Hsp101-1 Orchestrates Thermotolerance in Rice via Pre-Activated Transcriptional Networks and Modular Cross-Tissue Coordination
Abstract
1. Introduction
2. Materials and Methods
2.1. Cultivation, Heat Shock Treatment, and Transcriptome Sampling of Rice Materials
2.2. Transcriptome Sequencing
2.3. Analysis of RNA Sequencing Data
2.4. WGCNA
3. Results
3.1. DEGs Between WT and OE Lines After Heat Treatment
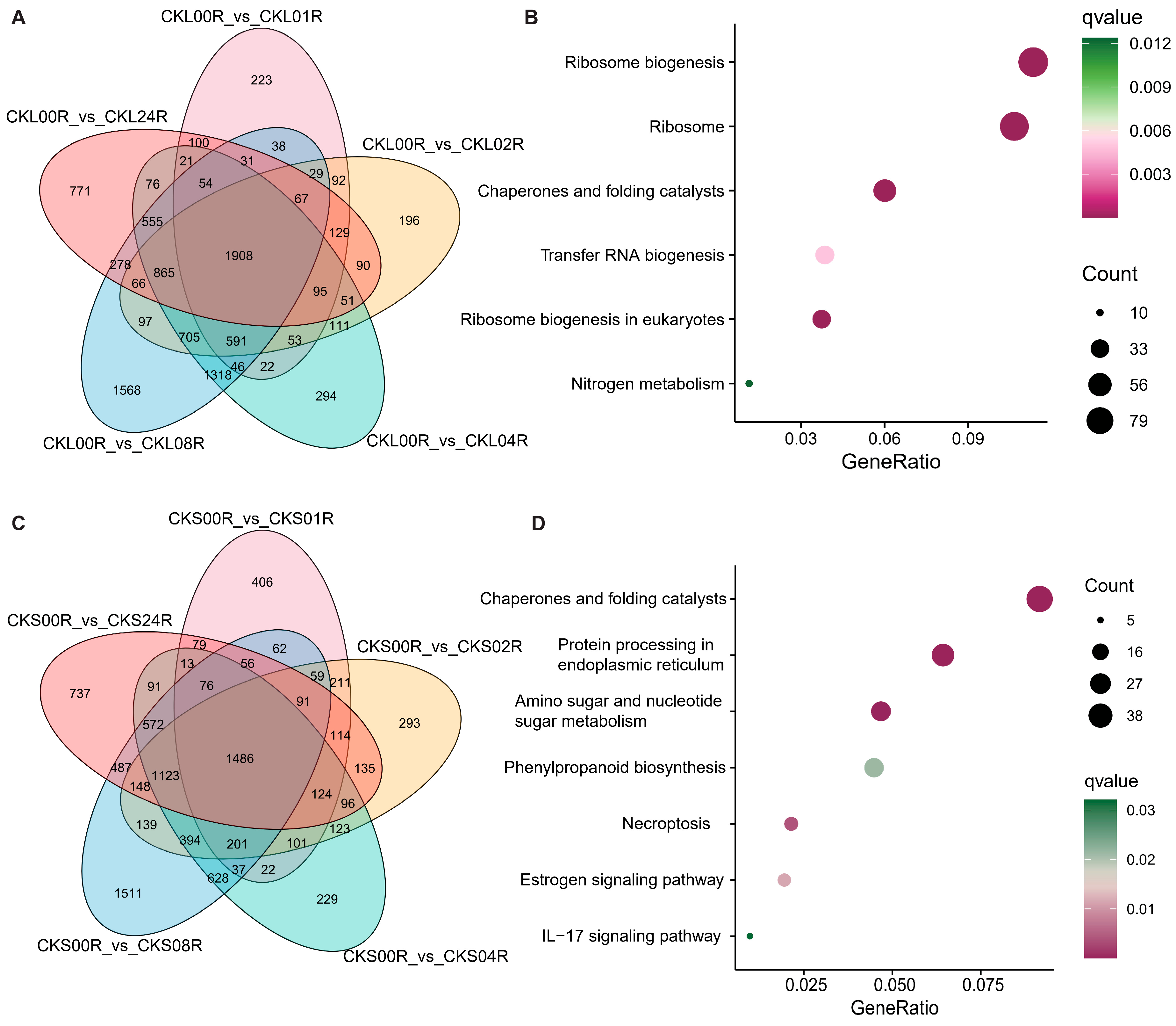
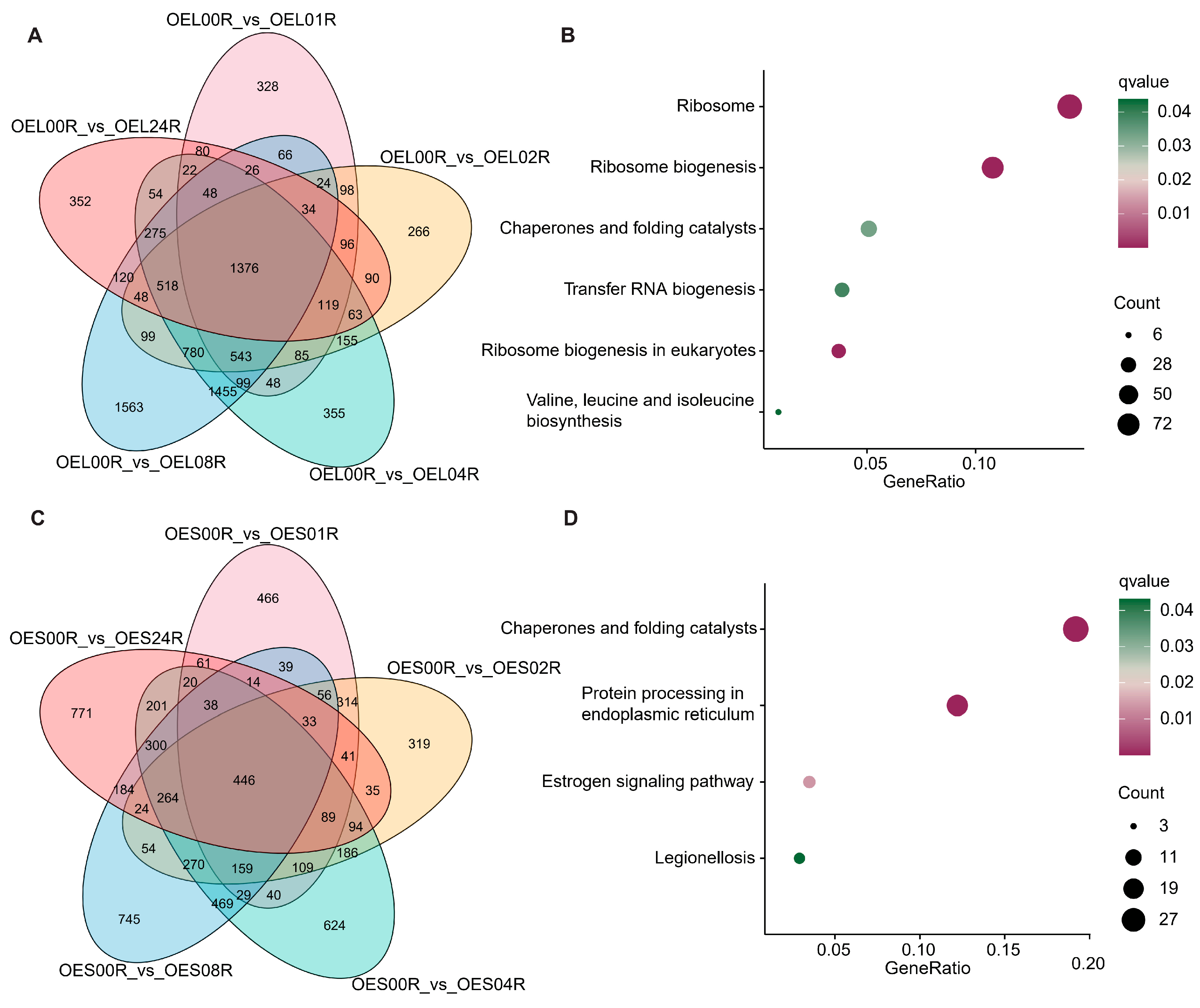
3.2. Functional Enrichment of DEGs
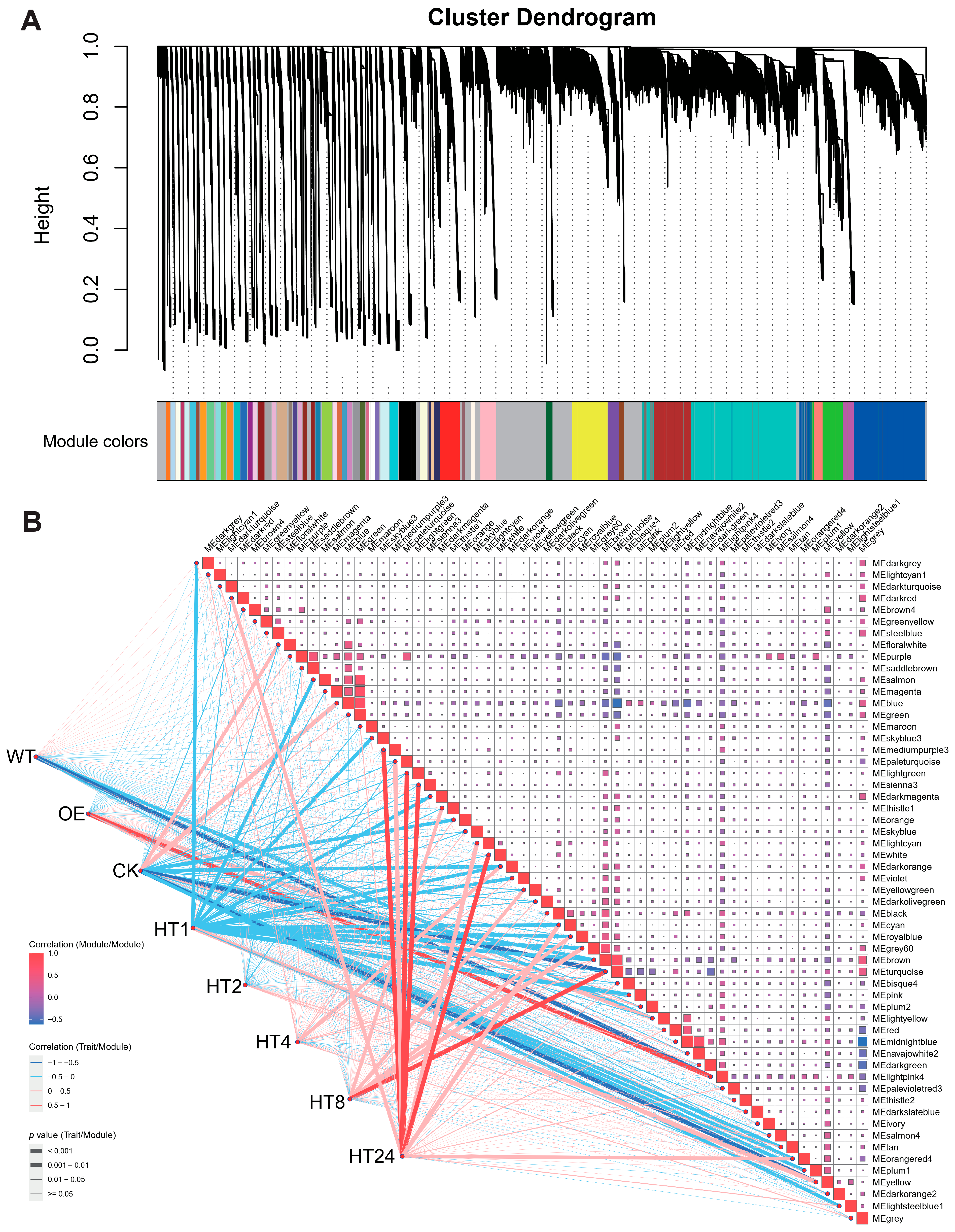
3.3. Identification of Co-Expression Modules by WGCNA
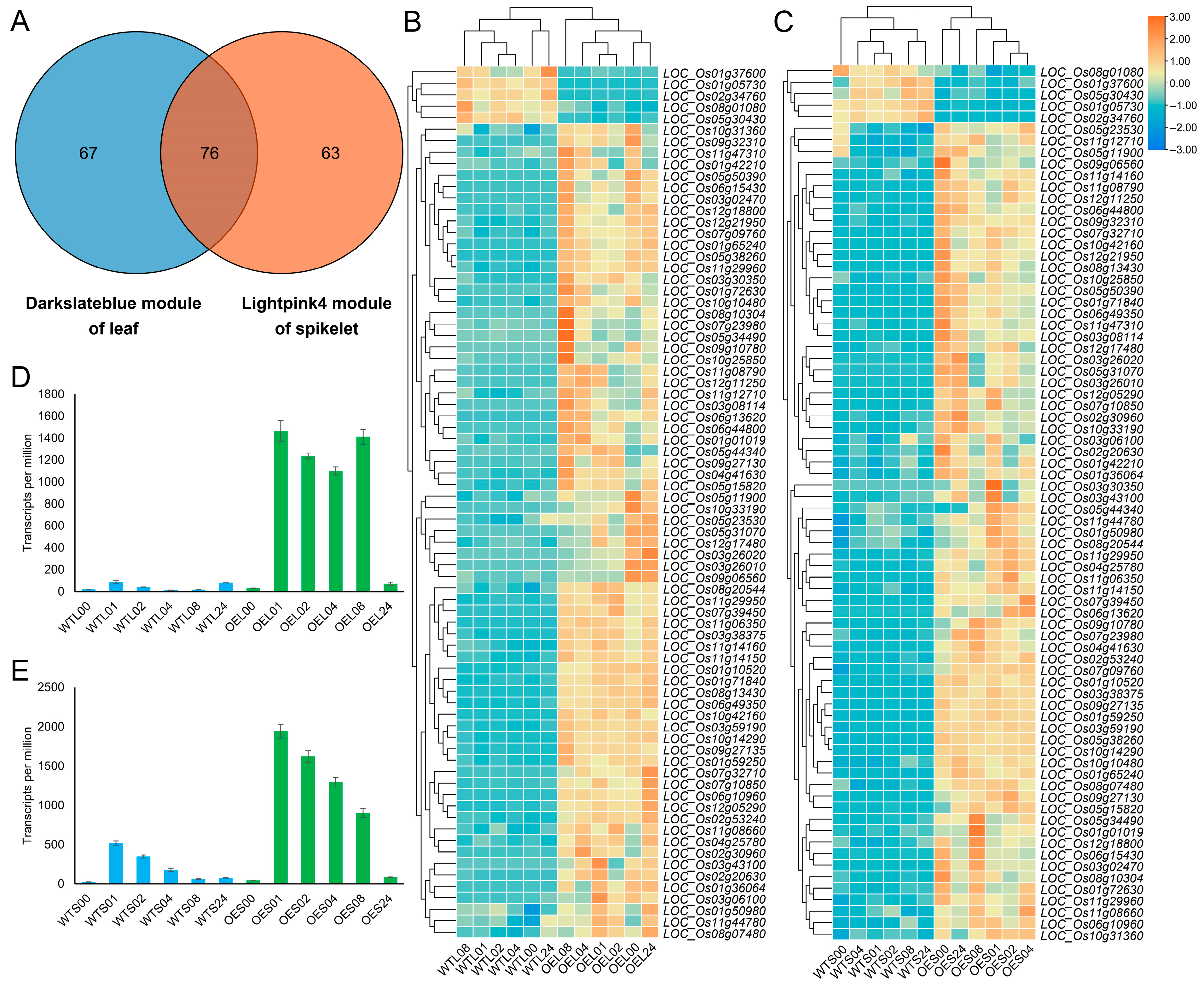
3.4. Gene Network of the Key Gene Modules Between WT and OE Lines
4. Discussion
4.1. Core Mechanisms of Hsp101-1 in Plant Thermotolerance
4.2. The Molecular Basis of Transcriptome Stability
4.3. Potential Heat Memory in Modules After Hsp101-1 Expression Decline
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Muthayya, S.; Sugimoto, J.D.; Montgomery, S.; Maberly, G.F. An overview of global rice production, supply, trade, and consumption. Ann. N. Y. Acad. Sci. 2014, 1324, 7–14. [Google Scholar] [CrossRef]
- Li, N.; Zhao, Y.; Han, J.; Yang, Q.; Liang, J.; Liu, X.; Wang, Y.; Huang, Z. Impacts of future climate change on rice yield based on crop model simulation-A meta-analysis. Sci. Total Environ. 2024, 949, 175038. [Google Scholar] [CrossRef]
- Itoh, H.; Yamashita, H.; Wada, K.C.; Yonemaru, J.I. Real-time emulation of future global warming reveals realistic impacts on the phenological response and quality deterioration in rice. Proc. Natl. Acad. Sci. USA 2024, 121, e2316497121. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.K.; Kumar, S.; Vatta, K.; Bheemanahalli, R.; Dhillon, J.; Reddy, K.N. Impact of recent climate change on corn, rice, and wheat in southeastern USA. Sci. Rep. 2022, 12, 16928. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S. Spatiotemporal Change of Heat Stress and Its Impacts on Rice Growth in the Middle and Lower Reaches of the Yangtze River. Agriculture 2022, 12, 1097. [Google Scholar] [CrossRef]
- Yu, J.; Du, T.; Zhang, P.; Ma, Z.; Chen, X.; Cao, J.; Li, H.; Li, T.; Zhu, Y.; Xu, F.; et al. Impacts of High Temperatures on the Growth and Development of Rice and Measures for Heat Tolerance Regulation: A Review. Agronomy 2024, 14, 2811. [Google Scholar] [CrossRef]
- Ren, H.; Bao, J.; Gao, Z.; Sun, D.; Zheng, S.; Bai, J. How rice adapts to high temperatures. Front. Plant Sci. 2023, 14, 1137923. [Google Scholar] [CrossRef]
- Hu, C.; Yang, J.; Qi, Z.; Wu, H.; Wang, B.; Zou, F.; Mei, H.; Liu, J.; Wang, W.; Liu, Q. Heat shock proteins: Biological functions, pathological roles, and therapeutic opportunities. MedComm (2020) 2022, 3, e161. [Google Scholar] [CrossRef]
- Wu, C.; Cui, K.; Wang, W.; Li, Q.; Fahad, S.; Hu, Q.; Huang, J.; Nie, L.; Peng, S. Heat-induced phytohormone changes are associated with disrupted early reproductive development and reduced yield in rice. Sci. Rep. 2016, 6, 34978. [Google Scholar] [CrossRef]
- Lohani, N.; Singh, M.B.; Bhalla, P.L. Deciphering the Vulnerability of Pollen to Heat Stress for Securing Crop Yields in a Warming Climate. Plant Cell Environ. 2025, 48, 2549–2580. [Google Scholar] [CrossRef] [PubMed]
- Kampinga, H.H.; Hageman, J.; Vos, M.J.; Kubota, H.; Tanguay, R.M.; Bruford, E.A.; Cheetham, M.E.; Chen, B.; Hightower, L.E. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones 2009, 14, 105–111. [Google Scholar] [CrossRef]
- Richter, K.; Haslbeck, M.; Buchner, J. The heat shock response: Life on the verge of death. Mol. Cell 2010, 40, 253–266. [Google Scholar] [CrossRef]
- Charng, Y.Y.; Liu, H.C.; Liu, N.Y.; Chi, W.T.; Wang, C.N.; Chang, S.H.; Wang, T.T. A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 2007, 143, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.Y.; Chai, K.H.; Ko, S.S.; Kuang, L.Y.; Lur, H.S.; Charng, Y.Y. A positive feedback loop between HEAT SHOCK PROTEIN101 and HEAT STRESS-ASSOCIATED 32-KD PROTEIN modulates long-term acquired thermotolerance illustrating diverse heat stress responses in rice varieties. Plant Physiol. 2014, 164, 2045–2053. [Google Scholar] [CrossRef]
- Yu, H.; Li, Q.; Li, Y.; Yang, H.; Lu, Z.; Wu, J.; Zhang, Z.; Shahid, M.Q.; Liu, X. Genomics Analyses Reveal Unique Classification, Population Structure and Novel Allele of Neo-Tetraploid Rice. Rice 2021, 14, 16. [Google Scholar] [CrossRef]
- Guan, Y.; Chen, Y.; Huang, Q.; He, Y.; Li, X.; Zhu, Z.; Xiong, Y.; Ouyang, J.; Jiang, G.; Zhang, Y.; et al. Exploring heat stress responses and heat tolerance in rice in the reproductive stage: A dual omics approach. Plant Growth Regul. 2025, 1–19. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, J.P.; Li, C.; Buell, C.R. The rice genome annotation project: An updated database for mining the rice genome. Nucleic Acids Res. 2025, 53, D1614–D1622. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef]
- Merret, R.; Carpentier, M.C.; Favory, J.J.; Picart, C.; Descombin, J.; Bousquet-Antonelli, C.; Tillard, P.; Lejay, L.; Deragon, J.M.; Charng, Y.Y. Heat Shock Protein HSP101 Affects the Release of Ribosomal Protein mRNAs for Recovery after Heat Shock. Plant Physiol. 2017, 174, 1216–1225. [Google Scholar] [CrossRef]
- Ma, Z.; Lv, J.; Wu, W.; Fu, D.; Lü, S.; Ke, Y.; Yang, P. Regulatory network of rice in response to heat stress and its potential application in breeding strategy. Mol. Breed. 2023, 43, 68. [Google Scholar] [CrossRef]
- Cao, G.; Huang, H.; Yang, Y.; Xie, B.; Tang, L. Analysis of drought and heat stress response genes in rice using co-expression network and differentially expressed gene analyses. PeerJ 2024, 12, e17255. [Google Scholar] [CrossRef]
- Hu, Y.; Zhu, N.; Wang, X.; Yi, Q.; Zhu, D.; Lai, Y.; Zhao, Y. Analysis of rice Snf2 family proteins and their potential roles in epigenetic regulation. Plant Physiol. Biochem. 2013, 70, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Brzezinka, K.; Altmann, S.; Czesnick, H.; Nicolas, P.; Gorka, M.; Benke, E.; Kabelitz, T.; Jähne, F.; Graf, A.; Kappel, C.; et al. Arabidopsis FORGETTER1 mediates stress-induced chromatin memory through nucleosome remodeling. eLife 2016, 5, e17061. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.Y.; Juan, Y.T.; Hsu, Y.H.; Wu, S.H.; Liao, H.T.; Fung, R.W.; Charng, Y.Y. Interplay between heat shock proteins HSP101 and HSA32 prolongs heat acclimation memory posttranscriptionally in Arabidopsis. Plant Physiol. 2013, 161, 2075–2084. [Google Scholar] [CrossRef] [PubMed]


| Treatment | Up_Flag Leaf a | Down_Flag Leaf b | Up_Spikelet c | Down_Spikelet d |
|---|---|---|---|---|
| HT_00h e | 545 | 676 | 910 | 1315 |
| HT_01h e | 116 | 36 | 127 | 34 |
| HT_02h e | 177 | 226 | 102 | 94 |
| HT_04h e | 108 | 28 | 93 | 24 |
| HT_08h e | 199 | 90 | 110 | 10 |
| HT_24h e | 189 | 88 | 167 | 21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, H.; Jiang, L.; Sun, B.; Liu, Q.; Mao, X.; Zhang, J.; Chen, P.; Chen, W.; Li, C.; Lyu, S. Hsp101-1 Orchestrates Thermotolerance in Rice via Pre-Activated Transcriptional Networks and Modular Cross-Tissue Coordination. Genes 2025, 16, 1039. https://doi.org/10.3390/genes16091039
Yu H, Jiang L, Sun B, Liu Q, Mao X, Zhang J, Chen P, Chen W, Li C, Lyu S. Hsp101-1 Orchestrates Thermotolerance in Rice via Pre-Activated Transcriptional Networks and Modular Cross-Tissue Coordination. Genes. 2025; 16(9):1039. https://doi.org/10.3390/genes16091039
Chicago/Turabian StyleYu, Hang, Liqun Jiang, Bingrui Sun, Qing Liu, Xingxue Mao, Jing Zhang, Pingli Chen, Wenfeng Chen, Chen Li, and Shuwei Lyu. 2025. "Hsp101-1 Orchestrates Thermotolerance in Rice via Pre-Activated Transcriptional Networks and Modular Cross-Tissue Coordination" Genes 16, no. 9: 1039. https://doi.org/10.3390/genes16091039
APA StyleYu, H., Jiang, L., Sun, B., Liu, Q., Mao, X., Zhang, J., Chen, P., Chen, W., Li, C., & Lyu, S. (2025). Hsp101-1 Orchestrates Thermotolerance in Rice via Pre-Activated Transcriptional Networks and Modular Cross-Tissue Coordination. Genes, 16(9), 1039. https://doi.org/10.3390/genes16091039






