Prime Editing for Crop Improvement: A Systematic Review of Optimization Strategies and Advanced Applications
Abstract
1. Introduction
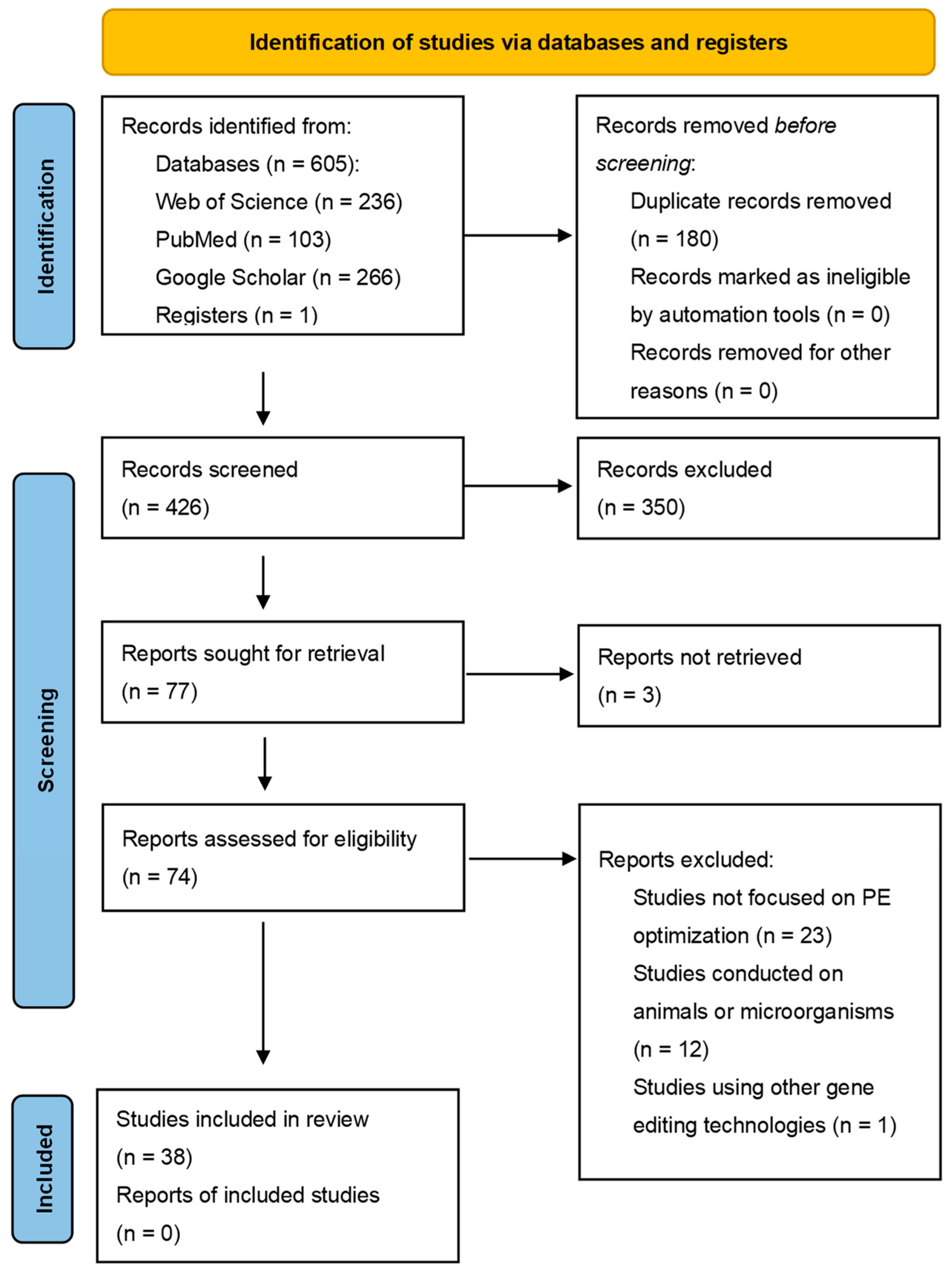
2. Methods
2.1. Study Selection
2.2. Optimization of PE Protein Components
2.3. Data Management and Extraction
2.4. Review Quality Assessment
3. Results
3.1. Working Mechanism and Foundational Functional Validation of Prime Editing
3.2. Core Challenge of PE: Overcoming the Bottleneck of Low Efficiency
3.2.1. Engineering Core Editing Components
- (1)
- Adjusting its spatial conformation and structure. Xu et al. (2022) found that relocating the RT enzyme from the traditional C-terminal fusion to an N-terminal fusion position favored the reverse transcription process in plant cells. This modification increased editing efficiency from 0–3.5% to 2.6–14.3% across multiple rice sites [21]. Zong et al. (2022) developed the ePPE system, which enhanced complex stability and editing capability by deleting the RNase H domain of RT and fusing it with a viral nucleocapsid (NC) protein. This resulted in average efficiency improvements of 3.9-fold for base substitutions and 6.5-fold for deletions, and enabled long fragment insertions unattainable with traditional PPE [19];
- (2)
- Optimizing or replacing the RT enzyme. Introducing point mutations is an effective “fine-tuning” strategy. For example, Ni et al. (2023) introduced the V223A mutation into M-MLV RT, which boosted the efficiency of various editing tasks by an additional 1.2- to 5.3-fold [22]. Replacing the RT source is more complex. While attempts by Lin et al. (2020) using CaMV RT or retron-derived RT resulted in reduced efficiency [9], Cao et al. (2024) found that an optimized Tf1 RT could increase average efficiency by 3.5-fold [23]. Paradoxically, Xu et al. (2024) reported that using Tf1 RT decreased efficiency [24]. These seemingly contradictory results underscore the complexity of PE optimization, where outcomes are highly dependent on the specific system, target site, and edit type. Furthermore, Cao et al. (2024) demonstrated that employing dual RT modules could synergistically further enhance editing efficiency [23].
3.2.2. Expression Regulation and Efficient Delivery of PE Systems
3.2.3. Optimization and Regulation of the Editing Reaction Process
3.2.4. Enrichment and Efficient Screening of Edited Events
3.3. Functional Expansion and Advanced Applications of PE
3.3.1. Expanding Editing Scope and Enhancing Applicability
3.3.2. Achieving Complex and Multifunctional Editing
4. Discussion
4.1. Interpretation of Key Findings: From Research Strategy to Functional Evolution
4.2. Limitations of the Included Studies and Current Research Challenges
4.3. Future Perspectives and Outstanding Questions
- (1)
- Fusing endogenous small RNA-binding proteins (e.g., La) to stabilize pegRNA and boost editing activity [53];
- (2)
- Using AI-driven rational design to optimize reverse transcription templates (RTTs). This can also be used to develop PE systems with a reverse editing window, which would expand the editable region and improve precision [54];
- (3)
- Developing innovative delivery platforms based on pseudoviral particles to enable more efficient and safer delivery of editing tools [55];
- (4)
- Constructing inverse PE platforms based on circular RNA to circumvent limitations inherent in traditional editing orientations [56].
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PE | prime editing |
| Cas9 | associated protein 9 |
| RT | reverse transcriptase |
| CRISPR/Cas9 | Clustered Regularly Interspaced Short Palindromic Repeats/associated protein 9 |
| DSBs | DNA double-strand breaks |
| NHEJ | non-homologous end joining |
| indels | insertions or deletions |
| HDR | homology-directed repair |
| BE | base editing |
| nCas9 | Cas9 nickase |
| pegRNA | prime editing guide RNA |
| PAM | protospacer adjacent motif |
| PRISMA | Preferred reporting items for systematic reviews and meta-analyses |
| NLS | nuclear localization signal |
| NC | nucleocapsid |
| mPE | modular PE |
| RTT | reverse transcriptase template |
| MMR | mismatch repair |
References
- Gelvin, S.B. (Ed.) Agrobacterium Biology: From Basic Science to Biotechnology; Current Topics in Microbiology and Immunology; Springer International Publishing: Cham, Switzerland, 2018; ISBN 978-3-030-03256-2. [Google Scholar]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-Guided Human Genome Engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef]
- Wyman, C.; Kanaar, R. DNA Double-Strand Break Repair: All’s Well That Ends Well. Annu. Rev. Genet. 2006, 40, 363–383. [Google Scholar] [CrossRef]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable Editing of a Target Base in Genomic DNA without Double-Stranded DNA Cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable Base Editing of A•T to G•C in Genomic DNA without DNA Cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-Replace Genome Editing without Double-Strand Breaks or Donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Zong, Y.; Xue, C.; Wang, S.; Jin, S.; Zhu, Z.; Wang, Y.; Anzalone, A.V.; Raguram, A.; Doman, J.L.; et al. Prime Genome Editing in Rice and Wheat. Nat. Biotechnol. 2020, 38, 582–585. [Google Scholar] [CrossRef]
- Xu, W.; Zhang, C.; Yang, Y.; Zhao, S.; Kang, G.; He, X.; Song, J.; Yang, J. Versatile Nucleotides Substitution in Plant Using an Improved Prime Editing System. Mol. Plant 2020, 13, 675–678. [Google Scholar] [CrossRef]
- Jiang, Y.-Y.; Chai, Y.-P.; Lu, M.-H.; Han, X.-L.; Lin, Q.; Zhang, Y.; Zhang, Q.; Zhou, Y.; Wang, X.-C.; Gao, C.; et al. Prime Editing Efficiently Generates W542L and S621I Double Mutations in Two ALS Genes in Maize. Genome Biol. 2020, 21, 257. [Google Scholar] [CrossRef]
- Lu, Y.; Tian, Y.; Shen, R.; Yao, Q.; Zhong, D.; Zhang, X.; Zhu, J.-K. Precise Genome Modification in Tomato Using an Improved Prime Editing System. Plant Biotechnol. J. 2021, 19, 415–417. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ma, R.; Liu, D.; Wang, M.; Zhu, T.; Deng, Y. A Straightforward Plant Prime Editing System Enabled Highly Efficient Precise Editing of Rice Waxy Gene. Plant Sci. 2022, 323, 111400. [Google Scholar] [CrossRef]
- Xu, R.; Li, J.; Liu, X.; Shan, T.; Qin, R.; Wei, P. Development of Plant Prime-Editing Systems for Precise Genome Editing. Plant Commun. 2020, 1, 100043. [Google Scholar] [CrossRef]
- Biswas, S.; Bridgeland, A.; Irum, S.; Thomson, M.J.; Septiningsih, E.M. Optimization of Prime Editing in Rice, Peanut, Chickpea, and Cowpea Protoplasts by Restoration of GFP Activity. Int. J. Mol. Sci. 2022, 23, 9809. [Google Scholar] [CrossRef]
- Vu, T.V.; Nguyen, N.T.; Kim, J.; Das, S.; Lee, J.; Kim, J.-Y. The Obstacles and Potential Solution Clues of Prime Editing Applications in Tomato. BioDesign Res. 2022, 2022, 0001. [Google Scholar] [CrossRef]
- Li, J.; Chen, L.; Liang, J.; Xu, R.; Jiang, Y.; Li, Y.; Ding, J.; Li, M.; Qin, R.; Wei, P. Development of a Highly Efficient Prime Editor 2 System in Plants. Genome Biol. 2022, 23, 161. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Zong, Y.; Liu, Y.; Xue, C.; Li, B.; Li, X.; Wang, Y.; Li, J.; Liu, G.; Huang, X.; Cao, X.; et al. An Engineered Prime Editor with Enhanced Editing Efficiency in Plants. Nat. Biotechnol. 2022, 40, 1394–1402. [Google Scholar] [CrossRef] [PubMed]
- Hua, K.; Jiang, Y.; Tao, X.; Zhu, J.-K. Precision Genome Engineering in Rice Using Prime Editing System. Plant Biotechnol. J. 2020, 18, 2167–2169. [Google Scholar] [CrossRef]
- Xu, W.; Yang, Y.; Yang, B.; Krueger, C.J.; Xiao, Q.; Zhao, S.; Zhang, L.; Kang, G.; Wang, F.; Yi, H.; et al. A Design Optimized Prime Editor with Expanded Scope and Capability in Plants. Nat. Plants 2022, 8, 45–52. [Google Scholar] [CrossRef]
- Ni, P.; Zhao, Y.; Zhou, X.; Liu, Z.; Huang, Z.; Ni, Z.; Sun, Q.; Zong, Y. Efficient and Versatile Multiplex Prime Editing in Hexaploid Wheat. Genome Biol. 2023, 24, 156. [Google Scholar] [CrossRef]
- Cao, Z.; Sun, W.; Qiao, D.; Wang, J.; Li, S.; Liu, X.; Xin, C.; Lu, Y.; Gul, S.L.; Wang, X.-C.; et al. PE6c Greatly Enhances Prime Editing in Transgenic Rice Plants. J. Integr. Plant Biol. 2024, 66, 1864–1870. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Ma, C.; Sheng, J.; Zhu, J.; Wang, D.; Liu, X.; Wang, Q.; Li, J.; Qin, R.; Wei, P. Engineering PE6 Prime Editors to Efficiently Insert Tags in Rice. Plant Biotechnol. J. 2024, 22, 3383–3385. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Wu, Y.; Guo, Y.; Wei, S. Addition of the T5 Exonuclease Increases the Prime Editing Efficiency in Plants. J. Genet. Genom. 2023, 50, 582–588. [Google Scholar] [CrossRef]
- Lu, P.; Zuo, E.; Yan, J. Developing a Multi-Modular Assembled Prime Editing (mPE) System Improved Precise Multi-Base Insertion Efficiency in Dicots. J. Adv. Res. 2025, 71, 81–92. [Google Scholar] [CrossRef]
- Lin, Q.; Jin, S.; Zong, Y.; Yu, H.; Zhu, Z.; Liu, G.; Kou, L.; Wang, Y.; Qiu, J.-L.; Li, J.; et al. High-Efficiency Prime Editing with Optimized, Paired pegRNAs in Plants. Nat. Biotechnol. 2021, 39, 923–927. [Google Scholar] [CrossRef]
- Jiang, Y.; Chai, Y.; Qiao, D.; Wang, J.; Xin, C.; Sun, W.; Cao, Z.; Zhang, Y.; Zhou, Y.; Wang, X.-C.; et al. Optimized Prime Editing Efficiently Generates Glyphosate-Resistant Rice Plants Carrying Homozygous TAP-IVS Mutation in EPSPS. Mol. Plant 2022, 15, 1646–1649. [Google Scholar] [CrossRef]
- Lou, H.; Li, S.; Shi, Z.; Zou, Y.; Zhang, Y.; Huang, X.; Yang, D.; Yang, Y.; Li, Z.; Xu, C. Engineering Source-Sink Relations by Prime Editing Confers Heat-Stress Resilience in Tomato and Rice. Cell 2025, 188, 530–549.e20. [Google Scholar] [CrossRef]
- Qiao, D.; Wang, J.; Lu, M.; Xin, C.; Chai, Y.; Jiang, Y.; Sun, W.; Cao, Z.; Guo, S.; Wang, X.; et al. Optimized Prime Editing Efficiently Generates Heritable Mutations in Maize. J. Integr. Plant Biol. 2023, 65, 900–906. [Google Scholar] [CrossRef] [PubMed]
- Lu, P. Repeated High-Temperature Treatment Can Increase Prime Editing Efficiency in Dicot Model Species. ACS Agric. Sci. Technol. 2024, 4, 1179–1183. [Google Scholar] [CrossRef]
- Wang, L.; Kaya, H.B.; Zhang, N.; Rai, R.; Willmann, M.R.; Carpenter, S.C.D.; Read, A.C.; Martin, F.; Fei, Z.; Leach, J.E.; et al. Spelling Changes and Fluorescent Tagging With Prime Editing Vectors for Plants. Front. Genome Ed. 2021, 3, 617553. [Google Scholar] [CrossRef]
- Vu, T.V.; Nguyen, N.T.; Kim, J.; Song, Y.J.; Nguyen, T.H.; Kim, J.-Y. Optimized Dicot Prime Editing Enables Heritable Desired Edits in Tomato and Arabidopsis. Nat. Plants 2024, 10, 1502–1513. [Google Scholar] [CrossRef]
- Tang, X.; Sretenovic, S.; Ren, Q.; Jia, X.; Li, M.; Fan, T.; Yin, D.; Xiang, S.; Guo, Y.; Liu, L.; et al. Plant Prime Editors Enable Precise Gene Editing in Rice Cells. Mol. Plant 2020, 13, 667–670. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Gu, D.; Zhang, Y.; Jiang, Y.; Xiao, Z.; Xu, R.; Qin, R.; Li, J.; Wei, P. Conditional Knockdown of OsMLH1 to Improve Plant Prime Editing Systems without Disturbing Fertility in Rice. Genome Biol. 2024, 25, 131. [Google Scholar] [CrossRef]
- Bai, M.; Lin, W.; Peng, C.; Song, P.; Kuang, H.; Lin, J.; Zhang, J.; Wang, J.; Chen, B.; Li, H.; et al. Expressing a Human RNA Demethylase as an Assister Improves Gene-Editing Efficiency in Plants. Mol. Plant 2024, 17, 363–366. [Google Scholar] [CrossRef]
- Zou, J.; Meng, X.; Liu, Q.; Shang, M.; Wang, K.; Li, J.; Yu, H.; Wang, C. Improving the Efficiency of Prime Editing with epegRNAs and High-Temperature Treatment in Rice. Sci. China Life Sci. 2022, 65, 2328–2331. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhu, Z.; Li, S.; Li, J.; Yan, L.; Zhang, C.; Ma, Y.; Xia, L. Multiplex Precision Gene Editing by a Surrogate Prime Editor in Rice. Mol. Plant 2022, 15, 1077–1080. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, L.; Zhang, C.; Yang, Y.; Liu, H.; Li, L.; Zhang, S.; Li, X.; Liu, X.; Liu, Y.; et al. Developing an Efficient and Visible Prime Editing System to Restore Tobacco 8-Hydroxy-Copalyl Diphosphate Gene for Labdane Diterpene Z-Abienol Biosynthesis. Sci. China Life Sci. 2023, 66, 2910–2921. [Google Scholar] [CrossRef]
- Sun, C.; Lei, Y.; Li, B.; Gao, Q.; Li, Y.; Cao, W.; Yang, C.; Li, H.; Wang, Z.; Li, Y.; et al. Precise Integration of Large DNA Sequences in Plant Genomes Using PrimeRoot Editors. Nat. Biotechnol. 2024, 42, 316–327. [Google Scholar] [CrossRef]
- Li, X.; Zhang, S.; Wang, C.; Ren, B.; Yan, F.; Li, S.; Spetz, C.; Huang, J.; Zhou, X.; Zhou, H. Efficient in Situ Epitope Tagging of Rice Genes by Nuclease-Mediated Prime Editing. Plant Cell 2025, 37, koae316. [Google Scholar] [CrossRef]
- Zou, J.; Meng, X.; Hong, Z.; Rao, Y.; Wang, K.; Li, J.; Yu, H.; Wang, C. Cas9-PE: A Robust Multiplex Gene Editing Tool for Simultaneous Precise Editing and Site-Specific Random Mutation in Rice. Trends Biotechnol. 2025, 43, 433–446. [Google Scholar] [CrossRef]
- Lu, Y.; Naren, T.; Qiao, D.; Wang, J.; Lyu, T.; Cao, Z.; Sun, W.; Ji, X.; Chen, Q.; Jiang, L. One-Step Generation of Prime-Edited Transgene-Free Rice. Plant Commun. 2025, 6, 101227. [Google Scholar] [CrossRef]
- Li, J.; Ding, J.; Zhu, J.; Xu, R.; Gu, D.; Liu, X.; Liang, J.; Qiu, C.; Wang, H.; Li, M.; et al. Prime Editing-Mediated Precise Knockin of Protein Tag Sequences in the Rice Genome. Plant Commun. 2023, 4, 100572. [Google Scholar] [CrossRef]
- Li, F.; Hou, H.; Song, M.; Chen, Z.; Peng, T.; Du, Y.; Zhao, Y.; Li, J.; Miao, C. Targeted Insertion of Large DNA Fragments through Template-Jumping Prime Editing in Rice. Plant Biotechnol. J. 2025, 23, 2645. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhang, X.; Xu, W.; Kang, G.; Liu, Y.; Liu, X.; Ren, W.; Zhao, J.; Yang, J. Efficient and Precise Genomic Deletion in Rice Using Enhanced Prime Editing. aBIOTECH 2024, 5, 214–218. [Google Scholar] [CrossRef]
- Zhao, Y.; Huang, Z.; Zhou, X.; Teng, W.; Liu, Z.; Wang, W.; Tang, S.; Liu, Y.; Liu, J.; Wang, W.; et al. Precise Deletion, Replacement and Inversion of Large DNA Fragments in Plants Using Dual Prime Editing. Nat. Plants 2025, 11, 191–205. [Google Scholar] [CrossRef]
- Xu, R.; Liu, X.; Li, J.; Qin, R.; Wei, P. Identification of Herbicide Resistance OsACC1 Mutations via in Planta Prime-Editing-Library Screening in Rice. Nat. Plants 2021, 7, 888–892. [Google Scholar] [CrossRef]
- Gupta, A.; Liu, B.; Raza, S.; Chen, Q.-J.; Yang, B. Modularly Assembled Multiplex Prime Editors for Simultaneous Editing of Agronomically Important Genes in Rice. Plant Commun. 2024, 5, 100741. [Google Scholar] [CrossRef]
- Nguyen, C.X.; Nguyen, T.D.; Dinh, T.T.; Nguyen, L.T.; Ly, L.K.; Chu, H.H.; La, T.C.; Do, P.T. Prime Editing via Precise Sequence Insertion Restores Function of the Recessive rc Allele in Rice. Plant Cell Rep. 2025, 44, 57. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhao, Y.; Zheng, M.; Yu, S.; Gao, Y.; Zhu, G.; Zhu, J.-K.; Hua, K.; Wang, Z. A Natural Variation Contributes to Sugar Accumulation in Fruit during Tomato Domestication. Plant Biotechnol. J. 2024, 22, 3520–3522. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wheatley, M.; Meakem, V.; Galarneau, E.; Gutierrez, B.; Zhong, G.-Y. Editing VvDXS1 for the Creation of Muscat Flavour in Vitis vinifera cv. Scarlet Royal. Plant Biotechnol. J. 2024, 22, 1610–1621. [Google Scholar] [CrossRef]
- Yan, J.; Oyler-Castrillo, P.; Ravisankar, P.; Ward, C.C.; Levesque, S.; Jing, Y.; Simpson, D.; Zhao, A.; Li, H.; Yan, W.; et al. Improving Prime Editing with an Endogenous Small RNA-Binding Protein. Nature 2024, 628, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Fang, Q.; Li, M.; Zhang, J.; Li, R.; Zhou, T.; Wang, K.; Deng, J.; Wang, X.; Huang, C. Prime Editor with Rational Design and AI-Driven Optimization for Reverse Editing Window and Enhanced Fidelity. Nat. Commun. 2025, 16, 5144. [Google Scholar] [CrossRef] [PubMed]
- Halegua, T.; Risson, V.; Carras, J.; Rouyer, M.; Coudert, L.; Jacquier, A.; Schaeffer, L.; Ohlmann, T.; Mangeot, P.E. Delivery of Prime Editing in Human Stem Cells Using Pseudoviral NanoScribes Particles. Nat. Commun. 2025, 16, 397. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.; Wang, S.; Cai, Y.; Li, Z.; Li, K.M.; Wei, J.; Sun, C.; Zhu, H.; Chen, K.; Gao, C. Circular RNA-Mediated Inverse Prime Editing in Human Cells. Nat. Commun. 2025, 16, 5057. [Google Scholar] [CrossRef]
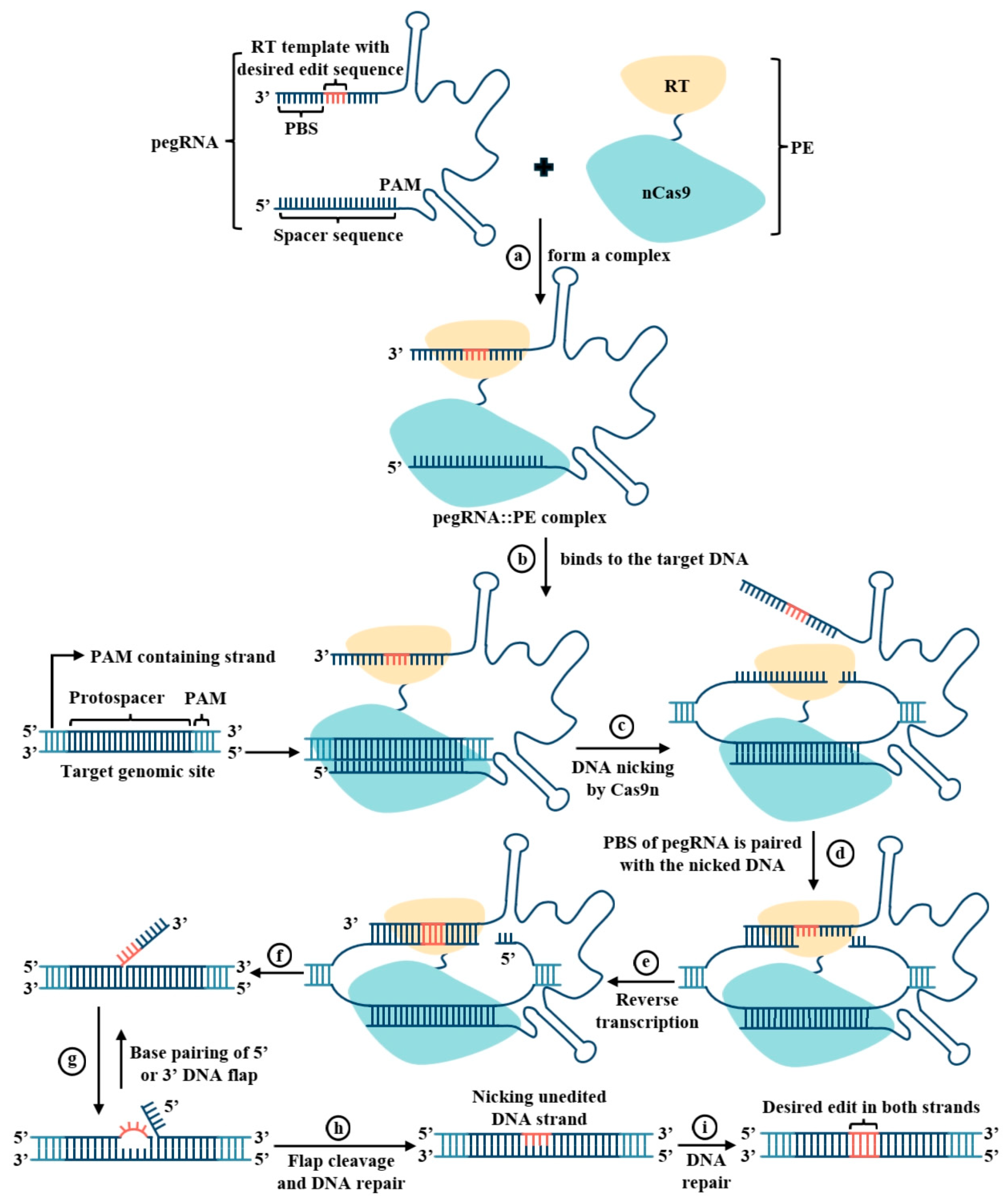
| Species | Target Gene | Edit Type | PE System | Experimental System | Editing Efficiency | Remarks | Reference |
|---|---|---|---|---|---|---|---|
| Rice | OsCDC48 | Deletion (6 bp) | PPE2 | Protoplast | 8.20% | Achieved 6-bp deletion in rice protoplasts | [9] |
| Rice | OsCDC48 | Insertion (3 bp) | PPE2 | Protoplast | 2.00% | Achieved 3-bp insertion in rice protoplasts | [9] |
| Rice | OsCDC48 | Base substitution (1 bp) | PPE2 | Protoplast | 5.70% | Highest efficiency among multiple single-base substitutions | [9] |
| Wheat | TaGASR7 | Base substitution (1 bp) | PPE2 | Protoplast | 1.40% | Highest efficiency among multiple single-base substitutions in wheat protoplasts | [9] |
| Rice | OsPDS | Insertion (3 bp) | pPE2 | Callus | 19.80% | Achieved small-fragment insertion in rice plants | [14] |
| Rice | OsWx | Base substitution (3 bp) | PE2 | Callus | 66.70% | High-efficiency base substitution in rice plants | [13] |
| Rice | OsWx | Insertion (6 bp) | PE2 | Regenerated plants | 36.80% | Achieved 6-bp insertion with relatively high efficiency | [13] |
| Rice | OsRDD1 | Deletion (18 bp) | PPE | Regenerated plants | 2.80% | Successfully achieved 18-bp deletion, albeit at low efficiency | [19] |
| Legume crops | Exogenous mutant GFP | Base substitution (1 bp) | PE2 | Protoplast | 0.00% | PE2 system failed to edit legume crop protoplasts | [15] |
| Tomato | SlWH9 | Base substitution (1 bp) | nCas9-RT | Callus | 0.11% | Highest editing efficiency in tomato callus, yet still suboptimal | [16] |
| Rice | OsCDC48 | Base substitution (1 bp) | pPE2 | Regenerated plants | 29.17% | Significant variation in PE2 efficiency across different target genes | [17] |
| Rice | OsACC | Base substitution (1 bp) | pPE2 | Regenerated plants | 0.00% | — | [17] |
| Rice | OsPDS | Base substitution (A to T) | pPE2 | Regenerated plants | 31.30% | Markedly different efficiencies for different edits at the same locus | [14] |
| Rice | OsPDS | Base substitution (A to C) | pPE2 | Regenerated plants | 0.00% | — | [14] |
| Rice | OsACC | Base substitution (G to C) | pPE2 | Regenerated plants | 14.60% | Efficiency variation using different pegRNAs for the same edit at the same locus | [14] |
| Rice | OsACC | Base substitution (G to C) | pPE2 | Regenerated plants | 3.10% | — | [14] |
| Rice | OsACC | Base substitution (G to C) | pPE2 | Regenerated plants | 1.00% | — | [14] |
| Rice | OsACC | Base substitution (G to C) | pPE2 | Regenerated plants | 0.00% | — | [14] |
| Component | Specific Strategy | Baseline | Edit Type | Key Effect | Species | Reference |
|---|---|---|---|---|---|---|
| nCas9 | PEmax architecture | pPE2 | Base sub (1-bp or 2-bp) | · Introduced R221K/N394K mutations enhancing pegRNA binding · Efficiency increased 3.80- to 5.35-fold | Rice | [17] |
| nCas9 | SaCas9 (N580A) | PE3 | Base sub (1-bp or 2-bp) | · Extremely low editing efficiency, virtually no effective edits | Rice | [20] |
| RT | N-terminal fusion | PE3 | Base sub (1-bp) | · Efficiency increased: OsGS3 (3.5%→14.3%), OsALS-1 (0%→2.6%), OsACC2 (0%→4.4%) | Rice | [21] |
| RT | RNase H domain deletion + NC fusion | PPE | Base sub (1/2-bp), Del (15–90 bp), Ins (18/24/34 bp) | · Avg base sub efficiency ×3.9 (max ×121.5) · Avg del efficiency ×6.5 · Ins efficiency: 18 bp (3.1%), 24 bp (0.2%), 34 bp (0.3%) | Rice | [19] |
| RT | V223A mutation | ePPE | Base sub (1-bp), Del (1/5/6-bp) | · Editing efficiency ×1.2–5.3 (avg ×2.8) · No increase in byproducts | Wheat | [22] |
| RT | CaMV RT or retron-derived RT | PPE3b | Base sub (2-bp) | · Efficiency lower than M-MLV RT | Rice | [9] |
| RT | Ec48 RT (PE6a), Tf1 RT (PE6b), Opt Tf1 RT (PE6c), Opt M-MLV RT (PE6d) | PE3 | Base sub (1/2/3-bp), Del (84-bp), Ins (30-bp) | · All PE6 editors except PE6a increased efficiency · PE6c highest: avg editing ×3.5, homozygous rate significantly increased | Rice | [23] |
| RT | Tf1 RT | ePE2 | Base sub (1-bp), Ins (1/4/27-bp) | · Reduced efficiency for all edit types | Rice | [24] |
| RT | Dual RT module (Tf1 RT, Opt M-MLV RT) | PE3 | Base sub (1/2/3-bp), Del (84-bp), Ins (30-bp) | · Synergistic effect, efficiency higher than single modules | Rice | [23] |
| Auxiliary Module | T5 exonuclease upstream of Cas9 | PE2 | Base sub (1/2-bp), Del (4-bp), Ins (4-bp) | · Protoplast efficiency ×1.7–2.9 · Transgenic plant efficiency ×1.34, homozygous mutant ratio ×5 | Rice | [25] |
| Expression System | Modular split (mPE system) | PE2 | Base sub (1/2/4-bp), Ins (3/4/6/7-bp) | · Tobacco transient: Total efficiency 0.01%→0.26% (×26.4); Multi-base ins avg ×197.9 (max ×1288) | Tobacco | [26] |
| Category | Specific Strategy | Baseline | Edit Type | Key Effect | Species | Reference |
|---|---|---|---|---|---|---|
| Key Element Design | PBS Tm = 30 °C | PPE2 | Base sub (1/2-bp), Del (2/3-bp), Ins (1-bp) | · PBS Tm 30 °C optimal for rice · Efficiency follows normal distribution | Rice | [27] |
| Key Element Design | Dual-pegRNA | PPE2 | Base sub (1/2-bp), Del (1/2-bp), Ins (1-bp) | · Editing efficiency ×1.8–4.2 · No increase in byproduct ratio | Rice | [27] |
| Key Element Design | RTT termination principle | Unoptimized pegRNA | Base sub (1-bp) | · Terminate RTT 1–3 bp after C/GC/TGC · Completely eliminated byproducts | Rice | [28] |
| Key Element Design | Optimized secondary structure | PE2 | Base sub (3-bp) | · Avoid template hairpin/spacer complementarity + maintain gRNA conserved domains · Improper structure reduced efficiency from 66.7%→0% | Rice | [13] |
| Key Element Design | RT-M strategy | PE-P2–RT-S | Base sub (3/4-bp) | · Introduce primary mutation + adjacent synonymous mutation · Efficiency increased: OsALS-1 (0→4.3%), OsACC-2 (0.5%→4.4%), OsDEP1 (1.1%→2.6%), OsWaxy-1 (0→2.2%) | Rice | [21] |
| Key Element Design | Target selection principle | Unoptimized pegRNA | Ins (10-bp) | · Avoid functional elements/select open chromatin/near translation start site · Successfully applied to 4 tomato + 2 rice varieties, boosting yield under normal/heat stress | Tomato, Rice | [29] |
| Enhanced Stability | Add evopreQ1 RNA motif | pPE2 | Base sub (1/2-bp), Ins (1-bp) | · Mutation frequency ×2.35–29.22 | Rice | [17] |
| Enhanced Stability | Csy4 nuclease system | tRNA system | Simultaneous small-fragment edits | · Avg efficiency 13.8% · Synchronous editing of 4–10 genes: 7.4–10.3% | Wheat | [22] |
| Category | Specific Strategy | Baseline | Edit Type | Key Effect | Species | Reference |
|---|---|---|---|---|---|---|
| Enhanced Expression—PE Protein | Zmubi1 promoter | OsU6a promoter | Base sub (1/3/4-bp) | · Combined with hygromycin selection: Editing efficiency increased from 0–1.2%→2.6–26% | Rice | [10] |
| Enhanced Expression—PE Protein | AtRPS5A promoter | 35S promoter | Base sub (3-bp), Del (2-bp), Ins (4-bp) | · Avg editing efficiency 0.85%→2.6% | Tomato | [12] |
| Enhanced Expression—pegRNA | U6 composite promoter or increased pegRNA cassette number | PE3 | Base sub (1/2/3-bp) | · Editing efficiency 0.8–4.9%→1.9–7.1% · Doubling cassettes did not enhance efficiency | Maize | [11] |
| Enhanced Expression—pegRNA | Doubled epegRNA cassette number | ePE5max | Base sub (1 bp) | · Homozygous editing efficiency 0%→0.6–1.3% | Rice | [30] |
| Enhanced Expression—pegRNA | U6 composite promoter | pPE2max-evopreQ1 | Base sub (1/2-bp), Ins (1-bp) | · Efficiency ×1.66–15.60 | Rice | [17] |
| Enhanced Expression—pegRNA | CmYLCV promoter | CAMV 35S/OsU6 promoter | Base sub (1-bp) | · Successful editing only with CmYLCV promoter | Legume crops | [15] |
| Enhanced Expression—pegRNA | Pol II promoter system (tRNA processing) | Pol III promoter (AtU6) | Base sub (1/2-bp) | · tRNA processing system + AtUb10 promoter · Enabled editing in dicots (where Pol III failed) | Tobacco | [31] |
| Enhanced Expression—pegRNA | Pol II promoter system | Pol III promoter | Base sub (1/2/4-bp), Ins (3/4/6/7-bp) | · Unaffected by poly-T sequences in template · pegRNA expression ×20 · Cas9 cutting efficiency (indel rate) ×2–3 | Tobacco | [26] |
| Enhanced Delivery | pPEG system | pPPEM | Base sub (2 bp), Ins (25 bp) | · Co-transformation with an additional vector expressing pegRNA/sgRNA · No significant change in editing efficiency | Rice | [32] |
| Enhanced Delivery | Geminiviral replicon vector | T-DNA vector | N/A | · DNA cassette ×1.3 · RNA transcript ×1.9–2.0 · PE protein level ×4.5 · PE efficiency ×6.6–7.8 | Tomato | [33] |
| Category | Specific Strategy | Baseline | Edit Type | Key Effect | Species | Reference |
|---|---|---|---|---|---|---|
| DNA Repair Pathway | PE3/PE3b system | PE2 | Base sub (1-bp), Del (6-bp), Ins (3-bp) | · Efficiency comparable to PE2 | Rice, Wheat | [9] |
| DNA Repair Pathway | PE3/PE3b system | PE2 | Ins (3-bp) | · Efficiency comparable to PE2 · PE3 prone to large deletions · PE3b reduced deletions but introduced other byproducts | Rice | [34] |
| DNA Repair Pathway | PE3/PE3b system | PE2 | Base sub (1-bp) | · OsACC1 locus: PE2 (14.6%) vs. PE3 (18.8% + byproducts) vs. PE3b (6.3% no byproducts) | Rice | [14] |
| DNA Repair Pathway | PE3 system | PE2 | Base sub (3-bp), Del (2/4/18-bp), Ins (1/2/12 bp) | · Efficiency: PE3 (2.6–13%) < PE2 (30–66.7%) · PE3 induced NHEJ byproducts (26.3–38.9%) | Rice | [13] |
| DNA Repair Pathway | PE3 system | PE2 | Base sub (1/2/3-bp) | · Protoplast: Avg efficiency ×2.2 at most sites · Reduced byproducts | Rice | [28] |
| DNA Repair Pathway | Fusion hMLH1dn | pPE2max | Base sub (1/2-bp), Ins (1-bp) | · No significant enhancement | Rice | [17] |
| DNA Repair Pathway | Fusion of various OsMLH1dn | ePE3max | Base sub (3 bp) | · Did not significantly increase editing efficiency | Rice | [28] |
| DNA Repair Pathway | Fusion hMLH1dn, OsMLH1dn | ePE3 | Base sub (1-bp), Del (1-bp), Ins (1-bp) | · No enhancement; Efficiency at NRT1.1-T locus reduced by 52% | Rice | [35] |
| DNA Repair Pathway | Fusion ZmMLH1dn | ePE3max | Base sub (3 bp) | · Homozygous editing efficiency 2.2%→12% | Maize | [30] |
| DNA Repair Pathway | OsMLH1-specific ihpRNA introduction | ePE3 | Base sub (1-bp), Del (1-bp), Ins (1-bp) | · Efficiency ×1.30–2.11 (avg ×1.51), no increased off-targets · Edited plant ratio 71.53%→87.15% | Rice | [35] |
| Chromatin Opening | hFTO introduction | enpPE2 | Base sub (1-bp), Del (2-bp), Ins (1-bp) | · Editing efficiency 33.49%→52.48% · Homozygous mutation frequency 13.71%→26.88% · Mild increase in off-target editing frequency | Rice | [36] |
| Temperature | 37 °C | 26 °C | Base sub (1-bp), Ins (3-bp) | · Editing efficiency 3.9%→6.3% | Rice | [9] |
| Temperature | 42 °C treatment for 2 h | 34 °C | Base sub (1/3-bp) | · Efficiency ×3.1–3.7 | Rice | [37] |
| Temperature | 34 °C | 25 °C | N/A | · Efficiency ×2.9–3.2 | Tomato | [33] |
| Temperature | RHTT cycle | 25 °C | Base sub (1/2-bp) | · 37 °C heat shock for 2 h + 25 °C recovery for 6 h, cycle repeated for 96 h · Precise editing efficiency max ×16.3 | Tobacco | [31] |
| Category | Specific Strategy | Baseline | Edit Type | Key Effect | Species | Reference |
|---|---|---|---|---|---|---|
| Screening System | Hygromycin selection system | PE-P1 | Base sub (1/3/4-bp) | · Combined with Zmubi1 promoter: Editing efficiency increased from 0–1.2%→2.6–26% | Rice | [10] |
| Screening System | Hygromycin selection system | pPE2 | Base sub (1-bp) | · Efficiency: 0%→16.7% | Rice | [14] |
| Screening System | Dual selection system | PE3 | N/A | · Combined bispyribac-sodium + hygromycin selection outperformed single systems · Efficiency: 0–1%→3.2–54.2% | Rice | [38] |
| Screening System | Anthocyanin screening system | PE-Nt3 | Base sub (3/4-bp) | · PAP1 gene enables visual purple phenotype, facilitating efficient screening · Efficiency: 1.1–7.5%→1.3–16.3% | Tobacco | [39] |
| Category | Specific System/Strategy | Key Capability and Effect | Species | Reference |
|---|---|---|---|---|
| Expand PAM Range | SpG | · Targets NG PAM · Efficiency up to 1.9% | Rice | [27] |
| Expand PAM Range | SpG | · Targets NG PAM (NGC/NGA/NGG) · Efficiency range 0.4–7.5% | Rice | [19] |
| Expand PAM Range | SpG or SpRY | · Targets NG PAM · dual-pegRNA efficiency: NGC + NGC > NGC + NGT > NGT + NGT > NGC + NGA > NGT + NGA > NGA + NGA | Rice | [40] |
| Expand PAM Range | SpRY | · High PAM flexibility (NRN/NYN) · High self-editing rate (33–64%) · Tagging efficiency only 2.38–6.25% | Rice | [41] |
| Expand PAM Range | ScCas9 | · Targets NNG PAM · No self-editing issue · Tagging efficiency 20–70.83% · Targets nearly 100% of rice genes | Rice | [41] |
| Enhance Usability | Active Cas9 | · Simultaneous precise editing + random mutation, generating transgene-free T0 plants · Reduced precise editing efficiency | Rice | [42] |
| Enhance Usability | · Optimized PE architecture (Csy4 system, RT variants) · Agrobacterium with extra Vir genes · Pyroxsulam selection | · Achieved transient co-editing, generating transgene-free T0 plants | Rice | [43] |
| Achieve Complex Edits | ePPE | · Extended PPE capability: Specific insertion lengths: 18 bp (3.1%), 24 bp (0.2%), 34 bp (0.3%) | Rice | [19] |
| Achieve Complex Edits | NM-PE | · 44 bp insertion efficiency 55.00–56.25% | Rice | [41] |
| Achieve Complex Edits | PE6d | · Significantly increased byproducts for point mutations and small edits · Tag insertion (27–135 bp), but knock-in capacity sharply decreases with tag size | Rice | [24] |
| Achieve Complex Edits | GRAND editing | · Replaced 57 bp, 90 bp, or 186 bp sequences with a 72 bp sequence at 8.33%–25% efficiency | Rice | [44] |
| Achieve Complex Edits | TJ-PE | · Inserted up to 1002 bp at 12.6% efficiency · Combining Csy4 system + re-added RNase H further improved efficiency | Rice | [45] |
| Achieve Complex Edits | PRIME-Del | · Enabled 50 bp–2000 bp deletions · Editing efficiency 37.5–84.2% · Homozygous editing efficiency 14.3–63% | Rice | [46] |
| Achieve Complex Edits | PrimeRoot | · Achieved 1.4 kb, 4.9 kb insertions in regenerated plants · Up to 11.1 kb insertion in protoplasts | Rice | [40] |
| Achieve Complex Edits | DualPE | · Generated specific deletions (~500 bp to 2 Mb) in protoplasts and plants; · Direct replacement of fragments up to 258 kb; · Precise inversion of a 205.4 kb fragment in plants | Wheat | [47] |
| Achieve Complex Edits | DualPE | · Large-fragment DNA editing efficiency up to 72.7% | Tobacco, Tomato | [47] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, S.; Yao, L.; Zhang, Y.; Rao, X.; Zhu, H. Prime Editing for Crop Improvement: A Systematic Review of Optimization Strategies and Advanced Applications. Genes 2025, 16, 965. https://doi.org/10.3390/genes16080965
Tian S, Yao L, Zhang Y, Rao X, Zhu H. Prime Editing for Crop Improvement: A Systematic Review of Optimization Strategies and Advanced Applications. Genes. 2025; 16(8):965. https://doi.org/10.3390/genes16080965
Chicago/Turabian StyleTian, Shuangrui, Lan Yao, Yuhong Zhang, Xiaoyu Rao, and Hongliang Zhu. 2025. "Prime Editing for Crop Improvement: A Systematic Review of Optimization Strategies and Advanced Applications" Genes 16, no. 8: 965. https://doi.org/10.3390/genes16080965
APA StyleTian, S., Yao, L., Zhang, Y., Rao, X., & Zhu, H. (2025). Prime Editing for Crop Improvement: A Systematic Review of Optimization Strategies and Advanced Applications. Genes, 16(8), 965. https://doi.org/10.3390/genes16080965







