Genome-Wide Identification of the Dendrocalamus latiflorus IDD Gene Family and Its Functional Role in Bamboo Shoot Development
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome-Wide Identification of DlIDD Family Members
2.2. Phylogenetic Reconstruction and Sequence Analyses Across Four Plant Species
2.3. Conserved Motif and Gene Structure Analysis
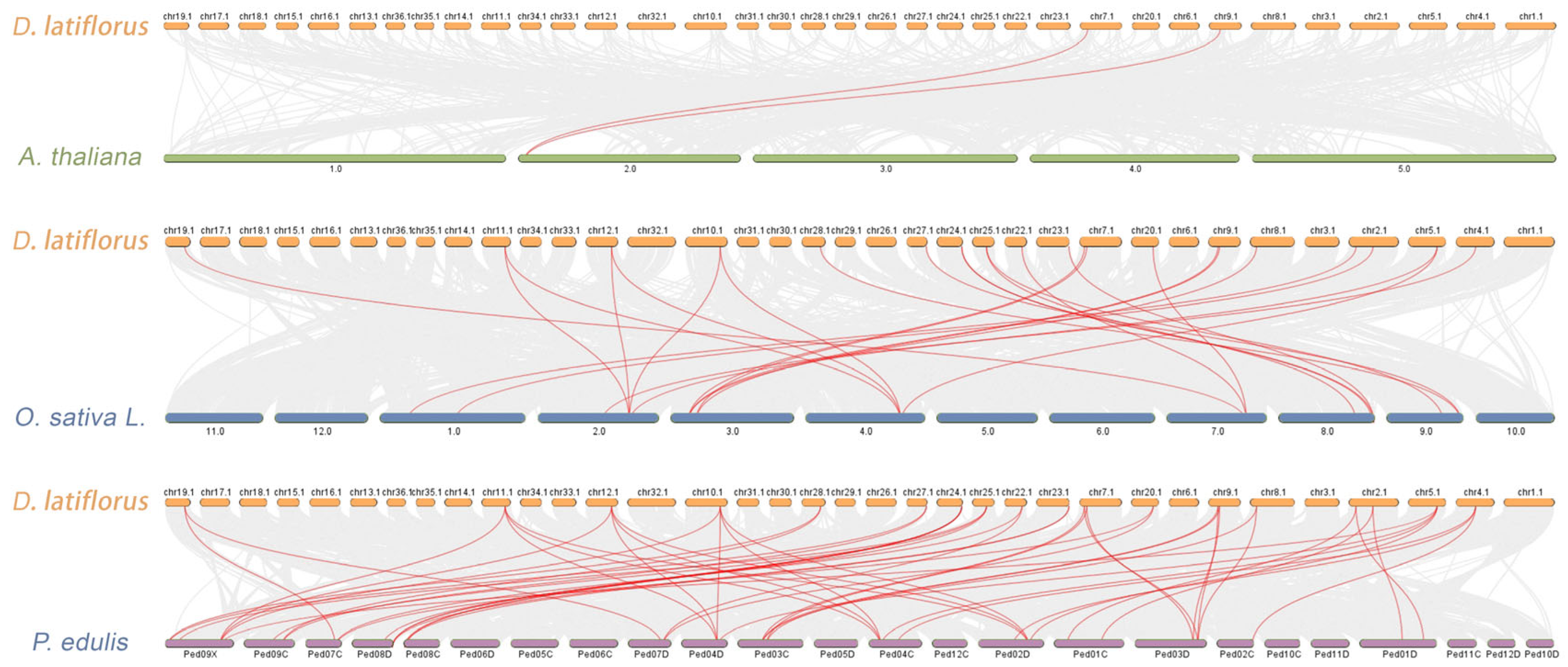
2.4. Chromosomal Mapping, Synteny, and Gene Duplication Analysis
2.5. Promoter Cis-Acting Element Analysis
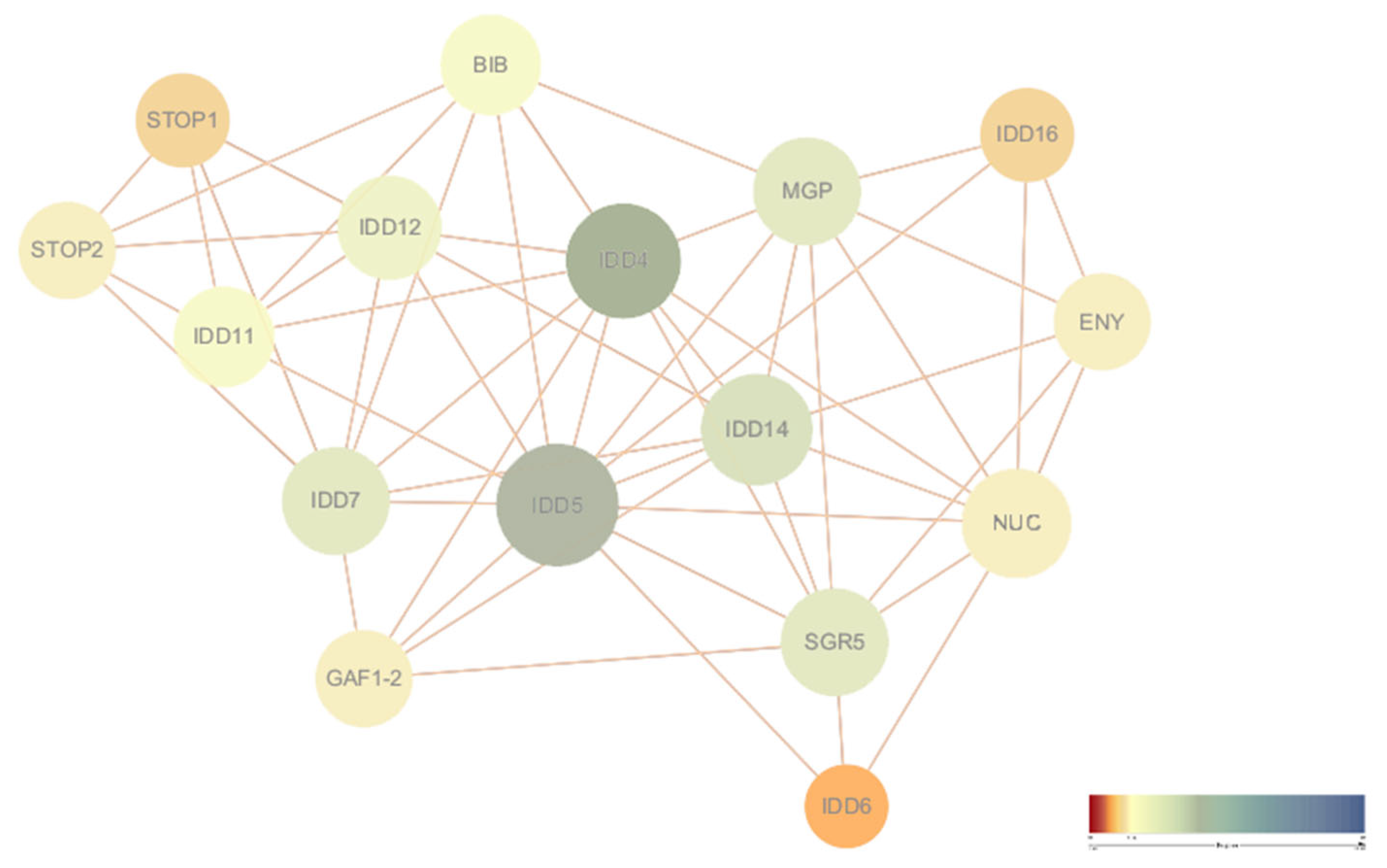
2.6. Protein–Protein Interaction (PPI) Network Construction
2.7. Plant Materials and Growth Conditions
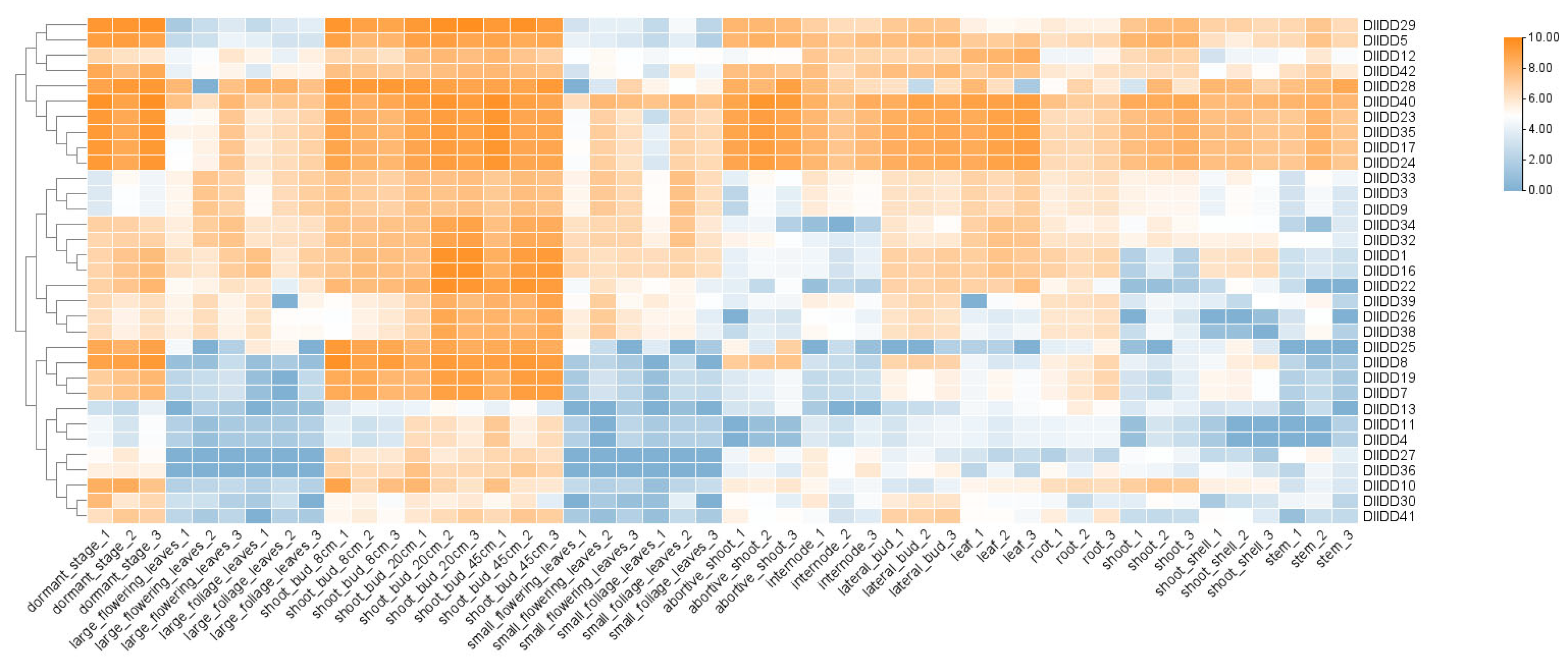
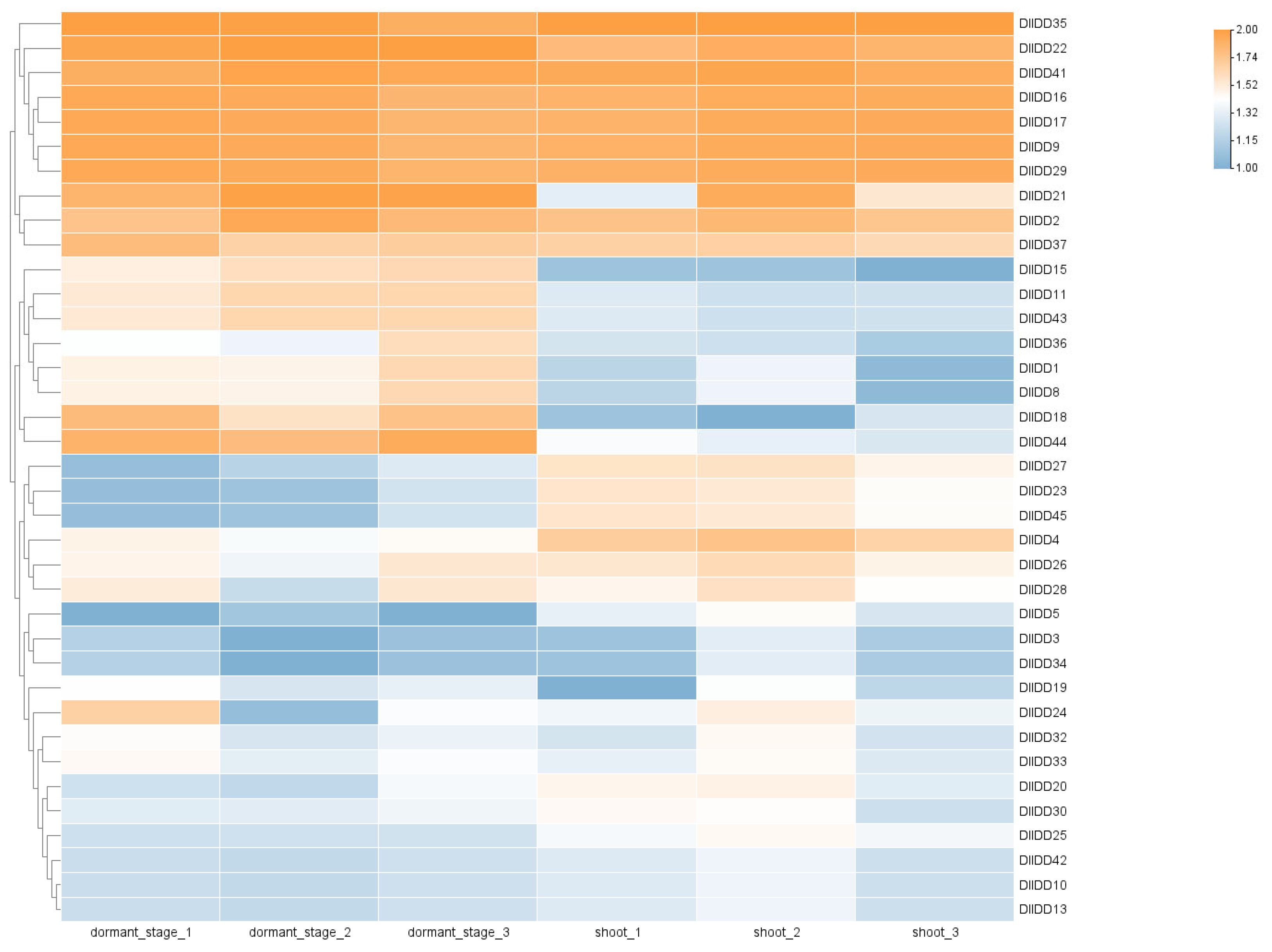
2.8. Transcriptome Sequencing and Expression Profiling of DlIDD Genes
2.9. Quantitative Real-Time PCR (qRT-PCR) Validation
3. Results
3.1. Identification of DlIDD Genes and Physicochemical Characterization of Encoded Proteins
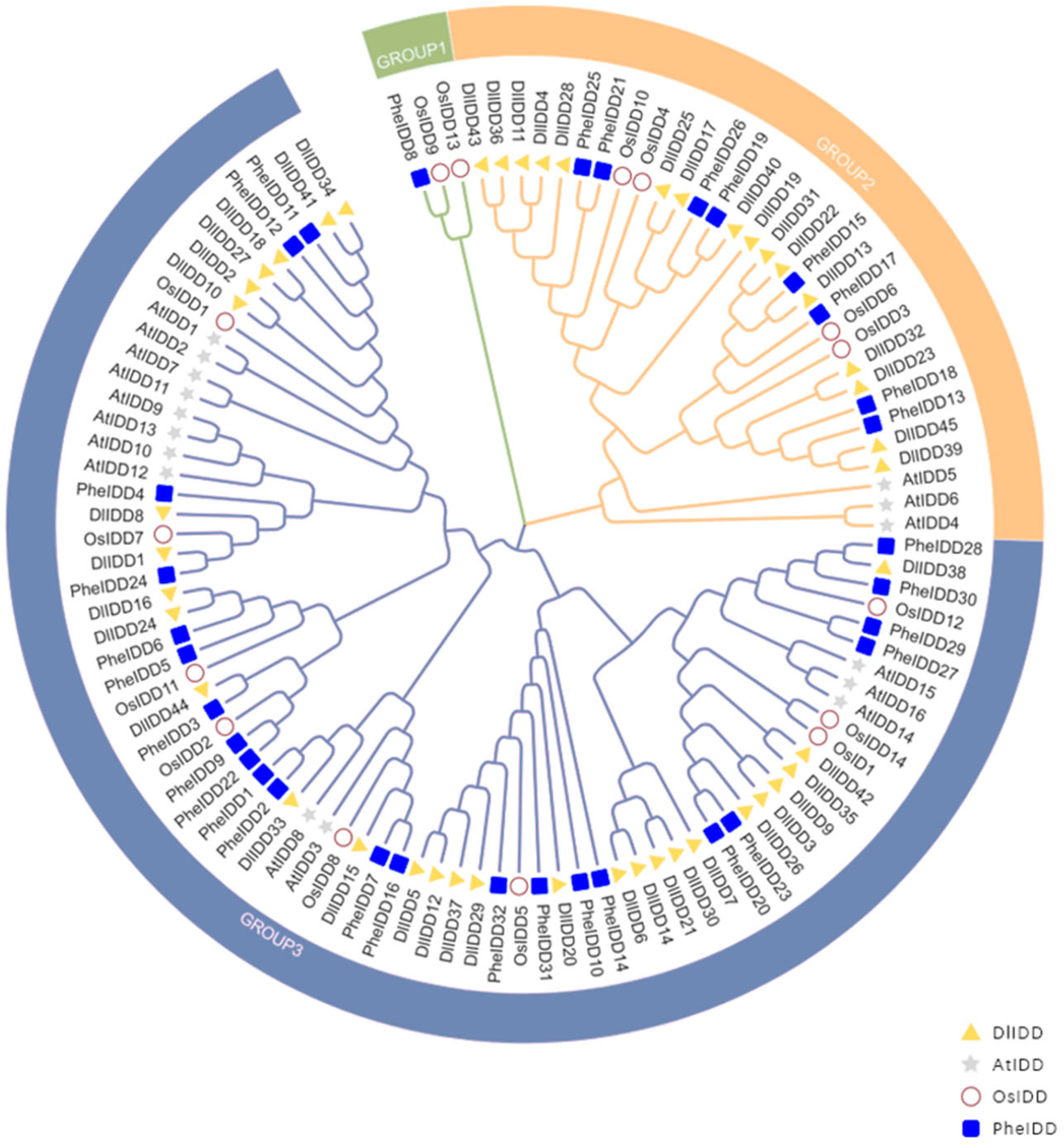
3.2. Phylogenetic Analysis of IDD Gene Families Across Four Plant Species
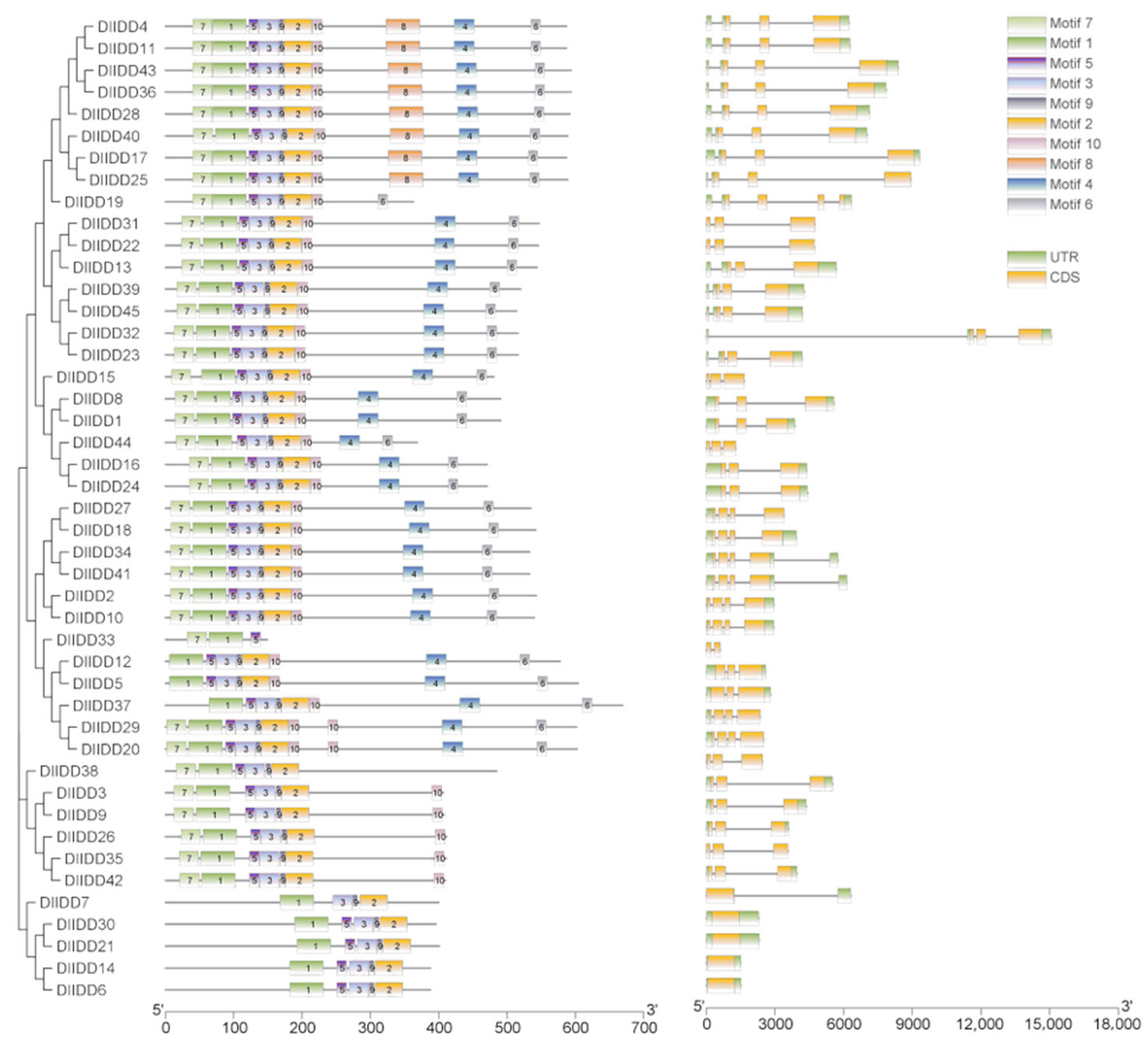
3.3. Gene Structure and Conserved Protein Motif Analysis of DlIDD Genes
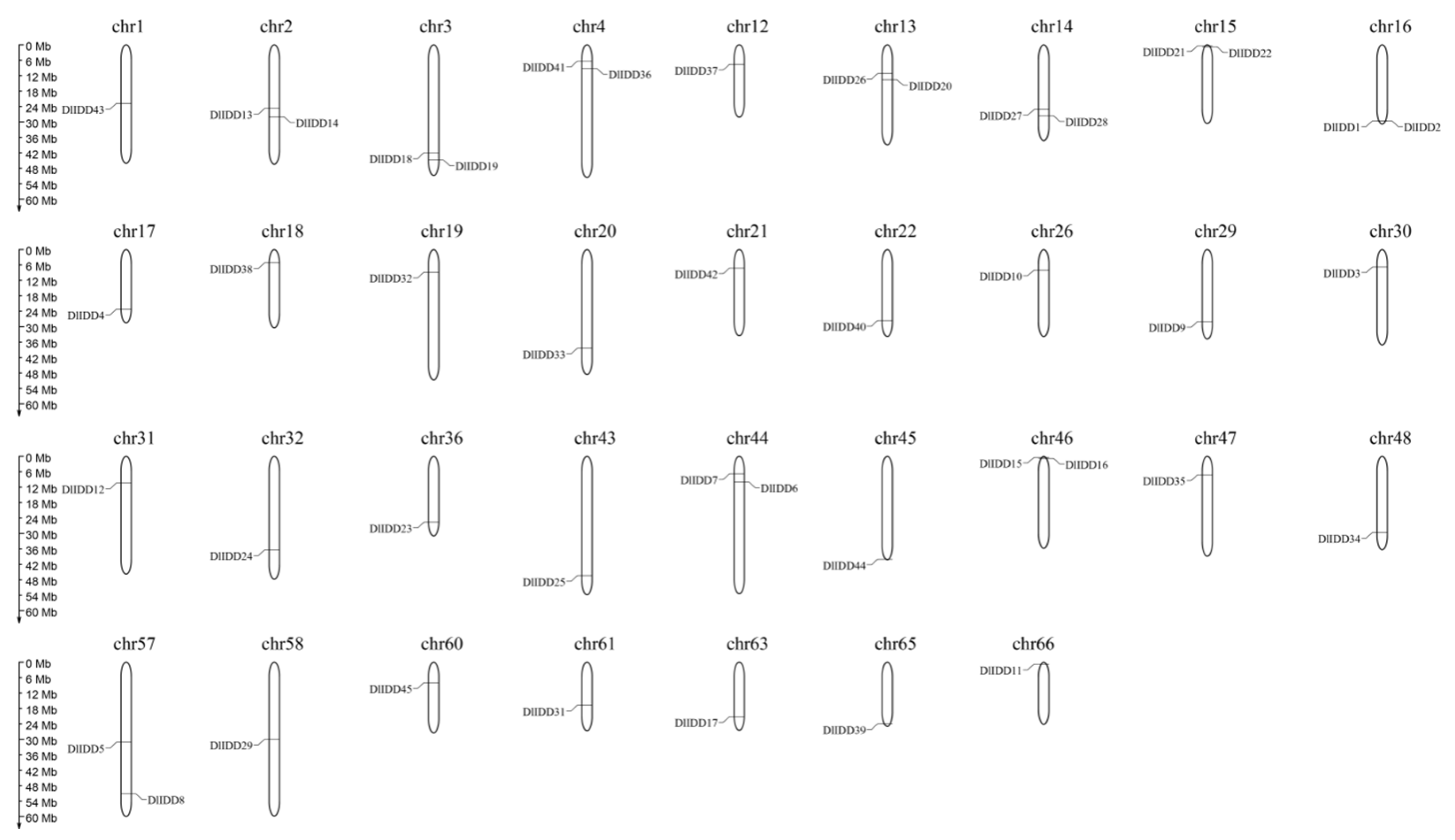
3.4. Chromosomal Distribution and Gene Duplication Events of DlIDD Genes
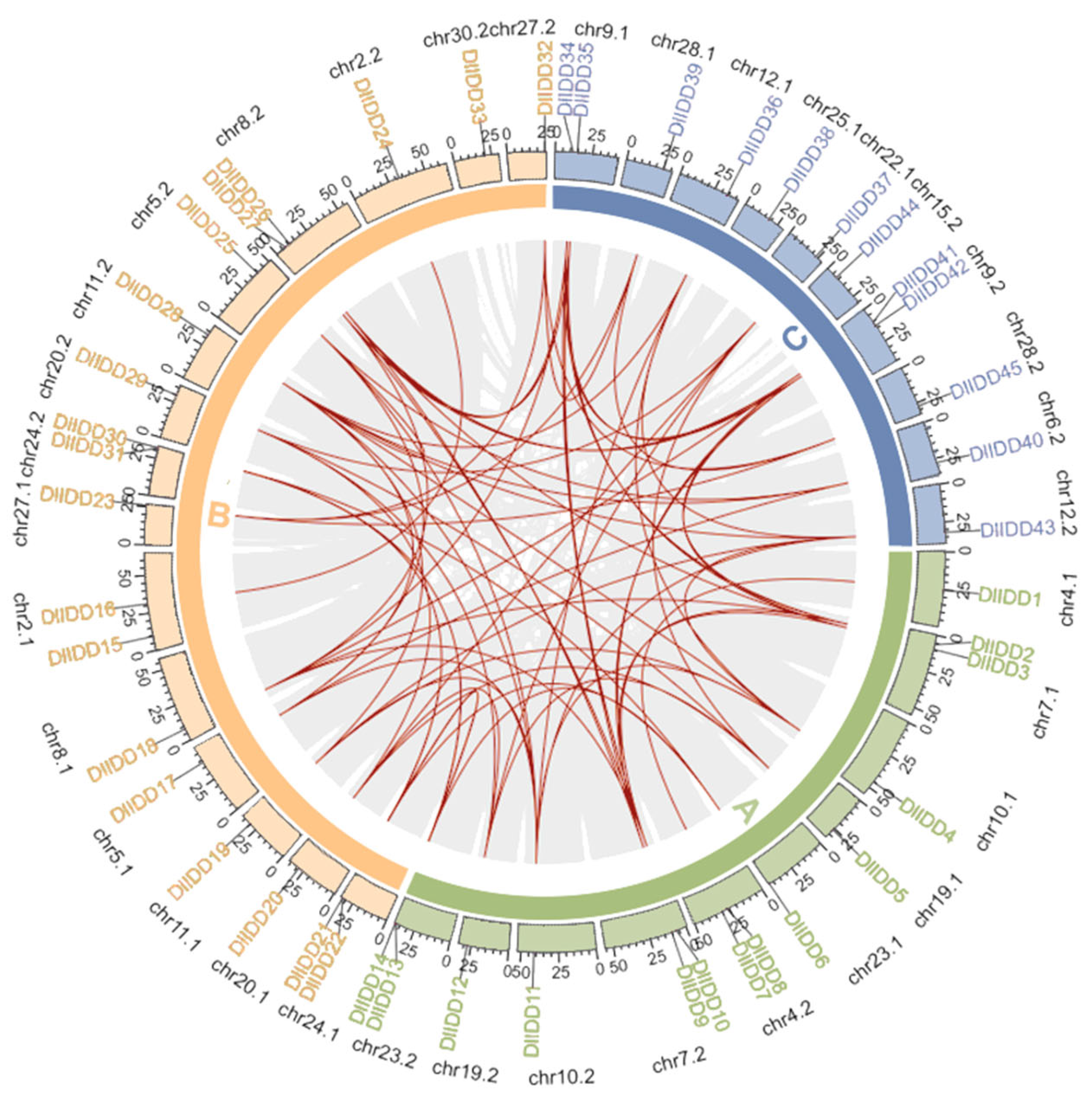
3.5. Synteny and Positive Selection Analysis of IDD Gene Families
3.6. Identification and Analysis of Cis-Regulatory Elements in DlIDD Gene Promoters
3.7. Protein–Protein Interaction Network Construction and Functional Interpretation of DlIDD Proteins
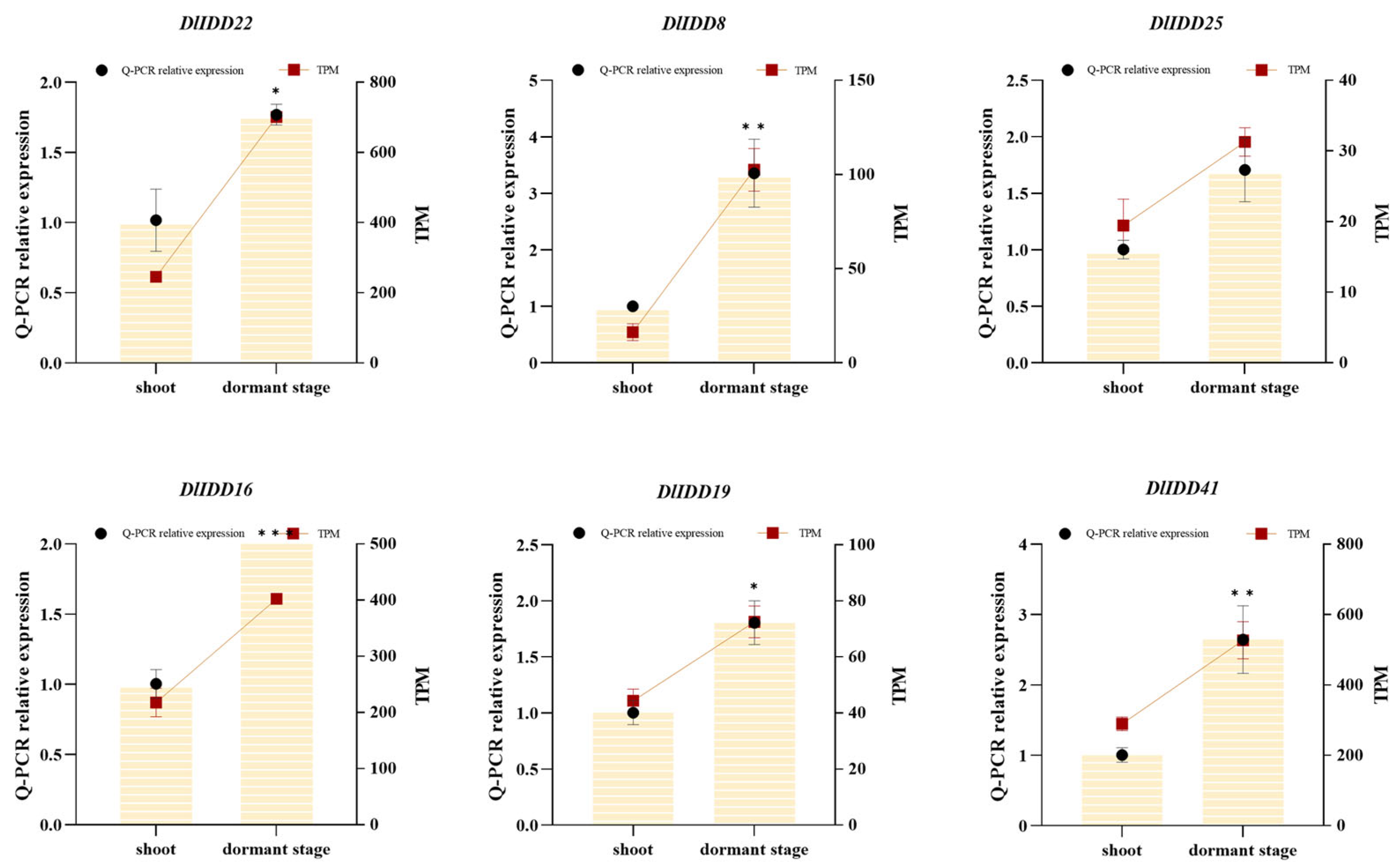
3.8. Expression Profiling of IDDs in Germinating Buds, Dormant Buds, and Distinct Organs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis Transcription Factors: Genome-Wide Comparative Analysis Among Eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Feng, Z.; Meng, Y.; Bian, L.; Xie, H.; Mysore, K.S.; Liang, J. SLENDER RICE1 and Oryza sativa INDETERMINATE DOMAIN2 Regulating OsmiR396 Are Involved in Stem Elongation. Plant Physiol. 2020, 182, 2213–2227. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-F.; Wang, J.-X.; Du, Y.-F.; Zou, H.-W.; Zhang, Z.-X. Identification of Indeterminate Domain Protein Family Genes Associated with Flowering Time in Maize. Acta Agron. Sin. 2019, 45, 499–507. [Google Scholar] [CrossRef]
- Wang, D.; Yao, S.; Agassin, R.H.; Zhang, M.; Lou, X.; Huang, Z.; Ji, K. Transcriptome-Wide Identification of CCCH-Type Zinc Finger Proteins Family in Pinus Massoniana and RR-TZF Proteins in Stress Response. Genes 2022, 13, 1639. [Google Scholar] [CrossRef]
- Shalmani, A.; Fan, S.; Jia, P.; Li, G.; Muhammad, I.; Li, Y.; Sharif, R.; Dong, F.; Zuo, X.; Li, K.; et al. Genome Identification of B-BOX Gene Family Members in Seven Rosacea Species and Their Expression Analysis in Response to Flower Induction in Malus domestica. Molecules 2018, 23, 1763. [Google Scholar] [CrossRef]
- Colasanti, J.; Tremblay, R.; Wong, A.Y.; Coneva, V.; Kozaki, A.; Mable, B.K. The Maize INDETERMINATE1 Flowering Time Regulator Defines a Highly Conserved Zinc Finger Protein Family in Higher Plants. BMC Genom. 2006, 7, 158. [Google Scholar] [CrossRef]
- Aoyanagi, T.; Ikeya, S.; Kobayashi, A.; Kozaki, A. Gene Regulation via the Combination of Transcription Factors in the INDETERMINATE DOMAIN and GRAS Families. Genes 2020, 11, 613. [Google Scholar] [CrossRef]
- Yoshida, H.; Hirano, K.; Sato, T.; Mitsuda, N.; Nomoto, M.; Maeo, K.; Koketsu, E.; Mitani, R.; Kawamura, M.; Ishiguro, S.; et al. DELLA Protein Functions as a Transcriptional Activatothrough the DNA Binding of the INDETERMINATE DOMAIN Family Proteins. Proc. Natl. Acad. Sci. USA 2014, 111, 7861–7866. [Google Scholar] [CrossRef]
- Jöst, M.; Hensel, G.; Kappel, C.; Druka, A.; Sicard, A.; Hohmann, U.; Beier, S.; Himmelbach, A.; Waugh, R.; Kumlehn, J.; et al. The INDETERMINATE DOMAIN Protein BROAD LEAF1 Limits Barley Leaf Width by Restricting Lateral Proliferation. Curr. Biol. 2016, 26, 903–909. [Google Scholar] [CrossRef]
- Akhtar, K.; Ain, N.U.; Prasad, P.V.V.; Naz, M.; Aslam, M.M.; Djalovic, I.; Riaz, M.; Ahmad, S.; Varshney, R.K.; He, B.; et al. Physiological, molecular, and environmental insights into plant nitrogen uptake, and metabolism under abiotic stresses. Plant Genome 2024, 17, e20461. [Google Scholar] [CrossRef]
- Meeks, T. Investigating the Function of the Arabidopsis Thaliana INDETERMINATE DOMAIN2 Gene. Master’s Thesis, University of Guelph, Guelph, ON, Canada, 2015. [Google Scholar]
- Liu, J.; Shu, D.; Tan, Z.; Ma, M.; Yang, H. The IDD Transcription Factors: Their Functions in Plant Development and Environmental Response. Phyton Int. J. Exp. Bot. 2024, 93, 63–79. [Google Scholar] [CrossRef]
- Fukazawa, J.; Teramura, H.; Murakoshi, S.; Nasuno, K.; Nishida, N.; Ito, T.; Yoshida, M.; Kamiya, Y.; Yamaguchi, S.; Takahashi, Y. DELLAs Function as Coactivators of GAI-ASSOCIATED FACTOR1 in Regulation of Gibberellin Homeostasis and Signaling in Arabidopsis. Plant Cell 2014, 26, 2920–2938. [Google Scholar] [CrossRef]
- Feurtado, J.A.; Huang, D.; Wicki-Stordeur, L.; Hemstock, L.E.; Potentier, M.S.; Tsang, E.W.T.; Cutler, A.J. The Arabidopsis C2H2 Zinc Finger INDETERMINATE DOMAIN1/ENHYDROUS Promotes the Transition to Germination by Regulating Light and Hormonal Signaling during Seed Maturation. Plant Cell 2011, 23, 1772–1794. [Google Scholar] [CrossRef] [PubMed]
- Qiao, G.; Liu, M.; Song, K.; Li, H.; Yang, H.; Yin, Y.; Zhuo, R. Phenotypic and Comparative Transcriptome Analysis of Different Ploidy Plants in Dendrocalamus latiflorus Munro. Front. Plant Sci. 2017, 8, 1371. [Google Scholar] [CrossRef]
- Chen, J. The Correlation Between IDD Gene Family and Development of Shoot/Bud in Phyllostachys Edulis. Master’s Thesis, Zhejiang Agricultural and Forestry University, Hangzhou, China, 2020. Available online: https://link.cnki.net/doi/10.27756/d.cnki.gzjlx.2020.000089 (accessed on 13 July 2025).
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple Sequence Alignment Using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2003. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Wang, W. The Molecular Detection of Corynespora Cassiicola on Cucumber by PCR Assay Using DNAman Software and NCBI. In Computer and Computing Technologies in Agriculture IX, Proceedings of the CCTA 2015, Beijing, China, 27–30 September 2015; Li, D., Li, Z., Eds.; IFIP Advances in Information and Communication Technology 479; Springer: Cham, Switzerland, 2016. [Google Scholar] [CrossRef]
- Monroe, J.D.; Moolani, S.A.; Irihamye, E.N.; Lett, K.E.; Hebert, M.D.; Gibert, Y.; Smith, M.E. Cisplatin and phenanthriplatin modulate long-noncoding RNA expression in A549 and IMR90 cells revealing regulation of microRNAs, Wnt/β-catenin and TGF-β signaling. Sci. Rep. 2021, 11, 10408. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: ProteinProtein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Majeed, A.; Mukhtar, S. Protein-Protein Interaction Network Exploration Using Cytoscape. Methods Mol. Biol. 2023, 2690, 419–427. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Vera Alvarez, R.; Pongor, L.S.; Mariño-Ramírez, L.; Landsman, D. TPMCalculator: One-step software to quantify mRNA abundance of genomic features. Bioinformatics 2019, 35, 1960–1962. [Google Scholar] [CrossRef] [PubMed]
- Ding, A.; Ding, A.; Li, P.; Wang, J.; Cheng, T.; Bao, F.; Zhang, Q. Genome-Wide Identification and Low-Temperature Expression Analysis of bHLH Genes in Prunus mume. Front. Genet. 2021, 12, 762135. [Google Scholar] [CrossRef]
- Rajurkar, M.; Parikh, A.R.; Solovyov, A.; You, E.; Kulkarni, A.S.; Chu, C.; Xu, K.H.; Jaicks, C.; Taylor, M.S.; Wu, C.; et al. Reverse Transcriptase Inhibition Disrupts Repeat Element Life Cycle in Colorectal Cancer. Cancer Discov. 2022, 12, 1462–1481. [Google Scholar] [CrossRef]
- Guo, W.; Cherubini, P.; Zhang, J.; Li, M.H.; Qi, L. Leaf stomatal traits rather than anatomical traits regulate gross primary productivity of moso bamboo (Phyllostachys edulis) stands. Front. Plant Sci. 2023, 14, 1117564. [Google Scholar] [CrossRef]
- Gourgas, O.; Lemire, G.; Eaton, A.J.; Alshahrani, S.; Duker, A.L.; Li, J.; Carroll, R.S.; Mackenzie, S.; Nikkel, S.M.; Care4Rare Canada Consortium; et al. Specific heterozygous variants in MGP lead to endoplasmic reticulum stress and cause spondyloepiphyseal dysplasia. Nat. Commun. 2023, 14, 7054. [Google Scholar] [CrossRef]
- Cui, K.; He, C.; Zhang, J.; Liao, S. Characteristics of temporal and spatial tissue development during the rapidly growing stage of Moso bamboo culms. For. Res. 2012, 25, 425–431. [Google Scholar]
- Shao, D.; Villet, O.; Zhang, Z.; Choi, S.W.; Yan, J.; Ritterhoff, J.; Gu, H.; Djukovic, D.; Christodoulou, D.; Kolwicz, S.C.; et al. Glucose promotes cell growth by suppressing branched-chain amino acid degradation. Nat. Commun. 2018, 9, 2935. [Google Scholar] [CrossRef]
- Kozaki, A. INDETERMINATE DOMAIN Transcription Factors in Crops: Plant Architecture, Disease Resistance, Stress Response, Flowering, and More. Int. J. Mol. Sci. 2024, 25, 10277. [Google Scholar] [CrossRef]
- Kailash, S.C.; Jyoti, S.P.; Laxmipreeya, B.; Trupti, D. Understanding the BLAST (Basic Local Alignment Search Tool) Programand a Step-by-step Guide for its use in Life Science Research. Bhartiya Krishi Anusandhan Patrika 2021, 36, 55–61. [Google Scholar] [CrossRef]
- Prochetto, S.; Reinheimer, R. Step by step evolution of Indeterminate Domain (IDD) transcriptional regulators: From algae to angiosperms. Ann. Bot. 2020, 126, 85101. [Google Scholar] [CrossRef]
- Liu, Y.; Zhu, C.; Yue, X.; Lin, Z.; Li, H.; Di, X.; Wang, J.; Gao, Z. Evolutionary relationship of moso bamboo forms and a multihormone regulatory cascade involving culm shape variation. Plant Biotechnol. J. 2024, 22, 2578–2592. [Google Scholar] [CrossRef]
- Kumar, M.; Le, D.T.; Hwang, S.; Seo, P.J.; Kim, H.U. Role of the INDETERMINATE DOMAIN Genes in Plants. Int. J. Mol. Sci. 2019, 20, 2286. [Google Scholar] [CrossRef] [PubMed]
- Coelho, C.P.; Huang, P.; Lee, D.-Y.; Brutnell, T.P. Making Roots, Shoots, and Seeds: IDD Gene Family Diversification in Plants. Trends Plant Sci. 2018, 23, 66–78. [Google Scholar] [CrossRef] [PubMed]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene Duplication and Evolution in Recurring Polyploidization-Diploidization Cycles in Plants. Genome Biol. 2019, 20, 38. [Google Scholar] [CrossRef]
- Birchler, J.A.; Yang, H. The multiple fates of gene duplications: Deletion, hypofunctionalization, subfunctionalization, neofunctionalization, dosage balance constraints, and neutral variation. Plant Cell 2022, 34, 2466–2474. [Google Scholar] [CrossRef] [PubMed]
- Cui, D.; Zhao, J.; Jing, Y.; Fan, M.; Liu, J.; Wang, Z.; Xin, W.; Hu, Y. The Arabidopsis IDD14, IDD15, and IDD16 Cooperatively Regulate Lateral Organ Morphogenesis and Gravitropism by Promoting Auxin Biosynthesis and Transport. PLoS Genet. 2013, 9, e1003759. [Google Scholar] [CrossRef]
- Mabry, M.E.; Brose, J.M.; Blischak, P.D.; Sutherland, B.; Dismukes, W.T.; Bottoms, C.A.; Edger, P.P.; Washburn, J.D.; An, H.; Hall, J.C.; et al. Phylogeny and multiple independent whole-genome duplication events in the Brassicales. Am. J. Bot. 2020, 107, 1148–1164. [Google Scholar] [CrossRef]
- Gao, B.; Chen, M.; Li, X.; Liang, Y.; Zhu, F.; Liu, T.; Zhang, D.; Wood, A.J.; Oliver, M.J.; Zhang, J. Evolution by duplication: Paleopolyploidy events in plants reconstructed by deciphering the evolutionary history of VOZ transcription factors. BMC Plant Biol. 2018, 18, 256. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Chen, Q.; Wu, W.; Wang, J.; Li, G.; Xu, S.; Shao, S.; Liu, M.; Zhong, C.; Wu, C.-I.; et al. Genomic evidence for rediploidization and adaptive evolution following the whole-genome triplication. Nat. Commun. 2024, 15, 1635. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Zhu, W.; Zhou, J.; Li, H.; Xu, X.; Zhang, B.; Gao, X. Repetitive DNA sequence detection and its role in the human genome. Commun. Biol. 2023, 6, 954. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Shi, A.; Meinhardt, L.W.; Mou, B. Genome-wide characterization and evolutionary analysis of the AP2/ERF gene family in lettuce (Lactuca sativa). Sci. Rep. 2023, 13, 21990. [Google Scholar] [CrossRef]
- Ezer, D.; Shepherd, S.J.K.; Brestovitsky, A.; Dickinson, P.; Cortijo, S.; Charoensawan, V.; Box, M.S.; Biswas, S.; Jaeger, K.E.; Wigge, P.A. The G-Box Transcriptional Regulatory Code in Arabidopsis. Plant Physiol. 2017, 175, 628–640. [Google Scholar] [CrossRef]
- Ji, S.; Chen, F.; Stein, P.; Wang, J.; Zhou, Z.; Wang, L.; Zhao, Q.; Lin, Z.; Liu, B.; Xu, K.; et al. OBOX regulates mouse zygotic genome activation and early development. Nature 2023, 620, 1047–1053. [Google Scholar] [CrossRef]
- Xu, X.; Mo, Q.; Cai, Z.; Jiang, Q.; Zhou, D.; Yi, J. Promoters, Key Cis-Regulatory Elements, and Their Potential Applications in Regulation of Cadmium (Cd) in Rice. Int. J. Mol. Sci. 2024, 25, 13237. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef]
- Han, B.; Di Wu, D.; Zhang, Y.; Li, D.-Z.; Xu, W.; Liu, A. Epigenetic regulation of seed-specific gene expression by DNA methylation valleys in castor bean. BMC Biol. 2022, 20, 57. [Google Scholar] [CrossRef]
- Rajendran, S.; Kang, Y.M.; Yang, I.B.; Eo, H.B.; Baek, K.L.; Jang, S.; Eybishitz, A.; Kim, H.C.; Je, B.I.; Park, S.J.; et al. Functional characterization of plant specific Indeterminate Domain (IDD) transcription factors in tomato (Solanum lycopersicum L.). Sci. Rep. 2024, 14, 8015. [Google Scholar] [CrossRef] [PubMed]
- Ban, E.; Song, E.J. Considerations and Suggestions for the Reliable Analysis of miRNA in Plasma Using qRT-PCR. Genes 2022, 13, 328. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Zheng, Y.; Yang, D.; Rong, J.; Chen, L.; Zhu, Q.; He, T.; Chen, L.; Ye, J.; Fan, L.; Gao, Y.; et al. Alleleaware Chromosomescale Assembly of the Allopolyploid Genome of Hexaploid Ma Bamboo (Dendrocalamus latiflorus Munro). J. Integr. Plant Biol. 2022, 64, 649–670. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, İ.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Liu, Z.; Pan, J.; Liu, S.; Yang, Z.; Zhang, H.; Yu, T.; He, S. Integrated Transcriptome and Metabolome Analysis Provides Insights into the Low-Temperature Response in Sweet Potato (Ipomoea batatas L.). Genes 2025, 16, 899. [Google Scholar] [CrossRef]
- Wang, J.; Huang, J.; Jia, X.; Hao, Z.; Yang, Y.; Tian, R.; Liang, Y. Characterization and Expression Analysis of β-Glucosidase Gene Under Abiotic Stresses in Pepper (Capsicum annuum L.). Genes 2025, 16, 889. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Bi, C. Comparative Genomic Analysis of Lactiplantibacillus plantarum: Insights into Its Genetic Diversity, Metabolic Function, and Antibiotic Resistance. Genes 2025, 16, 869. [Google Scholar] [CrossRef]
- Yang, J.; Ma, L.; Jiang, W.; Yao, Y.; Tang, Y.; Pang, Y. Comprehensive identification and characterization of abiotic stress and hormone responsive glycosyl hydrolase family 1 genes in Medicago truncatula. Plant Physiol. Biochem. 2021, 158, 21–33. [Google Scholar] [CrossRef]
- Dong, X.; Jiang, Y.; Hur, Y. Genome-Wide Analysis of Glycoside Hydrolase Family 1 β-glucosidase Genes in Brassica rapa and Their Potential Role in Pollen Development. Int. J. Mol. Sci. 2019, 20, 1663. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, M.; Zhang, X.; Deng, X.; Li, J.; Wang, M. Genome-wide identification and characterization of active ingredients related β-Glucosidases in Dendrobium catenatum. BMC Genom. 2022, 23, 612. [Google Scholar] [CrossRef]
- Gao, C.; Zhang, Z.; Zhu, Y.; Tian, J.; Yu, K.; Hou, J.; Luo, D.; Cai, J.; Zhu, Y. Genome-Wide Analysis of HIPP Gene Family in Maize Reveals Its Role in the Cadmium Stress Response. Genes 2025, 16, 770. [Google Scholar] [CrossRef]
- Wang, T.; Wang, C.; Liu, Y.; Zou, K.; Guan, M.; Wu, Y.; Yue, S.; Hu, Y.; Yu, H.; Zhang, K.; et al. Genome-Wide Identification of the Maize Chitinase Gene Family and Analysis of Its Response to Biotic and Abiotic Stresses. Genes 2024, 15, 1327. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Zhang, X.; Cao, M.; Yin, L.; Gao, A.; An, K.; Gao, S.; Guo, S.; Yin, H. Genome-Wide Identification, Expression and Interaction Analyses of PP2C Family Genes in Chenopodium quinoa. Genes 2024, 15, 41. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Wan, Y.; Jiang, D.; Gong, M.; Lin, J.; Xia, M.; Shi, C.; Xing, H.; Li, H.-L. Genome-Wide Identification, Characterization, and Expression Analysis of GRAS Gene Family in Ginger (Zingiber officinale Roscoe). Genes 2023, 14, 96. [Google Scholar] [CrossRef] [PubMed]
- Waschburger, E.L.; Guzman, F.; Turchetto-Zolet, A.C. Genome-Wide Identification and Analysis of DOF Gene Family in Eugenia uniflora L. (Myrtaceae). Genes 2022, 13, 2235. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, Y.-H.; Zhu, P.-K.; Zeng, M.-Y.; Gao, X.-R.; He, T.-Y.; Rong, J.-D.; Zheng, Y.-S.; Chen, L.-Y. Genome-Wide Identification of the Dendrocalamus latiflorus IDD Gene Family and Its Functional Role in Bamboo Shoot Development. Genes 2025, 16, 1036. https://doi.org/10.3390/genes16091036
Lin Y-H, Zhu P-K, Zeng M-Y, Gao X-R, He T-Y, Rong J-D, Zheng Y-S, Chen L-Y. Genome-Wide Identification of the Dendrocalamus latiflorus IDD Gene Family and Its Functional Role in Bamboo Shoot Development. Genes. 2025; 16(9):1036. https://doi.org/10.3390/genes16091036
Chicago/Turabian StyleLin, Yu-Han, Peng-Kai Zhu, Mei-Yin Zeng, Xin-Ru Gao, Tian-You He, Jun-Dong Rong, Yu-Shan Zheng, and Ling-Yan Chen. 2025. "Genome-Wide Identification of the Dendrocalamus latiflorus IDD Gene Family and Its Functional Role in Bamboo Shoot Development" Genes 16, no. 9: 1036. https://doi.org/10.3390/genes16091036
APA StyleLin, Y.-H., Zhu, P.-K., Zeng, M.-Y., Gao, X.-R., He, T.-Y., Rong, J.-D., Zheng, Y.-S., & Chen, L.-Y. (2025). Genome-Wide Identification of the Dendrocalamus latiflorus IDD Gene Family and Its Functional Role in Bamboo Shoot Development. Genes, 16(9), 1036. https://doi.org/10.3390/genes16091036





