Interaction Between Vitamin D Metabolism Genetic Variants: Association with Hypovitaminosis D, Rheumatoid Arthritis, and Its Clinical Disease Activity
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Ethical Considerations
2.3. Clinical and Biochemical Evaluation
2.4. Calcidiol Quantification
2.5. Genotyping of SNVs in Vitamin D Metabolism Genes
2.6. Statistical Analysis
3. Results
3.1. Anthropometric, Biochemical, Clinical, and Vitamin D Variables from RA Patients and CS
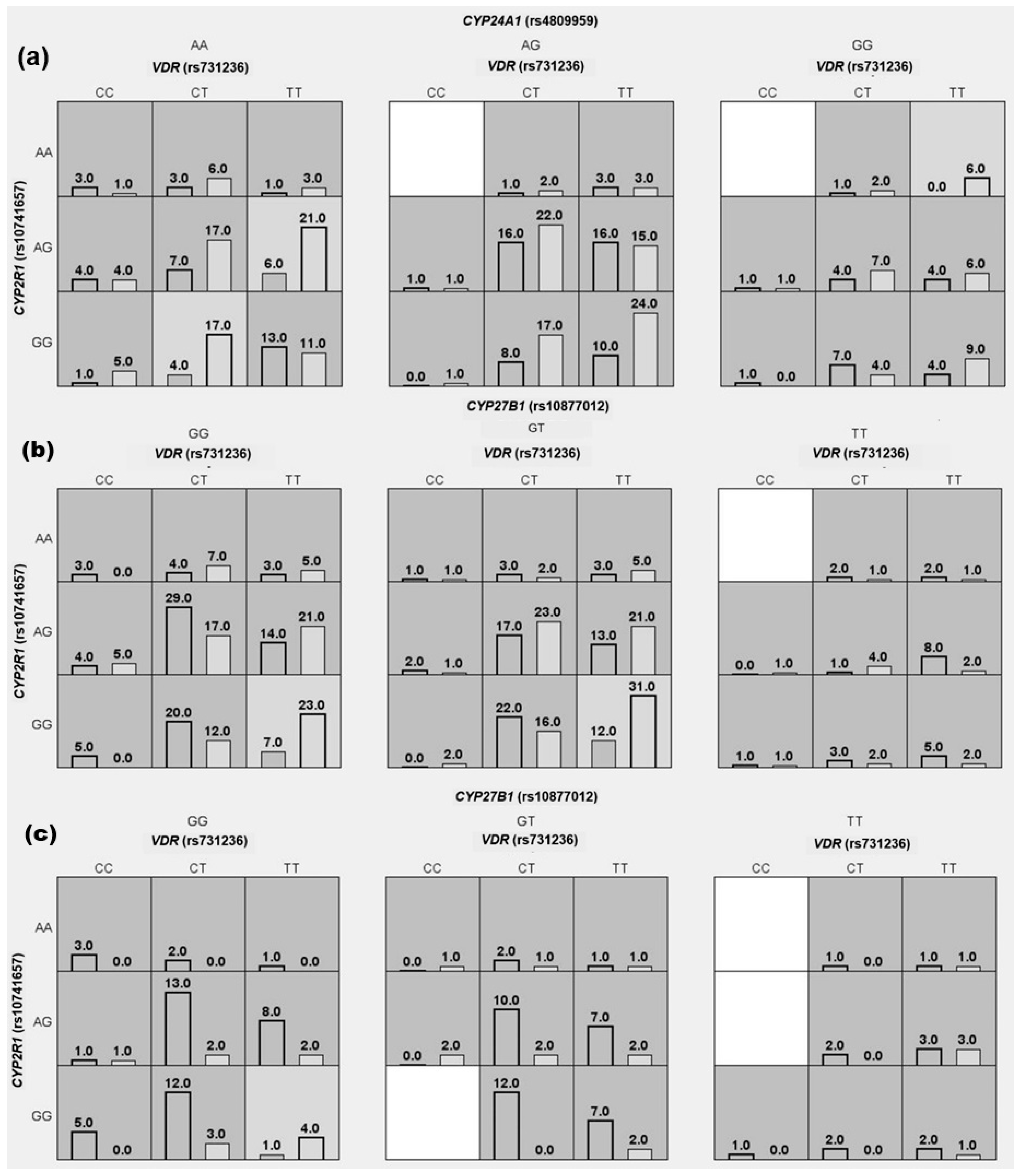
3.2. Interaction of Vitamin D Metabolism SNVs with Hypovitaminosis D, RA, and Its Clinical Disease Activity Susceptibility
3.3. Genotypic and Allelic Frequencies of SNV from Vitamin D Metabolism Genes in RA Patients and CS
3.4. Association of SNVs from Vitamin D Metabolism Genes with Hypovitaminosis D, RA, and Its Clinical Disease Activity Susceptibility
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACR | American College of Rheumatology |
| ACPAs | Anti-citrullinated peptide antibodies |
| BMI | Body mass index |
| CI | Confidence interval |
| CRP | C-reactive protein |
| CVC | Cross-validation consistency |
| CYP2R1 | Cytochrome P450 family 2 subfamily R member 1 |
| CYP24A1 | Cytochrome P450 family 24 subfamily A member 1 |
| CYP27B1 | Cytochrome P450 family 27 subfamily B member 1 |
| DAS28 | Disease activity score in 28 joints |
| ESR | Erythrocyte sedimentation rate |
| HDL-C | High-density lipoprotein cholesterol |
| CS | Control subjects |
| LDL-C | Low-density lipoprotein cholesterol |
| MDR | Multifactor dimensionality reduction |
| OR | Odds ratio |
| PCR | Polymerase chain reaction |
| RA | Rheumatoid arthritis |
| RF | Rheumatoid factor |
| SNV | Single nucleotide variant |
| TA | Testing accuracy |
| UTR | Untranslated region |
| VDR | Vitamin D receptor |
| VDRE | Vitamin D response element |
References
- Smolen, J.S.; Aletaha, D.; Barton, A.; Burmester, G.R.; Emery, P.; Firestein, G.S.; Kavanaugh, A.; McInnes, I.B.; Solomon, D.H.; Strand, V.; et al. Rheumatoid Arthritis. Nat. Rev. Dis. Primers 2018, 4, 18001. [Google Scholar] [CrossRef]
- Lee, Y.H.; Bae, S.-C. Vitamin D Level in Rheumatoid Arthritis and Its Correlation with the Disease Activity: A Meta-Analysis. Clin. Exp. Rheumatol. 2016, 34, 827–833. [Google Scholar]
- Ruiz-Ballesteros, A.I.; Meza-Meza, M.R.; Vizmanos-Lamotte, B.; Parra-Rojas, I.; de la Cruz-Mosso, U. Association of Vitamin D Metabolism Gene Polymorphisms with Autoimmunity: Evidence in Population Genetic Studies. Int. J. Mol. Sci. 2020, 21, 9626. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Zhao, Y.; Pan, J.; Fang, K.; Wang, Y.; Li, Z.; Chang, X. Vitamin D-Binding Protein (Group-Specific Component) Has Decreased Expression in Rheumatoid Arthritis. Clin. Exp. Rheumatol. 2012, 30, 525–533. [Google Scholar] [PubMed]
- Yoshida, S.; Ikari, K.; Furuya, T.; Toyama, Y.; Taniguchi, A.; Yamanaka, H.; Momohara, S. A GC Polymorphism Associated with Serum 25-Hydroxyvitamin D Level Is a Risk Factor for Hip Fracture in Japanese Patients with Rheumatoid Arthritis: 10-Year Follow-up of the Institute of Rheumatology, Rheumatoid Arthritis Cohort Study. Arthritis Res. Ther. 2014, 16, R75. [Google Scholar] [CrossRef]
- Jeffery, L.E.; Burke, F.; Mura, M.; Zheng, Y.; Qureshi, O.S.; Hewison, M.; Walker, L.S.K.; Lammas, D.A.; Raza, K.; Sansom, D.M. 1,25-Dihydroxyvitamin D3 and IL-2 Combine to Inhibit T Cell Production of Inflammatory Cytokines and Promote Development of Regulatory T Cells Expressing CTLA-4 and FoxP3. J. Immunol. 2009, 183, 5458–5467. [Google Scholar] [CrossRef] [PubMed]
- Dankers, W.; Colin, E.M.; van Hamburg, J.P.; Lubberts, E. Vitamin D in Autoimmunity: Molecular Mechanisms and Therapeutic Potential. Front. Immunol. 2017, 7, 697. [Google Scholar] [CrossRef]
- Meza-Meza, M.R.; Ruiz-Ballesteros, A.I.; de la Cruz-Mosso, U. Functional Effects of Vitamin D: From Nutrient to Immunomodulator. Crit. Rev. Food Sci. Nutr. 2022, 62, 3042–3062. [Google Scholar] [CrossRef]
- Sepulveda-Villegas, M.; Elizondo-Montemayor, L.; Trevino, V. Identification and Analysis of 35 Genes Associated with Vitamin D Deficiency: A Systematic Review to Identify Genetic Variants. J. Steroid Biochem. Mol. Biol. 2020, 196, 105516. [Google Scholar] [CrossRef]
- Clifton-Bligh, R.J.; Nguyen, T.V.; Au, A.; Bullock, M.; Cameron, I.; Cumming, R.; Chen, J.S.; March, L.M.; Seibel, M.J.; Sambrook, P.N. Contribution of a Common Variant in the Promoter of the 1-α-Hydroxylase Gene (CYP27B1) to Fracture Risk in the Elderly. Calcif. Tissue Int. 2011, 88, 109–116. [Google Scholar] [CrossRef]
- Ma, X.; Xie, Z.; Qin, J.; Luo, S.; Zhou, Z. Association of Vitamin D Pathway Gene CYP27B1 and CYP2R1 Polymorphisms with Autoimmune Endocrine Disorders: A Meta-Analysis. J. Clin. Endocrinol. Metab. 2020, 105, 3575–3587. [Google Scholar] [CrossRef]
- Wu, Y.; Zeng, Y.; Wu, W.; Lin, J.; Ou, Q. Polymorphisms of CYP 27B1 Are Associated with IFN Efficacy in HB eAg-positive Patients. Clin. Lab. Anal. 2018, 32, e22367. [Google Scholar] [CrossRef]
- Zacharioudaki, M.; Messaritakis, I.; Galanakis, E. Vitamin D Receptor, Vitamin D Binding Protein and CYP27B1 Single Nucleotide Polymorphisms and Susceptibility to Viral Infections in Infants. Sci. Rep. 2021, 11, 13835. [Google Scholar] [CrossRef]
- Uitterlinden, A.G.; Fang, Y.; Van Meurs, J.B.J.; Pols, H.A.P.; Van Leeuwen, J.P.T.M. Genetics and Biology of Vitamin D Receptor Polymorphisms. Gene 2004, 338, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Kerr Whitfield, G.; Remus, L.S.; Jurutka, P.W.; Zitzer, H.; Oza, A.K.; Dang, H.T.L.; Haussler, C.A.; Galligan, M.A.; Thatcher, M.L.; Dominguez, C.E.; et al. Functionally Relevant Polymorphisms in the Human Nuclear Vitamin D Receptor Gene. Mol. Cell. Endocrinol. 2001, 177, 145–159. [Google Scholar] [CrossRef]
- Hahn, L.W.; Ritchie, M.D.; Moore, J.H. Multifactor Dimensionality Reduction Software for Detecting Gene–Gene and Gene–Environment Interactions. Bioinformatics 2003, 19, 376–382. [Google Scholar] [CrossRef]
- Aletaha, D.; Neogi, T.; Silman, A.J.; Funovits, J.; Felson, D.T.; Bingham, C.O.; Birnbaum, N.S.; Burmester, G.R.; Bykerk, V.P.; Cohen, M.D.; et al. 2010 Rheumatoid Arthritis Classification Criteria: An American College of Rheumatology/European League Against Rheumatism Collaborative Initiative. Ann. Rheum. Dis. 2010, 69, 1580–1588. [Google Scholar] [CrossRef] [PubMed]
- Gómez, A. Nuevos criterios de clasificación de artritis reumatoide. Reumatol. Clínica 2011, 6, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Cortés, G.; Salazar-Flores, J.; Haro-Guerrero, J.; Rubi-Castellanos, R.; Velarde-Félix, J.S.; Muñoz-Valle, J.F.; López-Casamichana, M.; Carrillo-Tapia, E.; Canseco-Avila, L.M.; Bravi, C.M.; et al. Maternal Admixture and Population Structure in Mexican–Mestizos Based on mtDNA Haplogroups. Am. J. Phys. Anthropol. 2013, 151, 526–537. [Google Scholar] [CrossRef] [PubMed]
- Wells, G.; Becker, J.-C.; Teng, J.; Dougados, M.; Schiff, M.; Smolen, J.; Aletaha, D.; van Riel, P.L.C.M. Validation of the 28-Joint Disease Activity Score (DAS28) and European League Against Rheumatism Response Criteria Based on C-Reactive Protein against Disease Progression in Patients with Rheumatoid Arthritis, and Comparison with the DAS28 Based on Erythrocyte Sedimentation Rate. Ann. Rheum. Dis. 2009, 68, 954–960. [Google Scholar] [CrossRef]
- Holick, M.F. Vitamin D Deficiency. N. Engl. J. Med. 2007, 357, 266–281. [Google Scholar] [CrossRef]
- Miller, S.A.; Dykes, D.D.; Polesky, H.F. A Simple Salting out Procedure for Extracting DNA from Human Nucleated Cells. Nucleic Acids Res. 1988, 16, 1215. [Google Scholar] [CrossRef]
- Dean, A.G.; Sullivan, K.M.; Soe, M.M. Open Source Epidemiologic Statistics for Public Health, Version. Available online: www.OpenEpi.com (accessed on 15 July 2025).
- The 1000 Genomes Project Consortium; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A Global Reference for Human Genetic Variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- Meza-Meza, M.R.; Muñoz-Valle, J.F.; Ruiz-Ballesteros, A.I.; Vizmanos-Lamotte, B.; Parra-Rojas, I.; Martínez-López, E.; Oregon-Romero, E.; Márquez-Sandoval, Y.F.; Cerpa-Cruz, S.; De La Cruz-Mosso, U. Association of High Calcitriol Serum Levels and Its Hydroxylation Efficiency Ratio with Disease Risk in SLE Patients with Vitamin D Deficiency. J. Immunol. Res. 2021, 2021, 2808613. [Google Scholar] [CrossRef]
- Rivera-Escoto, M.; Campos-López, B.; Pesqueda-Cendejas, K.; Ruiz-Ballesteros, A.I.; Mora-García, P.E.; Meza-Meza, M.R.; Parra-Rojas, I.; Oregon-Romero, E.; Cerpa-Cruz, S.; De La Cruz-Mosso, U. Analysis of Potential Vitamin D Molecule Biomarkers: Association of Calcitriol and Its Hydroxylation Efficiency Ratio with Cardiovascular Disease Risk in Rheumatoid Arthritis Patients. Biomedicines 2024, 12, 273. [Google Scholar] [CrossRef]
- Ahmad, H.M.; Zaki, Z.M.; Mohamed, A.S.; Ahmed, A.E. Genetic Risk of Rheumatoid Arthritis: A Case Control Study. Biochem. Genet. 2024, 62, 3624–3641. [Google Scholar] [CrossRef] [PubMed]
- Di Spigna, G.; Del Puente, A.; Covelli, B.; Abete, E.; Varriale, E.; Salzano, S.; Postiglione, L. Vitamin D Receptor Polymorphisms as Tool for Early Screening of Severe Bone Loss in Women Patients with Rheumatoid Arthritis. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 4664–4669. [Google Scholar] [PubMed]
- Punceviciene, E.; Gaizevska, J.; Sabaliauskaite, R.; Venceviciene, L.; Puriene, A.; Vitkus, D.; Jarmalaite, S.; Butrimiene, I. Vitamin D and VDR Gene Polymorphisms’ Association with Rheumatoid Arthritis in Lithuanian Population. Medicina 2021, 57, 346. [Google Scholar] [CrossRef] [PubMed]
- Jóźwicki, W.; Brożyna, A.; Siekiera, J.; Slominski, A. Expression of Vitamin D Receptor (VDR) Positively Correlates with Survival of Urothelial Bladder Cancer Patients. Int. J. Mol. Sci. 2015, 16, 24369–24386. [Google Scholar] [CrossRef]
- Smolders, J.; Schuurman, K.G.; Strien, M.E.V.; Melief, J.; Hendrickx, D.; Hol, E.M.; Eden, C.V.; Luchetti, S.; Huitinga, I. Expression of Vitamin D Receptor and Metabolizing Enzymes in Multiple Sclerosis—Affected Brain Tissue. J. Neuropathol. Exp. Neurol. 2013, 72, 91–105. [Google Scholar] [CrossRef]
| Variable | RA Patients (n = 204) | CS (n = 204) | p Value * |
|---|---|---|---|
| Anthropometric | |||
| Age (years) a | 48 (28–65) | 34 (19–59) | <0.001 |
| BMI (kg/m2) a | 27.2 (19.8–37.9) | 24.0 (18.8–34.9) | <0.001 |
| Biochemical | |||
| Glucose (mg/dL) a | 87 (69.9–127) | 88.2 (74.4–114.2) | 0.9 |
| Cholesterol (mg/dL) a | 168.2 (119.9–232) | 174.3 (125.3–243.8) | 0.2 |
| Triglycerides (mg/dL) a | 98.5 (47.0–195) | 76.0 (39.4–198.7) | <0.001 |
| HDL-C (mg/dL) a | 48.9 (29.9–74.9) | 51.4 (34.7–75.9) | 0.02 |
| LDL-C (mg/dL) a | 92.1 (40.8–142) | 95.2 (61.6–157.2) | <0.01 |
| Uric acid (mg/dL) a | 3.8 (2.1–6.8) | 4.4 (3.1–7.3) | <0.001 |
| Albumin (g/dL) a | 3.8 (3.3–4.5) | 3.8 (3.4–4.42) | 0.5 |
| Clinical | |||
| Disease duration (years) a | 7 (1–25) | - | |
| Tender joints a | 1.5 (0–10) | - | |
| Swollen joints a | 1 (0–8) | - | |
| DAS28 (ESR) a | 3.5 (1.8–6.1) | ||
| Remission (DAS28-ESR <2.6) b | 21 (29/136) | - | |
| Activity (DAS28-ESR >2.6 )b | 79 (107/136) | - | |
| CRP (mg/L) a | 4.9 (0.5–35.2) | 1.2 (0–12.6) | <0.001 |
| ESR (mm/h) a | 32.5 (8–79) | - | |
| ACPAs (UI/mL) a | 256 (1.5–1173) | - | |
| RF (UI/mL) a | 121.5 (8–607) | - | |
| Vitamin D metabolites | |||
| Calcidiol (ng/mL) a | 23.7 (9.4–53.9) | 22.9 (10.9–39.4) | 0.3 |
| Without hypovitaminosis D b | 37 (48/119) | 36 (71/119) | 0.87 |
| With hypovitaminosis D b | 63 (81/206) | 64 (125/206) | |
| Treatment | |||
| NSAIDs b | 68 (135/198) | - | |
| Glucocorticoids b | 18 (35/193) | - | |
| Azathioprine b | 4 (7/194) | - | |
| Methotrexate b | 85 (170/199) | - | |
| Chloroquine b | 29 (58/199) | - | |
| Hydroxychloroquine b | 8 (15/177) | - | |
| Sulfasalazine b | 71 (142/198) | - | |
| Vitamin D supplements (Cholecalciferol) | 23 (45/197) | - |
| Susceptibility to Hypovitaminosis D | ||||||
|---|---|---|---|---|---|---|
| Total (n = 324) | Without Hypovitaminosis D (n = 119) | With Hypovitaminosis D (n = 205) | TA | CVC | OR (95% CI) | p-Value |
| Model 1 VDR (rs731236), CYP2R1 (rs10741657), and CYP24A1 (rs4809959) | TT or CT in VDR (rs731236), plus any genotype in CYP2R1 (rs10741657), and AA or GG in CYP24A1 (rs4809959) | Any other combination | 0.51 | 4/10 | 2.9 (1.4–6.4) | <0.01 |
| Model 2 CYP27B1 (rs10877012) and CYP24A1 (rs4809959) | GT in CYP27B1 (rs10877012), and AA in CYP24A1 (rs4809959) | Any other combination | 0.50 | 5/10 | 2.2 (1.1–4.1) | 0.02 |
| Model 3 CYP27B1 (rs10877012) | GT in CYP27B1 (rs10877012) | GG or TT in CYP27B1 | 0.57 | 10/10 | 1.7 (1.1–2.7) | 0.02 |
| Susceptibility to rheumatoid arthritis | ||||||
| Total (n = 402) | Low (n = 199) | High (n = 204) | TA | CVC | OR (95% CI) | p-Value |
| Model 1 VDR (rs731236), CYP2R1 (rs10741657), and CYP27B1 (rs10877012) | TT in VDR (rs731236), plus GG in CYP2R1 (rs10741657), and plus GG or GT in CYP27B1 (rs10877012) | Any other combination | 0.57 | 7/10 | 2.9 (1.7–4.9) | <0.001 |
| Model 2 VDR (rs731236), and CYP27B1 (rs10877012) | TT in VDR (rs731236) plus GG and GT in CYP27B1 (rs10877012) | Any other combination | 0.61 | 10/10 | 2.5 (1.6–3.8) | <0.001 |
| Model 3 VDR (rs731236) | TT in VDR (rs731236) | CT or CC in VDR | 0.58 | 10/10 | 1.9 (1.3–2.8) | <0.01 |
| Susceptibility to high clinical disease activity (DAS28-ESR) | ||||||
| Total (n = 134) | Disease remission (n = 105) | Disease activity (n = 29) | TA | CVC | OR (95% CI) | p-Value |
| Model 1 VDR (rs731236), CYP2R1 (rs10741657), and CYP27B1 (rs10877012) | TT in VDR (rs731236), plus GG in CYP2R1 (rs10741657), and GG in CYP27B1 (rs10877012) | Any other combination | 0.57 | 10/10 | 10.7 (1.9–58.7) | <0.01 |
| Model 2 VDR (rs731236) and CYP2R1 (rs10741657) | TT in VDR (rs731236) and GG in CYP2R1 (rs10741657) | Any other combination | 0.50 | 7/10 | 2.9 (1.1–8.12) | 0.03 |
| Model 3 VDR (rs731236) | TT in VDR (rs731236) | CT or CC in VDR | 0.62 | 10/10 | 2.8 (1.2–6.6) | 0.01 |
| Hypovitaminosis D (Both Groups) | RA Disease Susceptibility | Clinical Disease Activity (DAS28-ESR) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNV | With Hypovitaminosis D (n = 119) | Without Hypovitaminosis D (n = 205) | p Value | OR (95% CI) | p Value | RA (n = 199) | CS (n = 204) | p Value | OR (95% CI) | p Value | Activity (n = 105) | Remission (n = 29) | p Value | OR (95% CI) | p Value |
| rs10741657 CYP2R1 | 0.8 | 0.8 | 0.8 | ||||||||||||
| AA § | 10 (12) | 11 (23) | 1 | 12 (23) | 10 (21) | 1 | 11 (12) | 14 (4) | 1 | ||||||
| AG | 50 (59) | 46 (94) | 1.2 (0.5–2.8) | 0.6 | 47 (94) | 47 (95) | 0.9 (0.4–1.8) | 0.8 | 45 (47) | 48 (14) | 1.1 (0.2–4.5) | 0.9 | |||
| GG | 40 (48) | 43 (88) | 1.0 (0.4–2.5) | 0.9 | 41 (82) | 43 (88) | 0.8 (0.4–1.7) | 0.6 | 44 (46) | 38 (11) | 1.4 (0.3–5.8) | 0.6 | |||
| rs10877012 CYP27B1 | 0.04 | 0.08 | 0.8 | ||||||||||||
| GG | 55 (66) | 41 (84) | 1.8 (1.1–3.0) | 0.01 | 47 (94) | 44 (89) | 1.3 (0.8–2.0) | 0.2 | 47 (49) | 45 (13) | 0.9 (0.3–2.6) | 0.9 | |||
| GT § | 37 (44) | 51 (104) | 1 | 41 (81) | 49 (101) | 1 | 41 (43) | 38 (11) | 1 | ||||||
| TT | 8 (9) | 8 (17) | 1.2 (0.4–3.2) | 0.6 | 12 (24) | 7 (14) | 2.1 (0.98–4.8) | 0.04 | 12 (13) | 17 (5) | 0.7 (0.2–2.9) | 0.5 | |||
| rs4809959 CYP24A1 | 0.5 | 0.2 | 0.7 | ||||||||||||
| AA § | 35 (42) | 41 (85) | 1 | 43 (85) | 36 (74) | 1 | 43 (45) | 45 (13) | 1 | ||||||
| AG | 46 (55) | 41 (85) | 1.3 (0.8–2.2) | 0.3 | 34 (67) | 42 (85) | 0.7 (0.4–1.1) | 0.1 | 33 (35) | 38 (11) | 0.9 (0.3–2.6) | 0.8 | |||
| GG | 18 (22) | 18 (35) | 1.2 (0.6–2.5) | 0.5 | 23 (47) | 22 (45) | 0.9 (0.5–1.5) | 0.7 | 24 (25) | 17 (5) | 1.4 (0.4–5.7) | 0.5 | |||
| rs731236 VDR | 0.6 | <0.01 | 0.02 | ||||||||||||
| CC | 9 (11) | 6 (13) | 1.4 (0.5–3.7) | 0.4 | 8 (16) | 5 (11) | 2.1 (0.9–5.4) | 0.06 | 10 (10) | 14 (4) | 1.2 (0.3–6.1) | 0.7 | |||
| CT | 43 (51) | 46 (94) | 0.9 (0.6–1.5) | 0.7 | 54 (107) | 41 (83) | 1.9 (1.2–2.9) | <0.01 | 57 (60) | 27 (8) | 3.6 (1.3–10.7) | <0.01 | |||
| TT § | 48 (57) | 48 (98) | 1 | 38 (75) | 54 (110) | 1 | 33 (35) | 59 (17) | 1 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campos-López, B.; Rivera-Escoto, M.; Ruiz-Ballesteros, A.I.; Pesqueda-Cendejas, K.; Mora-García, P.E.; Meza-Meza, M.R.; Parra-Rojas, I.; Moreno-Ortíz, J.M.; Turiján-Espinoza, E.; Vargas-Morales, J.M.; et al. Interaction Between Vitamin D Metabolism Genetic Variants: Association with Hypovitaminosis D, Rheumatoid Arthritis, and Its Clinical Disease Activity. Genes 2025, 16, 967. https://doi.org/10.3390/genes16080967
Campos-López B, Rivera-Escoto M, Ruiz-Ballesteros AI, Pesqueda-Cendejas K, Mora-García PE, Meza-Meza MR, Parra-Rojas I, Moreno-Ortíz JM, Turiján-Espinoza E, Vargas-Morales JM, et al. Interaction Between Vitamin D Metabolism Genetic Variants: Association with Hypovitaminosis D, Rheumatoid Arthritis, and Its Clinical Disease Activity. Genes. 2025; 16(8):967. https://doi.org/10.3390/genes16080967
Chicago/Turabian StyleCampos-López, Bertha, Melissa Rivera-Escoto, Adolfo I. Ruiz-Ballesteros, Karen Pesqueda-Cendejas, Paulina E. Mora-García, Mónica R. Meza-Meza, Isela Parra-Rojas, José M. Moreno-Ortíz, Eneida Turiján-Espinoza, Juan M. Vargas-Morales, and et al. 2025. "Interaction Between Vitamin D Metabolism Genetic Variants: Association with Hypovitaminosis D, Rheumatoid Arthritis, and Its Clinical Disease Activity" Genes 16, no. 8: 967. https://doi.org/10.3390/genes16080967
APA StyleCampos-López, B., Rivera-Escoto, M., Ruiz-Ballesteros, A. I., Pesqueda-Cendejas, K., Mora-García, P. E., Meza-Meza, M. R., Parra-Rojas, I., Moreno-Ortíz, J. M., Turiján-Espinoza, E., Vargas-Morales, J. M., Cerpa-Cruz, S., & De la Cruz-Mosso, U. (2025). Interaction Between Vitamin D Metabolism Genetic Variants: Association with Hypovitaminosis D, Rheumatoid Arthritis, and Its Clinical Disease Activity. Genes, 16(8), 967. https://doi.org/10.3390/genes16080967








