacBio N50 read length (nt) | 12,887 |
| Total Illumina read length (nt) | 1,782,256,322 |
| No. of Illumina paired reads | 11,803,022 |
| Illumina read length (nt) | 2 × 151 |
| Genome size (bp) | 7,273,959 |
| No. of protein-coding genes | 6949 |
| G+C content (%) | 62.14 |
| Genome coverage | 147× |
| No. of replicons | 7 |
| Replicon sizes (bp) | 24,038; 25,081; 61,874; 144,081; 605,378; 2,099,565; 4,313,942 |
| Replicon | Chromosome | pSfHH103e (p_e, pSymB)) | pSfHH103d (p_d, pSymA) | pSfHH103c (p_c) | pSfHH103b (p_b) | pSfHH103a2 (p_a2) | pSfHH103a1 (p_a1) | |
|---|---|---|---|---|---|---|---|---|
| Length (bp) | Previous | 4,305,723 | 2,096,125 | ca. 588,797 a | 144,082 | 61,880 | 25,081 | 24,036 |
| Updated | 4,313,942 | 2,099,565 | 605,378 | 144,081 | 61,874 | 25,081 | 24,038 | |
| GC content (%) | Previous | 62.61 | 62.38 | 59.59 | 58.68 | 58.47 | 58.02 | 58.21 |
| Updated | 62.60 | 62.38 | 59.56 | 58.68 | 58.47 | 58.03 | 58.21 | |
| CDS | Previous | 4008 | 1991 | 664 | 169 | 62 | 38 | 19 |
| Updated | 4013 | 1982 | 665 | 169 | 62 | 38 | 20 | |
| t-RNA genes | Previous | 53 | 0 | 1 | 0 | 0 | 0 | 0 |
| Updated | 53 | 0 | 1 | 0 | 0 | 0 | 0 | |
| rrn operons | Previous | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| Updated | 3 | 0 | 0 | 0 | 0 | 0 | 0 | |
| GenBank Accession number | Previous | HE616890 | HE616899 | CDSA010000001 to CDSA010000004 | HE616893 | HE616892 | LN735562 | HE616891 |
| Updated | CP183939.1 | CP183944.1 | CP183945.1 | CP183943.1 | CP183942.1 | CP183941.1 | CP183940.1 |
| Locus_Tag a | Replicon | Description |
|---|---|---|
| ACN6KE_000391 | Chromosome | Zinc metalloendopeptidase |
| ACN6KE_001521 | Chromosome | Hypothetical protein |
| ACN6KE_001826 | Chromosome | Antifreeze protein |
| ACN6KE_001897 | Chromosome | ABC transporter ATP-binding protein |
| ACN6KE_001899 | Chromosome | Hypothetical protein |
| ACN6KE_001932 | Chromosome | Hypothetical protein |
| ACN6KE_002195 | Chromosome | Hypothetical protein |
| ACN6KE_002415 | Chromosome | RTX toxin hemolysin-type protein |
| ACN6KE_003412 | Chromosome | IS21 family transposase |
| ACN6KE_003587 | Chromosome | Peptidoglycan-binding protein LysM |
| ACN6KE_003588 | Chromosome | Hypothetical protein |
| ACN6KE_003589 | Chromosome | Hypothetical protein |
| ACN6KE_0035890 | Chromosome | Imidazole glycerol phosphate synthase subunit HisF |
| ACN6KE_004165 | pSymA | TIR domain-containing protein |
| ACN6KE_004181 | pSymA | Hypothetical protein |
| ACN6KE_006263 | pSymB | DUF1059 domain-containing protein |
| S. fredii Strain | Genome Accession Number a | Genome Size (Mb) | Genome Structure (Sizes in Mb in Brackets) |
|---|---|---|---|
| CCBAU45436 | GCF_003100575.1 | 6.9 | 1 chromosome (4.16), four plasmids: a (0.42), b (1.96), d (0.20), e (0.17) |
| HH103 | GCF_048585425.1 | 7.3 | 1 chromosome (4.31), six plasmids: a1 (0.024), a2 (0.025, b (0.062), c (0.14), d (0.61), e (2.10) |
| NGR234 | GCF_000018545.1 | 6.9 | 1 chromosome (3.92), two plasmids: a (0.54), b (2.43) |
| SMH12 | GCF_024400375.1 | 7.0 | 1 chromosome (4.02), two plasmids: a (0.56), b (2.39) |
| USDA192 | GCF_041260365.1 | 6.9 | 4 contigs, not fully assembled (1 chromosome, three plasmids) |
| USDA193 | GCF_041262265.1 | 6.8 | 3 contigs, not fully assembled (1 chromosome, two plasmids) |
| USDA205T | GCF_001461695.1 | 6.6 | 209 contigs, non-assembled |
| USDA257 | GCF_024400375.1 | 7.0 | 1 chromosome (6.48) and one plasmid (19 contigs, non-assembled, 0.56) |
| Rhizobial Strain | Number of Genes Related to Mobile Elements 1 |
|---|---|
| R. leguminosarum SM52 | 90 |
| S. americanum CCGM7 | 113 |
| S. kummerowiae CCBAU71714 | 172 |
| S. medicae WSM419 | 191 |
| S. meliloti 1021 | 158 |
| S. meliloti Rm41 | 118 |
| S. sojae CCBAU05684 | 194 |
| S. terangae CB3126 | 108 |
| S. fredii_CCBAU45436 | 260 |
| S. fredii HH103 (updated) | 340 |
| S. fredii NGR234 | 240 |
| S. fredii SMH12 | 352 |
| S. fredii USDA192 | 286 |
| S. fredii USDA193 | 282 |
| S. fredii USDA205 | 356 |
| S. fredii USDA257 | 209 |
| Replicon | Number of Genes Related to Mobile Elements | Replicon Size (bp) | Genes Related to Mobile Elements per 10 kb |
|---|---|---|---|
| chromosome | 135 | 4,313,942 | 0.31 |
| pSfHH103a1 | 2 | 24,038 | 0.83 |
| pSfHH103a2 | 1 | 25,081 | 0.40 |
| pSfHH103b | 14 | 61,874 | 2.26 |
| pSfHH103c | 27 | 144,081 | 1.87 |
| pSymA | 110 | 605,378 | 1.82 |
| pSymB | 51 | 2,099,565 | 0.24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fuentes-Romero, F.; López-Baena, F.-J.; Vinardell, J.-M.; Acosta-Jurado, S. Updated Sequence and Annotation of the Broad Host Range Rhizobial Symbiont Sinorhizobium fredii HH103 Genome. Genes 2025, 16, 1094. https://doi.org/10.3390/genes16091094
Fuentes-Romero F, López-Baena F-J, Vinardell J-M, Acosta-Jurado S. Updated Sequence and Annotation of the Broad Host Range Rhizobial Symbiont Sinorhizobium fredii HH103 Genome. Genes. 2025; 16(9):1094. https://doi.org/10.3390/genes16091094
Chicago/Turabian StyleFuentes-Romero, Francisco, Francisco-Javier López-Baena, José-María Vinardell, and Sebastián Acosta-Jurado. 2025. "Updated Sequence and Annotation of the Broad Host Range Rhizobial Symbiont Sinorhizobium fredii HH103 Genome" Genes 16, no. 9: 1094. https://doi.org/10.3390/genes16091094
APA StyleFuentes-Romero, F., López-Baena, F.-J., Vinardell, J.-M., & Acosta-Jurado, S. (2025). Updated Sequence and Annotation of the Broad Host Range Rhizobial Symbiont Sinorhizobium fredii HH103 Genome. Genes, 16(9), 1094. https://doi.org/10.3390/genes16091094







