Association of Obesity-Related Genetic Variants with Android Fat Patterning and Cardiometabolic Risk in Women
Abstract
1. Introduction
2. Methods
2.1. Study Design and Population
2.2. Assessment of the Cardiovascular Risk Factors
2.3. Biochemical Risk Factors
2.4. Genetic Analysis
2.5. Statistical Analysis
Descriptive and Comparative Analysis
3. Results
3.1. Main Characteristics of the Population (Overweight and Obese Women) According to Fat Phenotype (Android or Gynoid)
3.2. Genetic Variants Associated with Obesity (Bivariate Analysis)
Multivariate Analysis
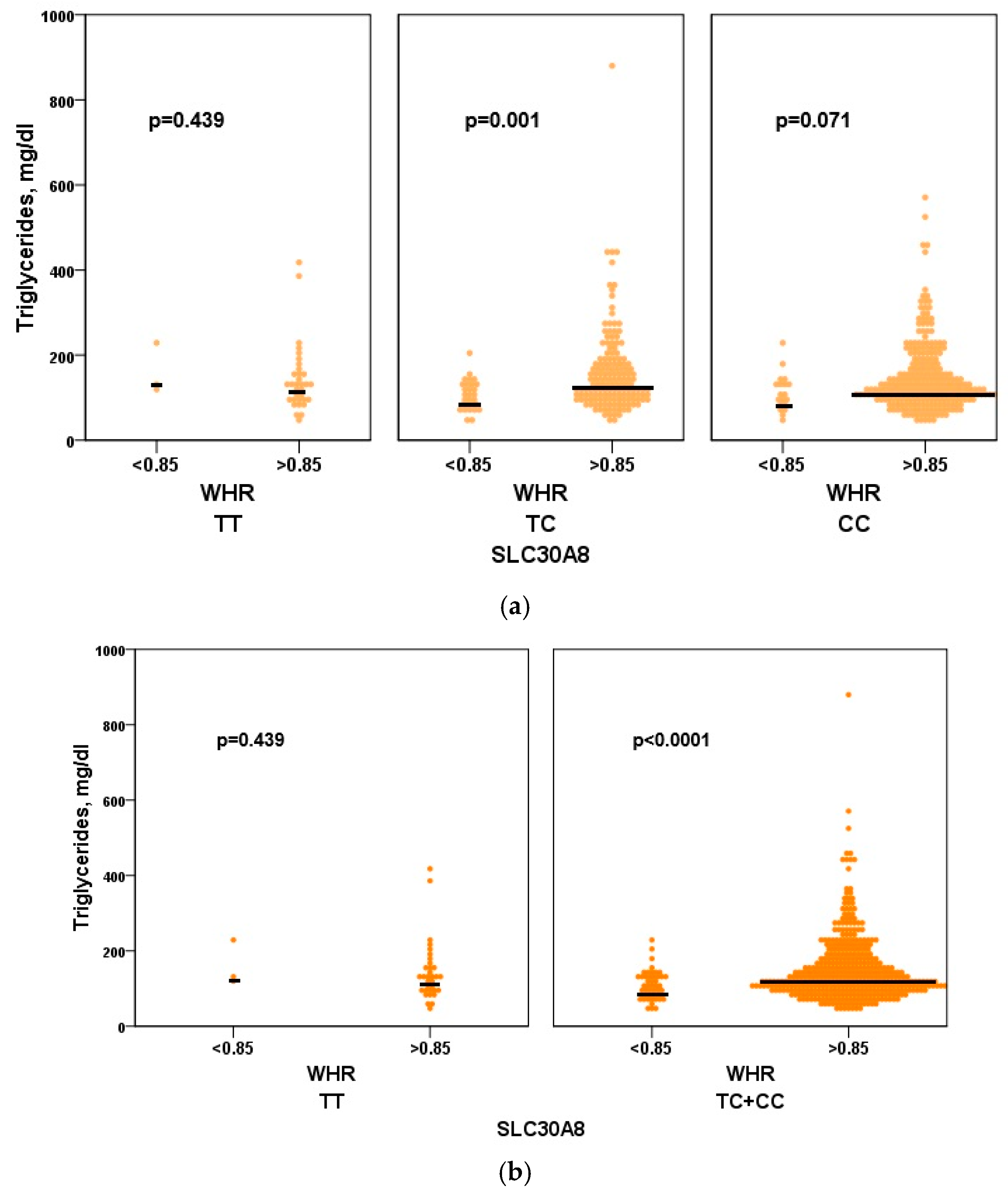
3.3. Biochemical Profile of the Genetic Variant SLC30A8 (CC) in a Cohort of Overweight and Obese Women
3.3.1. Fasting Glucose Profile
3.3.2. Lipid Profile
Triglyceride
HDL Cholesterol
4. Discussion
Strengths and Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Koliaki, C.; Dalamaga, M.; Lia, S. Update on the Obesity Epidemic: After the Sudden Rise, Is the Upward Trajectory Beginning to Flatten? Curr. Obes. Rep. 2023, 12, 514–527. [Google Scholar] [CrossRef]
- Darbandi, M.; Pasdar, Y.; Moradi, S.; Mohamed, H.; Hamzeh, B.; Salimi, Y. Discriminatory Capacity of Anthropometric Indices for Cardiovascular Disease in Adults: A Systematic Review and Meta-Analysis. Prev. Chronic Dis. 2020, 17, E131. [Google Scholar] [CrossRef]
- Cao, Q.; Yu, S.; Xiong, W.; Li, Y.; Li, H.; Li, J.; Li, F. Waist-hip ratio as a predictor of myocardial infarction risk: A systematic review and meta-analysis. Medicine 2018, 97, e11639. [Google Scholar] [CrossRef]
- Benites-Zapata, V.A.; Toro-Huamanchumo, C.J.; Urrunaga-Pastor, D.; Guarnizo-Poma, M.; Lazaro-Alcantara, H.; Paico-Palacios, S.; Pantoja-Torres, B.; Ranilla-Seguin, V.d.C. High waist-to-hip ratio levels are associated with insulin resistance markers in normal-weight women. Diabetes Metab. Syndr. 2019, 13, 636–642. [Google Scholar] [CrossRef]
- Widjaja, N.A.; Arifani, R.; Irawan, R. Value of waist-to-hip ratio as a predictor of metabolic syndrome in obese adolescents. Acta Biomed. 2023, 94, e2023076. [Google Scholar]
- Aghaei, M.; Joukar, F.; Hasanipour, S.; Ranjbar, Z.A.; Naghipour, M.; Mansour-Ghanaei, F. The association between waist-to-hip ratio (WHR) and diabetes in the PERSIAN Guilan cohort study population. BMC Endocr. Disord. 2024, 24, 113. [Google Scholar] [CrossRef] [PubMed]
- Pulit, S.L.; Stoneman, C.; Morris, A.P.; Wood, A.R.; Glastonbury, C.A.; Tyrrell, J.; Yengo, L.; Ferreira, T.; Marouli, E.; Ji, Y.; et al. Meta-analysis of genome-wide association studies for body fat distribution in 694,649 individuals of European ancestry. Hum. Mol. Genet. 2019, 28, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.; Li, X.; Liu, Z.; Liao, Y.; Xiao, Z.; Wei, Y.; Cao, Y. Association between Waist-Hip Ratio and Female Infertility in the United States: Data from National Health and Nutrition Examination Survey 2017–2020. Obes. Facts 2024, 17, 445–458. [Google Scholar] [CrossRef] [PubMed]
- Rose, K.M.; Newman, B.; Mayer-Davis, E.J.; Selby, J.V. Genetic and behavioural determinants of waist-hip ratio and waist circumference in women twins. Obes. Res. 1998, 6, 383–392. [Google Scholar] [CrossRef]
- Dash, S. Obesity and Cardiometabolic Disease: Insights from Genetic Studies. Can. J. Cardiol. 2025; online ahead of print. [Google Scholar] [CrossRef]
- Schleinitz, D.; Böttcher, Y.; Blüher, M.; Kov, P. The genetics of fat distribution. Diabetologia 2014, 57, 1276–1286. [Google Scholar] [CrossRef]
- Mendonça, M.I.; Henriques, E.; Borges, S. Genetic information improves the prediction of major adverse cardiovascular events in the GENEMACOR population. Genet. Mol. Biol. 2021, 44, e20200448. [Google Scholar] [CrossRef]
- Mendonça, M.I.; Pereira, A.; Monteiro, J. Impact of genetic information on Coronary Disease risk in Madeira: The GENEMACOR study. Rev. Port. Cardiol. 2022, 42, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Tirthani, E.; Said, M.; Rehman, A. Genetics and Obesity. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Després, J.P. Body Fat Distribution and Risk of Cardiovascular Disease: An Update. Circulation 2012, 126, 1301–1313. [Google Scholar] [CrossRef]
- Brettle, H.; Tran, V.; Drummond, G.R.; Franks, A.E.; Petrovski, S.; Vinh, A.; Jelinic, M. Sex hormones, intestinal inflammation, and the gut microbiome: Major influencers of the sexual dimorphisms in obesity. Front. Immunol. 2022, 13, 971048. [Google Scholar] [CrossRef] [PubMed]
- Loos, R.J.F.; Yeo, G.S.H. The genetics of obesity: From discovery to biology. Nat. Rev. Genet. 2022, 23, 120–133. [Google Scholar] [CrossRef] [PubMed]
- Kuryłowicz, A. Estrogens in Adipose Tissue Physiology and Obesity-Related Dysfunction. Biomedicines 2023, 11, 690. [Google Scholar] [CrossRef]
- Fox, C.S.; Liu, Y.; White, C.C. Genome-Wide Association for Abdominal Subcutaneous and Visceral Adipose Reveals a Novel Locus for Visceral Fat in Women. PLoS Genet. 2012, 8, e10026. [Google Scholar] [CrossRef]
- Richard, A.J.; White, U.; Elks, C.M.; Stephens, J.M. Adipose Tissue: Physiology to Metabolic Dysfunction. In Endotext; Feingold, K.R., Ahmed, S.F., Anawalt, B., Blackman, M.R., Boyce, A., Chrousos, G., Corpas, E., de Herder, W.W., Dhatariya, K., Dungan, K., Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2020; Bookshelf ID: NBK555602. [Google Scholar]
- Shungin, D.; Winkler, T.W.; Croteau-Chonka, D.C.; Ferreira, T.; Locke, A.E.; Mägi, R.; Strawbridge, R.J.; Pers, T.H.; Fischer, K.; Justice, A.E.; et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature 2015, 518, 187–196. [Google Scholar] [CrossRef]
- Angelini, S.; Rosticci, M.; Massimo, G.; Musti, M.; Ravegnini, G.; Consolini, N.; Sammarini, G.; D’Addato, S.; Rizzoli, E.; Botbayev, D.; et al. Relationship between Lipid Phenotypes, Overweight, Lipid Lowering Drug Response and KIF6 and HMG-CoA Genotypes in a Subset of the Brisighella Heart Study Population. Int. J. Mol. Sci. 2017, 19, 49. [Google Scholar] [CrossRef]
- Imamura, M.; Takahashi, A.; Yamauchi, T.; Hara, K.; Yasuda, K.; Grarup, N.; Zhao, W.; Wang, X.; Huerta-Chagoya, A.; Hu, C.; et al. Genome-wide association studies in the Japanese population identify seven novel loci for type 2 diabetes. Nat. Commun. 2016, 7, 10531. [Google Scholar] [CrossRef]
- Bonàs-Guarch, S.; Guindo-Martínez, M.; Miguel-Escalada, I.; Grarup, N.; Sebastian, D.; Rodriguez-Fos, E.; Sánchez, F.; Planas-Fèlix, M.; Cortes-Sánchez, P.; González, S.; et al. A re-analysis of public genetic data reveals a rare X-chromosomal variant associated with type 2 diabetes. Nat. Commun. 2018, 9, 321. [Google Scholar] [CrossRef]
- Zeggini, E.; Weedon, M.N.; Lindgren, C.M.; Frayling, T.M.; Elliott, K.S.; Lango, H.; Timpson, N.J.; Perry, J.R.B.; Rayner, N.W.; Freathy, R.M.; et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 2007, 316, 1336–1341, Corrected in Science 2007, 317, 1035–1036. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, F.; Serizawa, M.; Yamamoto, K.; Fujisawa, T.; Nakashima, E.; Ohnaka, K.; Ikegami, H.; Sugiyama, T.; Katsuya, T.; Miyagishi, M.; et al. Confirmation of multiple risk Loci and genetic impacts by a genome-wide association study of type 2 diabetes in the Japanese population. Diabetes 2009, 58, 1690–1699. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Huffman, J.E.; Rodriguez, A.; Conery, M.; Liu, M.; Ho, Y.-L.; Kim, Y.; Heise, D.A.; Guare, L.; Panickan, V.A.; et al. Diversity and scale: Genetic architecture of 2068 traits in the VA Million Veteran Program. Science 2024, 385, eadj118. [Google Scholar] [CrossRef]
- Franco, C.; Canzoniero, L.M.T. Zinc homeostasis and redox alterations in obesity. Front. Endocrinol. 2024, 14, 1273177. [Google Scholar] [CrossRef]
- Piché, M.-E.; Tchernof, A.; Després, J.-P. Obesity Phenotypes, Diabetes, and Cardiovascular Diseases. Circ. Res. 2020, 126, 1477–1500. [Google Scholar] [CrossRef] [PubMed]
- Liuzzi, J.P.; Masmouei, H.; Zarini, G.G.; Huffman, F.G. Association of the Slc30a8 rs13266634 polymorphism with type 2 diabetes and central obesity in a Cuban-American population. FASEB J. 2009, 23, LB517. [Google Scholar] [CrossRef]
- Klein, S.; Gastaldelli, A.; Yki-Järvinen, H.; Scherer, P.E. Why Does Obesity Cause Diabetes? Cell Metab. 2022, 34, 11–20. [Google Scholar] [CrossRef]
- Hussain, M.; Waheed, A.; Elahi, A.; Mustafa, G. Fat Mass and Obesity-Related (FTO) Gene Variant Is a Predictor of CVD in T2DM Patients. J. Diabetes Res. 2024, 2024, 5914316. [Google Scholar] [CrossRef]
- Lengton, R.; Dekker, F.W.; van Rossum, E.F.C.; de Fijter, J.W.; Rosendaal, F.R.; van Dijk, K.W.; Rabelink, T.J.; Le Cessie, S.; de Mutsert, R.; Hoogeveen, E.K. Hypertension and diabetes, but not leptin and adiponectin, mediate the relationship between body fat and chronic kidney disease. Endocrine 2024, 85, 1141–1153. [Google Scholar] [CrossRef]
- Girard, R.; Tremblay, S.; Noll, C.; St-Jean, S.; Jones, C.; Gélinas, Y.; Maloum-Rami, F.; Perreault, N.; Laplante, M.; Carpentier, A.C.; et al. The transcription factor hepatocyte nuclear factor 4A acts in the intestine to promote white adipose tissue energy storage. Nat. Commun. 2022, 13, 224. [Google Scholar] [CrossRef]
- Wooding, S.P.; Ramirez, V.A. Global population genetics and diversity in the TAS2R bitter taste receptor family. Front. Genet. 2022, 13, 952299. [Google Scholar] [CrossRef]
- Velmurugan, S.; Deshmukh, V.R.; Sontakke, B.R.; Chandrasekaran, K.; Subbaraj, G.K. Polymorphisms of IGF2BP2 and SIRT1 genes in type 2 diabetes mellitus: A comprehensive meta-analysis and statistical power analysis. World Acad. Sci. J. 2025, 7, 4. [Google Scholar] [CrossRef]
- Yu, K.; Li, L.; Zhang, L.; Guo, L.; Wang, C. Association between MC4R rs17782313 genotype and obesity: A meta-analysis. Gene 2020, 733, 144372. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Shriner, D.; Zhang, J.; Zhou, J.; Adikaram, P.; Doumatey, A.P.; Bentley, A.R.; Adeyemo, A.; Rotimi, C.N. Additive genetic effect of GCKR, G6PC2, and SLC30A8 variants on fasting glucose levels and risk of type 2 diabetes. PLoS ONE 2022, 17, e0269378. [Google Scholar] [CrossRef] [PubMed]
- Brumpton, B.M.; Fritsche, L.G.; Zheng, J.; Nielsen, J.B.; Mannilä, M.; Surakka, I.; Rasheed, H.; Åberge Vie, G.; Graham, S.E.; Gabrielsen, M.E.; et al. Variation in serum PCSK9 (Proprotein Convertase Subtilisin/Kexin type 9), Cardiovascular Disease Risk, and an Investigation of Potential Unanticipated Effects of PCSK9 Inhibition. Circ. Genom. Precis. Med. 2019, 12, e002335. [Google Scholar] [CrossRef]
- Jones, N.S.; Rebeck, G.W. The Synergistic Effects of APOE Genotype and Obesity on Alzheimer’s Disease Risk. Int. J. Mol. Sci. 2019, 20, 63. [Google Scholar] [CrossRef]
- Khorrami, M.S.; Sadabadi, F.; Pasdar, A.; Safarian-Bana, H.; Amerizadeh, F.; Esmaeily, H.; Moohebati, M.; Heidari-Bakavoli, A.; Ferns, G.; Ghayour-Mobarhan, M.; et al. A genetic variant in proline and serine rich coiled-coil 1 gene is associated with the risk of cardiovascular disease. Rep. Biochem. Mol. Biol. 2022, 10, 653–663. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Z.; Song, H. Association between KIF6 rs20455 polymorphism and the risk of coronary heart disease (CHD): A pooled analysis of 50 individual studies including 40,059 cases and 64,032 controls. Lipids Health Dis. 2018, 17, 4. [Google Scholar] [CrossRef]
- Volgman, A.S.; Koschinsky, M.L.; Mehta, A.; Rosenson, R.S. Genetics and pathophysiological mechanisms of lipoprotein(a)-associated cardiovascular risk. J. Am. Heart Assoc. 2024, 13, e033654. [Google Scholar] [CrossRef]
- Mirhafez, S.R.; Avan, A.; Pasdar, A.; Khatamianfar, S.; Hosseinzadeh, L.; Ganjali, S.; Movahedi, A.; Pirhoushiaran, M.; Gómez Mellado, V.; Rosace, D.; et al. Zinc Finger 259 gene polymorphism rs964184 is associated with serum triglyceride levels and metabolic syndrome. Int. J. Mol. Cell. Med. 2016, 5, 8–18. [Google Scholar] [PubMed]
- Li, S.; He, C.; Nie, H.; Pang, Q.; Wang, R.; Zeng, Z.; Song, Y. G allele of the rs1801282 polymorphism in PPARγ gene confers an increased risk of obesity and hypercholesterolemia, while T allele of the rs3856806 polymorphism displays a protective role against dyslipidemia: A systematic review and meta-analysis. Front. Endocrinol. 2022, 13, 919087. [Google Scholar] [CrossRef] [PubMed]
- Cropano, C.; Santoro, N.; Groop, L.; Dalla Man, C.; Cobelli, C.; Galderisi, A.; Kursawe, R.; Pierpont, B.; Goffredo, M.; Caprio, S.; et al. The rs7903146 variant in the TCF7L2 gene increases the risk of prediabetes/type 2 diabetes in obese adolescents by impairing β-cell function and hepatic insulin sensitivity. Diabetes Care 2017, 40, 1082–1089. [Google Scholar] [CrossRef] [PubMed]


| Variables | Total (n = 512) | WHR > 0.85 (n = 461) | WHR ≤ 0.85 (n = 51) | p Value |
|---|---|---|---|---|
| Age, years | 56.1 ± 6.4 | 56.1 ± 6.5 | 56.7 ± 6.4 | 0.540 |
| Smoking, n (%) | 91 (17.8) | 84 (18.2) | 7 (13.7) | 0.425 |
| AHT, n (%) | 352 (68.8) | 322 (69.8) | 30 (58.8) | 0.107 |
| CAD FH, n (%) | 118 (23.0) | 105 (22.8) | 13 (25.5) | 0.662 |
| T2DM, n (%) | 151 (29.5) | 145 (31.5) | 6 (11.8) | 0.003 |
| BMI ≥ 30 kg/m2, n (%) | 229 (44.7) | 211 (45.8) | 18 (35.3) | 0.153 |
| Dyslipidaemia, n (%) | 435 (85.0) | 396 (85.9) | 39 (76.5) | 0.074 |
| Physical inactivity, n (%) | 293 (57.2) | 265 (57.5) | 28 (54.9) | 0.724 |
| EAT, cm3 | 6.1 (2.3–12.0) | 6.2 (2.3–12.0) | 5.8 (2.4–8.6) | 0.159 |
| Fasting glucose, mg/dl | 102.0 (57.0–366.0) | 103.0 (70.0–366.0) | 96.0 (57.0–208.0) | 0.001 |
| Apo B, mg/dl | 92.4 (3.9–199.3) | 92.4 (3.9–171.1) | 90.2 (6.2–199.3) | 0.433 |
| Hcy, mg/dl | 11.2 (2.9–48.7) | 11.1 (2.9–48.7) | 11.9 (4.0–32.0) | 0.415 |
| Lp (a), mg/dl | 15.5 (0.9–241.0) | 15.5 (0.9–241.0) | 15.0 (3.0–179.2) | 0.982 |
| Triglycerides, mg/dl | 124.0 (42.0–880.0) | 127.0 (42.0–880.0) | 109.0 (42.0–231.0) | 0.001 |
| TC, mg/dl | 192.0 (98.0–341.0) | 193.0 (98.0–323.0) | 185.0 (131.0–341.0) | 0.378 |
| LDL-c, mg/dl | 115.3 (15.6–236.4) | 115.3 (15.6–236.4) | 105.5 (58.2–211.8) | 0.650 |
| HDL-c, mg/dl | 48.0 (21.7–110.0) | 47.0 (21.7–110.0) | 53.0 (33.0–92.0) | 0.021 |
| Non-HDL-c, mg/dl | 144.0 (59.0–269.0) | 145.0 (59.0–269.0) | 130.9 (83.0–258.0) | 0.143 |
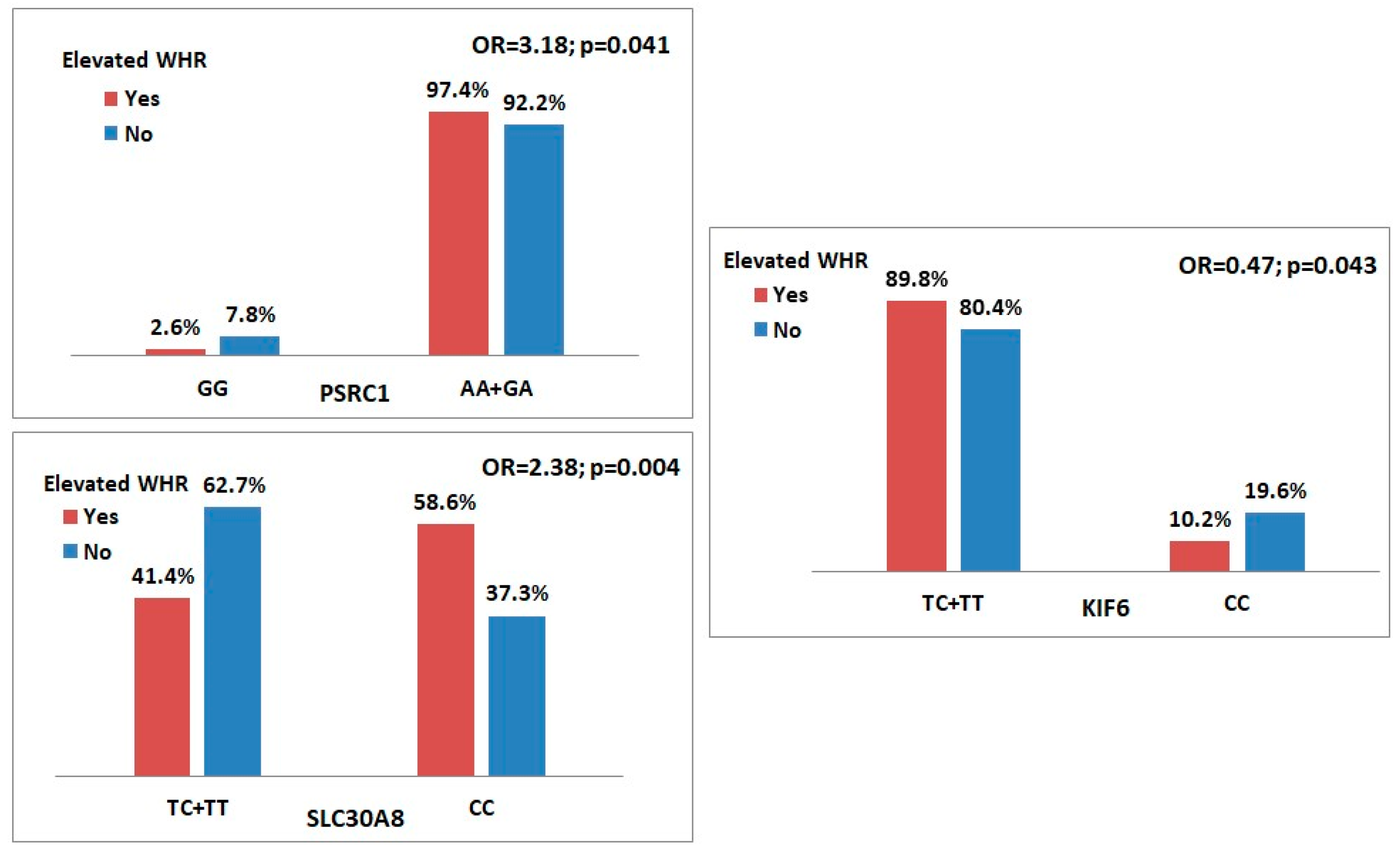
| Genetic Variants | WHR > 0.85 (n = 461) | WHR ≤ 0.85 (n = 51) | Odds Ratio (95% CI) | p Value | HWE p Value |
|---|---|---|---|---|---|
| PSRC1 rs599839 (G > A) | 0.057 | ||||
| G | 188 (20.4) | 30 (29.4) | 1.63 (1.03–2.56) | 0.035 | |
| A | 734 (79.6) | 72 (70.6) | |||
| Genotype | |||||
| GG | 12 (2.6) | 4 (7.8) | Reference | ||
| GA | 164 (35.6) | 22 (43.1) | 2.49 (0.74–8.38) | 0.131 | |
| AA | 285 (61.8) | 25 (49.0) | 3.80 (1.14–12.66) | 0.020 | |
| Best model | |||||
| Dominant (AA + GA vs. GG) | 3.18 (0.99–10.27) | 0.041 | |||
| SLC30A8 rs1326634 (T > C) | 0.448 | ||||
| T | 224 (24.3) | 35 (34.3) | 1.63 (1.05–2.52) | 0.027 | |
| C | 698 (75.7) | 67 (65.7) | |||
| Genotype | |||||
| TT | 33 (7.2) | 3 (5.9) | Reference | ||
| TC | 158 (34.3) | 29 (56.9) | 0.50 (0.14–1.72) | 0.261 | |
| CC | 270 (58.6) | 19 (37.3) | 1.29 (0.36–4.60) | 0.692 | |
| Best model | |||||
| Recessive (CC vs. TC + TT) | 2.38 (1.31–4.33) | 0.004 | |||
| KIF6 rs20455 (T > C) | 0.401 | ||||
| T | 630 (68.3) | 65 (63.7) | 0.81 (0.53–1.25) | 0.345 | |
| C | 292 (31.7) | 37 (36.3) | |||
| Genotype | |||||
| TT | 216 (46.9) | 24 (47.1) | Reference | ||
| TC | 198 (43.0) | 17 (33.3) | 1.29 (0.68–2.48) | 0.436 | |
| CC | 47 (10.2) | 10 (19.6) | 0.52 (0.23–1.17) | 0.108 | |
| Best model | |||||
| Recessive (CC vs. TC + TT) | 0.47 (0.22–0.99) | 0.043 |
| Variables | B | S.E. | Wald | df | Odds Ratio (95% CI) | p Value |
|---|---|---|---|---|---|---|
| Diabetes | 1.290 | 0.449 | 8.263 | 1 | 3.63 (1.51–8.75) | 0.004 |
| SLC30A8 (CC) | 0.917 | 0.308 | 8.876 | 1 | 2.50 (1.37–4.57) | 0.003 |
| Constant | 1.499 | 0.204 | 53.926 | 1 | 4.475 | <0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sá, D.; Mendonça, M.I.; Sousa, F.; Abreu, G.; Ferreira, M.; Henriques, E.; Freitas, S.; Rodrigues, M.; Borges, S.; Guerra, G.; et al. Association of Obesity-Related Genetic Variants with Android Fat Patterning and Cardiometabolic Risk in Women. Genes 2025, 16, 1019. https://doi.org/10.3390/genes16091019
Sá D, Mendonça MI, Sousa F, Abreu G, Ferreira M, Henriques E, Freitas S, Rodrigues M, Borges S, Guerra G, et al. Association of Obesity-Related Genetic Variants with Android Fat Patterning and Cardiometabolic Risk in Women. Genes. 2025; 16(9):1019. https://doi.org/10.3390/genes16091019
Chicago/Turabian StyleSá, Débora, Maria Isabel Mendonça, Francisco Sousa, Gonçalo Abreu, Matilde Ferreira, Eva Henriques, Sónia Freitas, Mariana Rodrigues, Sofia Borges, Graça Guerra, and et al. 2025. "Association of Obesity-Related Genetic Variants with Android Fat Patterning and Cardiometabolic Risk in Women" Genes 16, no. 9: 1019. https://doi.org/10.3390/genes16091019
APA StyleSá, D., Mendonça, M. I., Sousa, F., Abreu, G., Ferreira, M., Henriques, E., Freitas, S., Rodrigues, M., Borges, S., Guerra, G., Drumond, A., Sousa, A. C., & dos Reis, R. P. (2025). Association of Obesity-Related Genetic Variants with Android Fat Patterning and Cardiometabolic Risk in Women. Genes, 16(9), 1019. https://doi.org/10.3390/genes16091019






