Comparative Chloroplast Genomics of Ten Collabieae Species Including Three Novel Genomes
Abstract
1. Introduction
2. Materials and Methods
2.1. Material Sampling, DNA Isolation, and Sequencing
2.2. Complete Chloroplast Genome Assembly and Annotations
2.3. Relative Synonymous Codon Usage (RSCU), and Nucleotide Diversity Analysis
2.4. Comparative Chloroplast Genomes Analysis of Ten Species from Collabieae
2.5. Phylogenetic Analyses in Collabieae
3. Results
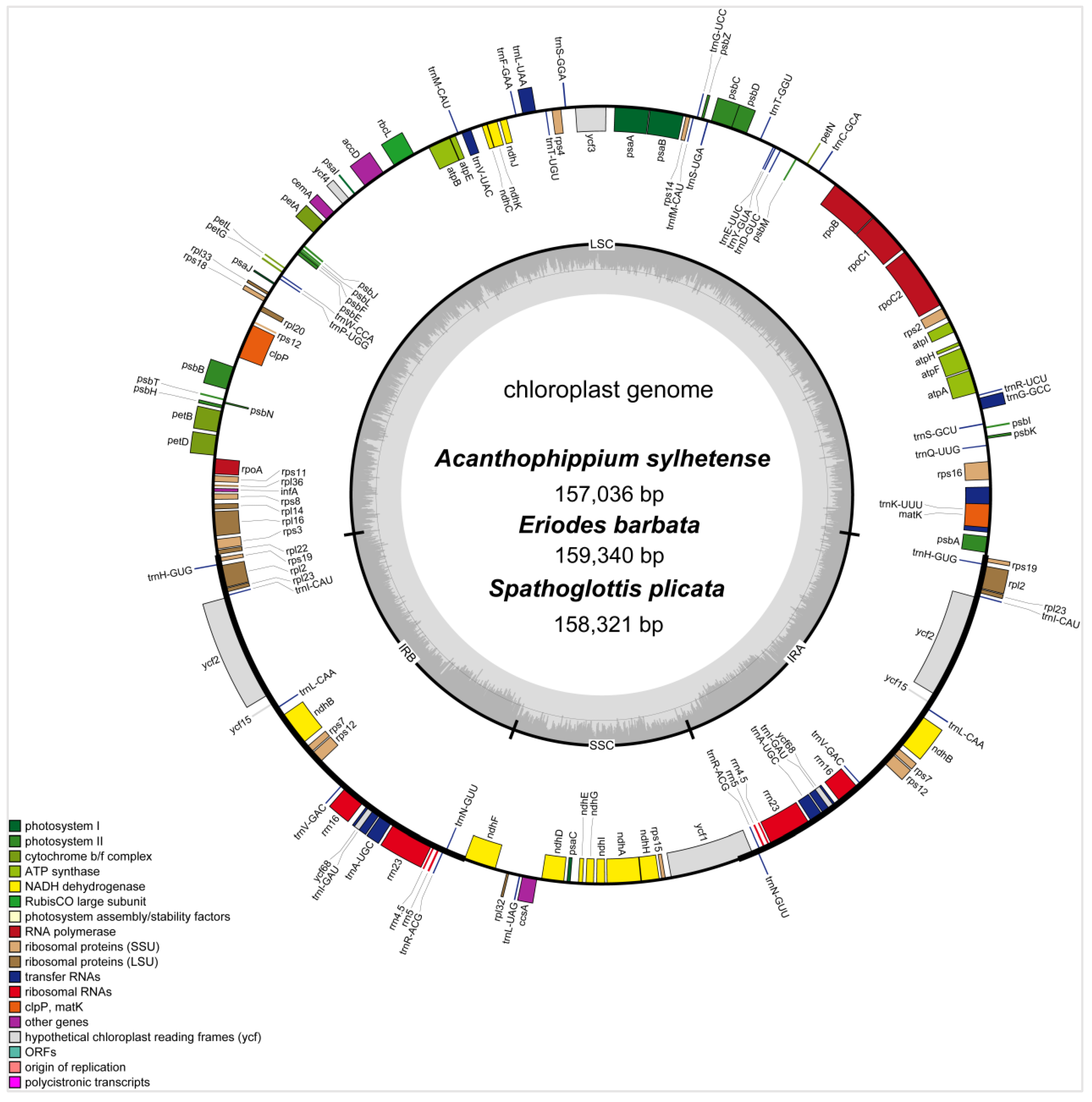
3.1. Plastome Structures of Three Collabieae Species
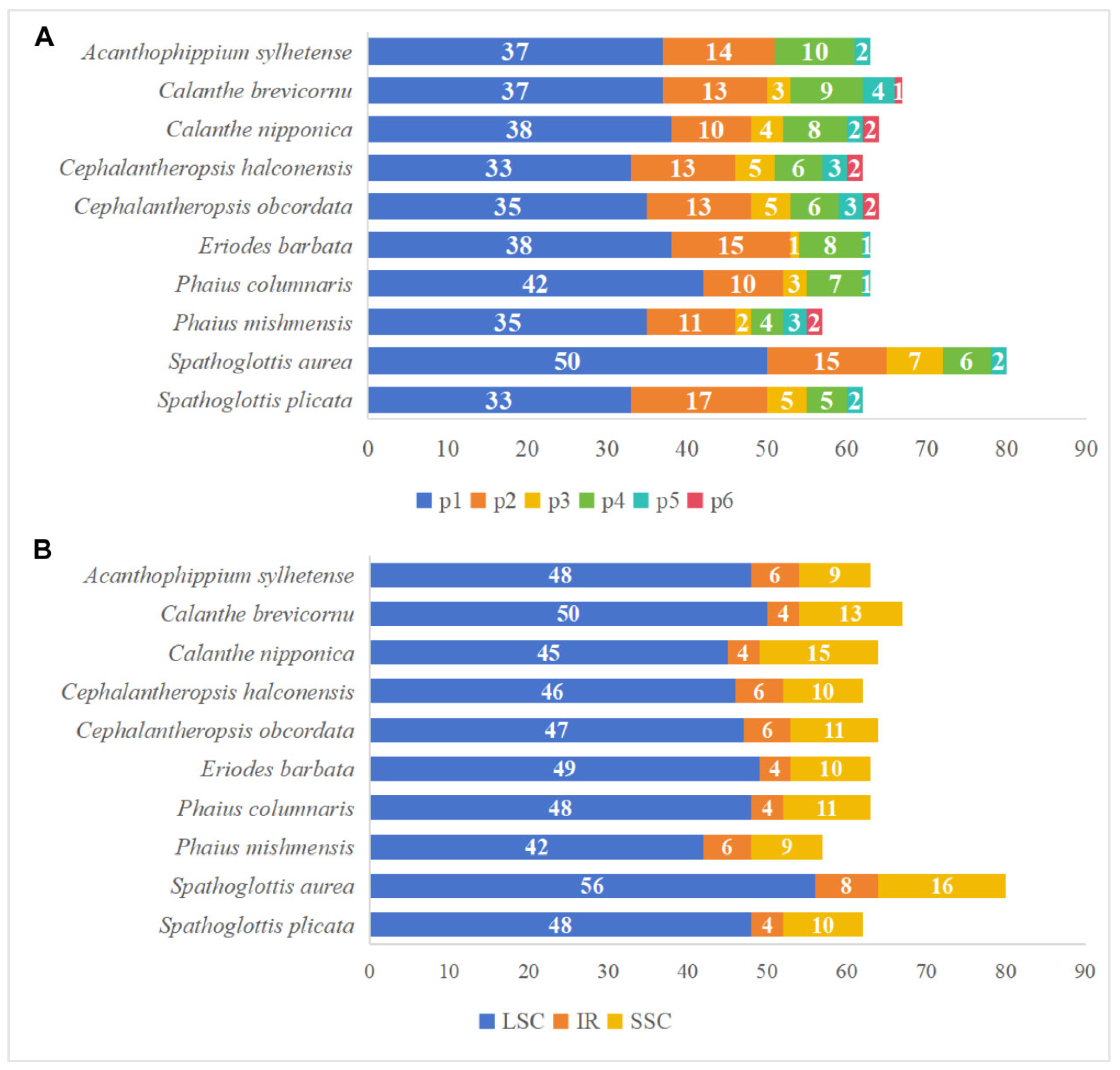
3.2. Analysis of Repeat Structures and SSRs
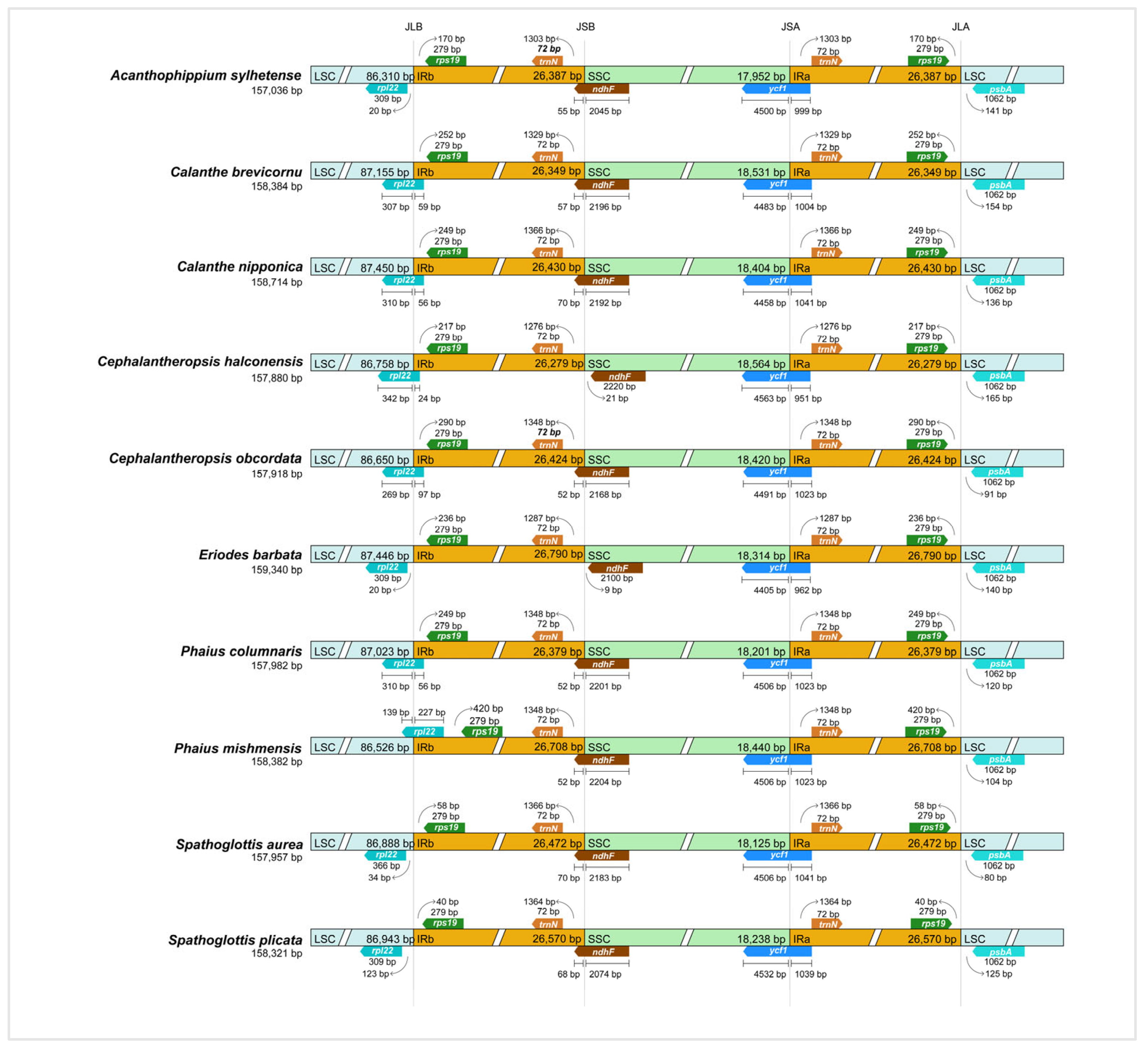
3.3. Contraction and Expansion Analysis of IR Regions
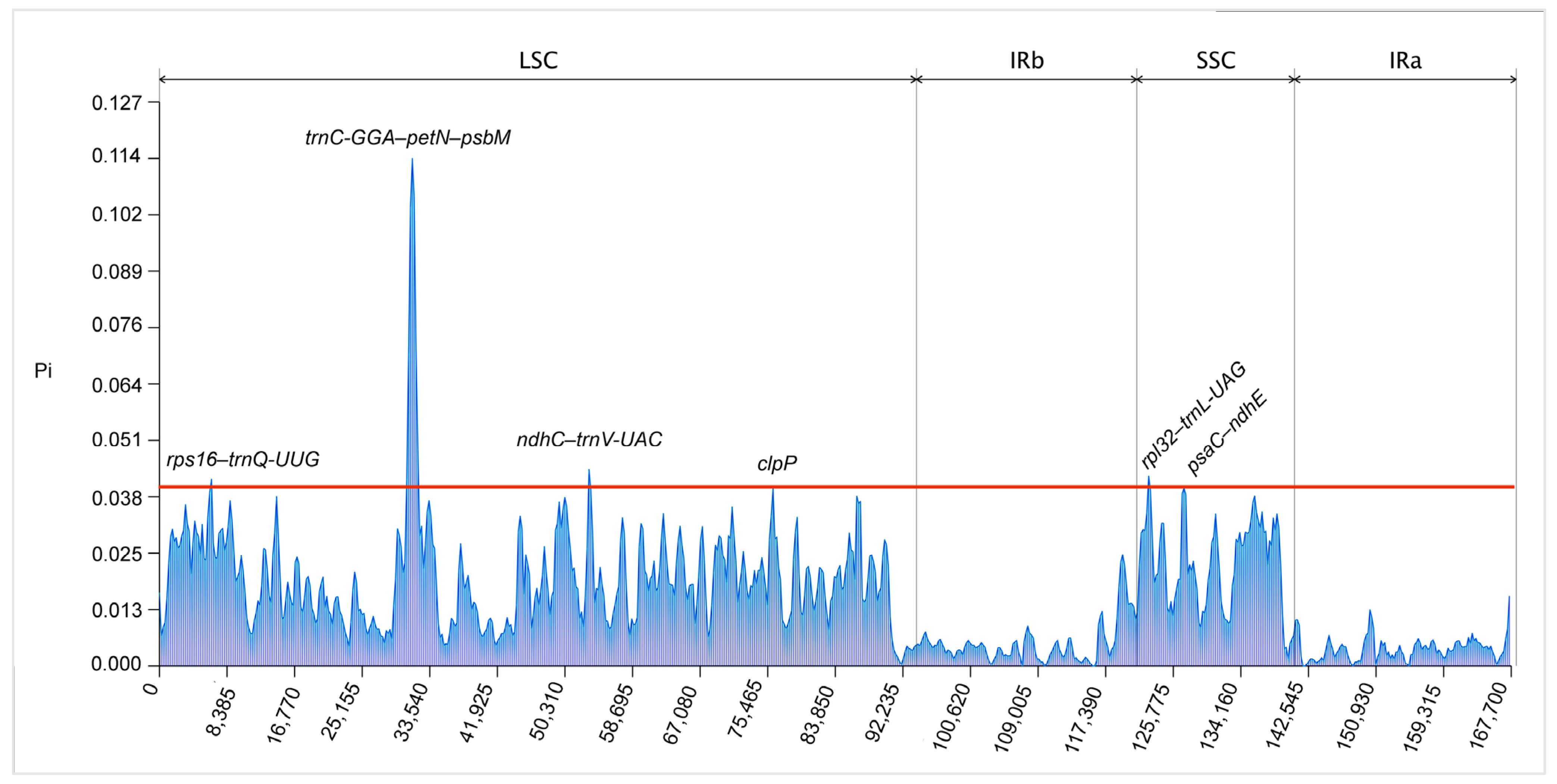
3.4. The Analysis of Sequence Divergence and Mutational Hotspots
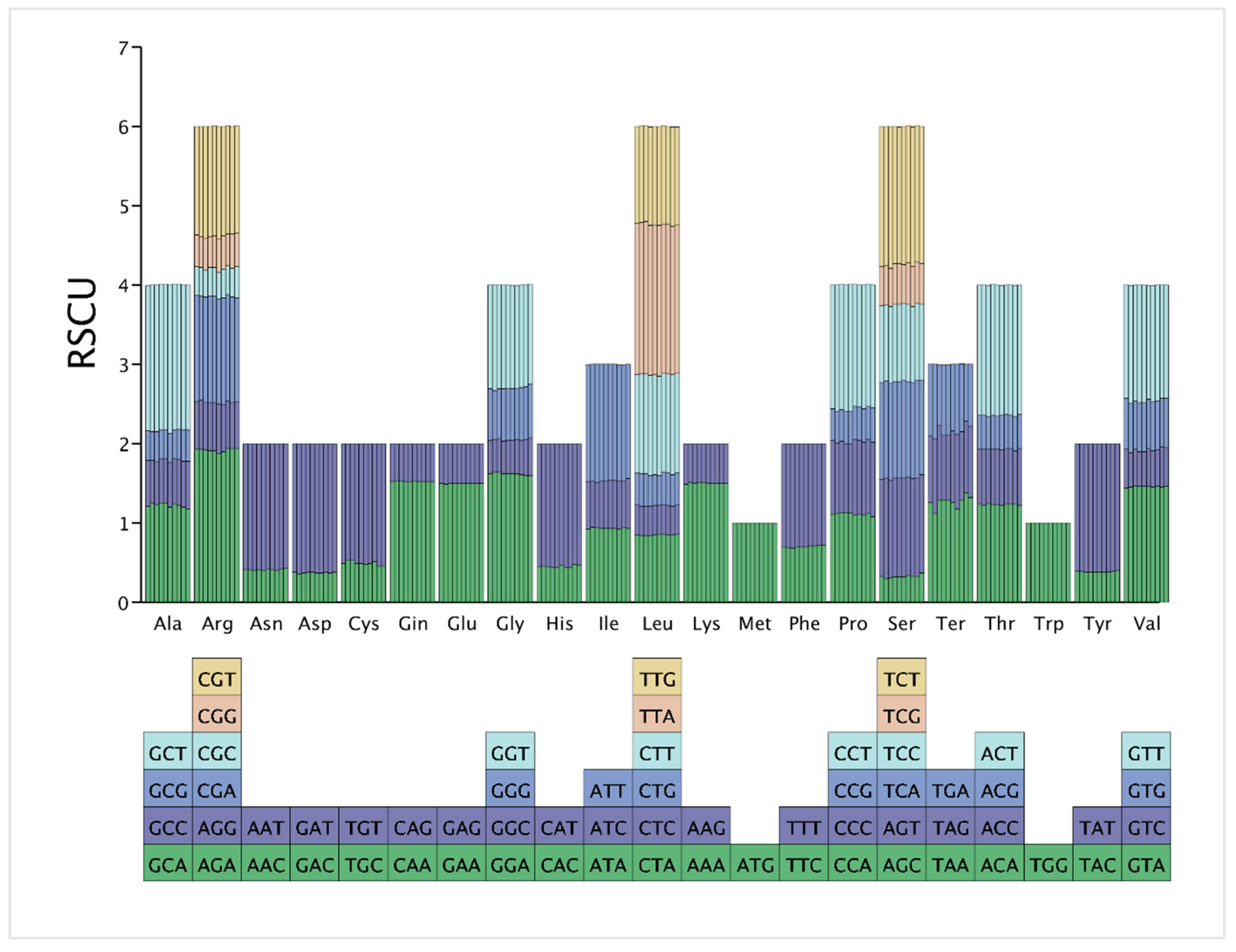
3.5. Codon Usage Analysis
3.6. Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chase, M.W.; Cameron, K.M.; Freudenstein, J.V.; Pridgeon, A.M.; Salazar, G.; Van den Berg, C.; Schuiteman, A. An updated classification of Orchidaceae. Bot. J. Linn. Soc. 2015, 177, 151–174. [Google Scholar] [CrossRef]
- Zhou, P.; Li, J.H.; Liu, Y.Z.; Zhu, Z.W.; Luo, Y.; Xiang, X.G. Species richness disparity in tropical terrestrial herbaceous floras: Evolutionary insight from Collabieae (Orchidaceae). Mol. Phylogenet. Evol. 2023, 186, 107860. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.Y.; Ye, C.; Chen, Y.Q.; Li, J.W.; Hidayat, A.; Miao, J.L.; Li, J.H.; Wu, J.Y.; Zhai, J.W.; Lan, S.R.; et al. Phylogenomics and biogeographical diversification of Collabieae (Orchidaceae) and its implication in the reconstruction of the dynamic history of Asian evergreen broadleaved forests. Mol. Phylogenet. Evol. 2024, 196, 108084. [Google Scholar] [CrossRef]
- Chase, M.; Christenhusz, M.; Schuiteman, A. Expansion of Calanthe to include the species of Cephalantheropsis, Gastrorchis and Phaius (Collabieae; Orchidaceae). Phytotaxa 2020, 472, 159–168. [Google Scholar] [CrossRef]
- Chen, X.Q. Orchidaceae. In Flora of China; Wu, Z.Y., Raven, P.H., Hong, D.Y., Eds.; Beijing and Missouri Botanical Garden Press: Beijing, China; St. Louis, MO, USA, 2009; Volume 25, pp. 285–310. [Google Scholar]
- Lindley, J. The Genera and Species of Orchidaceous Plants; Ridgways: London, UK, 1833; p. 177. [Google Scholar]
- Pridgeon, A.M.; Cribb, P.; Chase, M.W.; Rasmussen, F.N. Genera Orchidacearum, Epidendroideae (Part One); Oxford University Press: Oxford, UK, 2005; Volume 4, p. 269. [Google Scholar]
- Nordin, F.; Saibeh, K.; Go, R.; Mangsor, K.; Othman, A. Molecular Phylogenetics of the Orchid Genus Spathoglottis (Orchidaceae: Collabieae) in Peninsular Malaysia and Borneo. Forests 2022, 13, 2079. [Google Scholar] [CrossRef]
- Zhou, Z.H.; Shi, R.H.; Zhang, Y.; Xing, X.K.; Jin, X.H. Orchid conservation in China from 2000 to 2020: Achievements and perspectives. Plant Divers. 2021, 43, 343–349. [Google Scholar] [CrossRef]
- Shinozaki, K.; Ohme, M.; Tanaka, M.; Wakasugi, T.; Hayashida, N.; Matsubayashi, T.; Zaita, N.; Chunwongse, J.; Obokata, J.; Yamaguchi-Shinozaki, K.; et al. The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 1986, 5, 2043–2049. [Google Scholar] [CrossRef]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef]
- Yang, J.B.; Tang, M.; Li, H.T.; Zhang, Z.R.; Li, D.Z. Complete chloroplast genome of the genus Cymbidium: Lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol. Biol. 2013, 13, 84. [Google Scholar] [CrossRef]
- Luo, J.; Hou, B.W.; Niu, Z.T.; Liu, W.; Xue, Q.Y.; Ding, X.Y. Comparative chloroplast genomes of photosynthetic orchids: Insights into evolution of the Orchidaceae and development of molecular markers for phylogenetic applications. PLoS ONE 2014, 9, e99016. [Google Scholar] [CrossRef]
- Li, J.; Chen, C.; Wang, Z.Z. The complete chloroplast genome of the Dendrobium strongylanthum (Orchidaceae: Epidendroideae). Mitochondrial DNA Part A 2016, 27, 3048–3049. [Google Scholar] [CrossRef]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; DePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Huang, L.J.; Yu, H.X.; Wang, Z.; Xu, W.B. CPStools: A package for analyzing chloroplast genome sequences. iMetaOmics 2024, 1, e25. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.; Haeseler, A.V. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Talkah, N.S.M.; Jasim, J.H.M.; Nordin, F.A.; Othman, A.S. Complete chloroplast genome of a montane plant Spathoglottis aurea Lindl.: Comparative analyses and phylogenetic relationships among members of tribe Collabieae. PLoS ONE 2024, 19, e0291888. [Google Scholar] [CrossRef]
- Tao, K.F.; Tang, L.; Luo, Y.; Li, L. Complete chloroplast genome of eight Phaius (Orchidaceae) species from China: Comparative analysis and phylogenetic relationship. BMC Plant Biol. 2025, 25, 37. [Google Scholar] [CrossRef]
- Peng, X.B.; Ye, H.; Liu, H.Z.; Zhao, Z.X.; Hu, G.J.; Zhao, P. Characterization of the complete chloroplast genome of orchid family species Paphiopedilum bellatulum. MItochondrial DNA Part B 2022, 7, 1310–1312. [Google Scholar] [CrossRef]
- Yang, W.T.; Wu, K.L.; Fang, L.; Zeng, S.J.; Li, L. The complete chloroplast genomes of Blepharoglossum elegans and B. grossum and comparative analysis with related species (Orchidaceae, Malaxideae). Genes 2023, 14, 1069. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, W.Y.; Zhang, G.Q.; Wu, K.L.; Fang, L.; Li, M.Z.; Liu, Z.J.; Zeng, S.J. Comparative analyses and phylogenetic relationships of thirteen Pholidota species (Orchidaceae) inferred from complete chloroplast genomes. BMC Plant Biol. 2023, 23, 269. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.D.; Zhou, J.; Ma, S.H.; Chen, M.K.; Xie, T.X.; Chen, J.; Ai, Y. The complete chloroplast genome sequence of Tainia cordifolia (Orchidaceae). Mitochondrial DNA Part B 2019, 5, 1–2. [Google Scholar] [CrossRef]
- Xie, T.X.; Yu, X.; Zheng, Q.D.; Ma, S.H.; Liu, Z.J.; Ai, Y. The complete chloroplast genome of Tainia dunnii (Orchidaceae): Genome structure and evolution. Mitochondrial DNA Part B 2019, 5, 3–4. [Google Scholar] [CrossRef]
- Jiang, H.; Tian, J.; Yang, J.X.; Dong, X.; Zhong, Z.X.; Mwachala, G.; Zhang, C.; Hu, G.W.; Wang, Q.F. Comparative and phylogenetic analyses of six Kenya Polystachya (Orchidaceae) species based on the complete chloroplast genome sequences. BMC Plant Biol. 2022, 22, 177. [Google Scholar] [CrossRef]
- Liu, D.K.; Zhou, C.Y.; Tu, X.D.; Zhao, Z.; Chen, J.L.; Gao, X.Y.; Xu, S.W.; Zeng, M.Y.; Ma, L.; Ahmad, S.; et al. Comparative and phylogenetic analysis of Chiloschista (Orchidaceae) species and DNA barcoding investigation based on plastid genomes. BMC Genom. 2023, 24, 749. [Google Scholar] [CrossRef]
- Chen, J.L.; Wang, F.; Zhou, C.Y.; Ahmad, S.; Zhou, Y.Z.; Li, M.H.; Liu, Z.J.; Peng, D.H. Comparative Phylogenetic Analysis for Aerides (Aeridinae, Orchidaceae) based on six complete plastid genomes. Int. J. Mol. Sci. 2023, 24, 12473. [Google Scholar] [CrossRef]
- Raubeson, L.A.; Peery, R.; Chumley, T.W.; Dziubek, C.; Fourcade, H.M.; Boore, J.L.; Jansen, R.K. Comparative chloroplast genomics: Analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genom. 2007, 8, 174. [Google Scholar] [CrossRef]
- Kim, H.T.; Kim, J.S.; Moore, M.J.; Neubig, K.M.; Williams, N.H.; Whitten, W.M.; Kim, J.H. Seven new complete plastome sequences reveal rampant independent loss of the ndh gene family across orchids and associated instability of the inverted repeat/small single-copy region boundaries. PLoS ONE 2015, 10, e0142215. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.Y.; Li, L.K.; Wang, Z.L.; Yu, L.M.; Li, J.P.; Zeng, Y. Research progress on chloroplast genomes of Orchidaceae plants. Chin. Wild Plant Resour. 2023, 42, 73–79. [Google Scholar]
- Zheng, X.M.; Wu, J.R.; Li, F.; Liu, S.; Pang, H.B.; Lan, Q.; Li, J.; Sun, Y.; Qiao, W.H.; Zang, L.F.; et al. Inferring the evolutionary mechanism of the chloroplast genome size by comparing whole-chloroplast genome sequences in seed plants. Sci. Rep. 2017, 7, 1555. [Google Scholar] [CrossRef]
- Delannoy, E.; Fujii, S.; Francs-Small, C.C.; Brundrett, M.; Small, I. Rampant gene loss in the underground orchid rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol. Biol. Evol. 2011, 28, 2077–2086. [Google Scholar] [CrossRef]
- Greiner, S. Plastome mutants of higher plants. In Genomics of Chloroplasts and Mitochondria; Bock, R., Knoop, V., Eds.; Springer Netherlands: Dordrecht, The Netherlands, 2012; Volume 35, pp. 237–266. [Google Scholar]
- Powell, W.; Machray, G.C.; Provan, J. Polymorphism revealed by simple sequence repeats. Trends Plant Sci. 1996, 1, 215–222. [Google Scholar] [CrossRef]
- Li, Y.C.; Röder, M.S.; Fahima, T.; Kirzhner, V.M.; Beiles, A.; Korol, A.B.; Nevo, E. Climatic effects on microsatellite diversity in wild emmer wheat (Triticum dicoccoides) at the Yehudiyya microsite, Israel. Heredity 2002, 89, 127–132. [Google Scholar] [CrossRef]
- Nordin, F.A.; Raffi, A.; Go, R.; Yong, C.S.Y.; Saibeh, K.; Othman, A.S. Morphological systematics of Spathoglottis Blume (Orchidaceae: Collabieae) in Peninsular Malaysia and Borneo. Forests 2023, 14, 940. [Google Scholar] [CrossRef]
- Zhong, H.; Shen, L.M.; Liu, H.P.; Liu, Z.J.; Wu, S.S.; Zhai, J.W. The complete chloroplast genome of Calanthe arcuata, an endemic terrestrial orchid in China. Mitochondrial DNA Part B 2019, 4, 2629–2630. [Google Scholar] [CrossRef]
- Tan, C.J.; Yan, R.R.; Yu, P.; Lu, G.Q.; Wu, W.; Hu, G.X. The complete chloroplast genome of Chrysoglossum ornatum (Epidendroideae, Orchidaceae) and its phylogenetic analysis. Mitochondrial DNA Part B 2024, 9, 24–28. [Google Scholar] [CrossRef] [PubMed]
- Rokas, A.; Carroll, S.B. More genes or more taxa? the relative contribution of gene number and taxon number to phylogenetic accuracy. Mol. Biol. Evol. 2005, 22, 1337–1344. [Google Scholar] [CrossRef] [PubMed]
- Salamin, N.; Hodkinson, T.R.; Savolainen, V. Towards building the tree of life: A simulation study for all angiosperm genera. Syst. Biol. 2005, 54, 183–196. [Google Scholar] [CrossRef]
- Philippe, H.; Brinkmann, H.; Lavrov, D.V.; Littlewood, D.T.J.; Manuel, M.; Wörheide, G.; Baurain, D. Resolving difficult phylogenetic questions: Why more sequences are not enough. PLoS Biol. 2011, 9, e1000602. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, S.; Jiang, X.; Yang, W.; Wu, K.; Fang, L.; Zeng, S.; Zeng, J.; Li, L. Comparative Chloroplast Genomics of Ten Collabieae Species Including Three Novel Genomes. Genes 2025, 16, 1028. https://doi.org/10.3390/genes16091028
Xie S, Jiang X, Yang W, Wu K, Fang L, Zeng S, Zeng J, Li L. Comparative Chloroplast Genomics of Ten Collabieae Species Including Three Novel Genomes. Genes. 2025; 16(9):1028. https://doi.org/10.3390/genes16091028
Chicago/Turabian StyleXie, Shuangshuang, Xingyou Jiang, Wenting Yang, Kunlin Wu, Lin Fang, Songjun Zeng, Jingjue Zeng, and Lin Li. 2025. "Comparative Chloroplast Genomics of Ten Collabieae Species Including Three Novel Genomes" Genes 16, no. 9: 1028. https://doi.org/10.3390/genes16091028
APA StyleXie, S., Jiang, X., Yang, W., Wu, K., Fang, L., Zeng, S., Zeng, J., & Li, L. (2025). Comparative Chloroplast Genomics of Ten Collabieae Species Including Three Novel Genomes. Genes, 16(9), 1028. https://doi.org/10.3390/genes16091028





