Comparative Omics Analysis of Four Grape Varieties and Exploration of Their Anthocyanin Synthesis Mechanisms
Abstract
1. Introduction
2. Materials and Methods
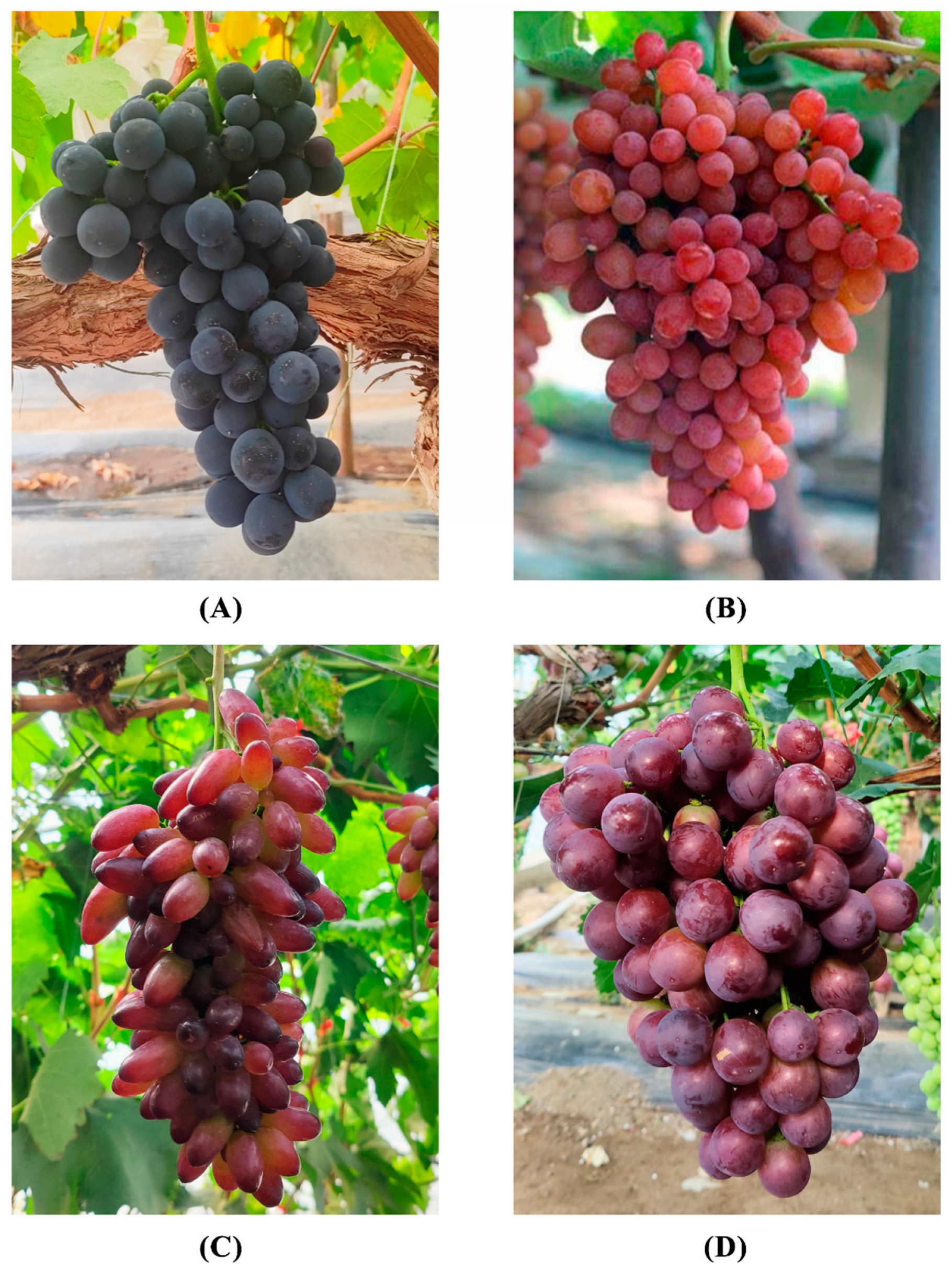
2.1. Sampling and Preparing
2.2. Metabolite Identification and Quantification
2.2.1. UPLC-MS/MS and GC-MS Conditions
2.2.2. Qualitative and Quantitative Analyses of Metabolites
2.2.3. Significantly Regulated Metabolites
2.3. RNA-Seq and Data Analysis
2.3.1. cDNA Library Construction and RNA Sequencing
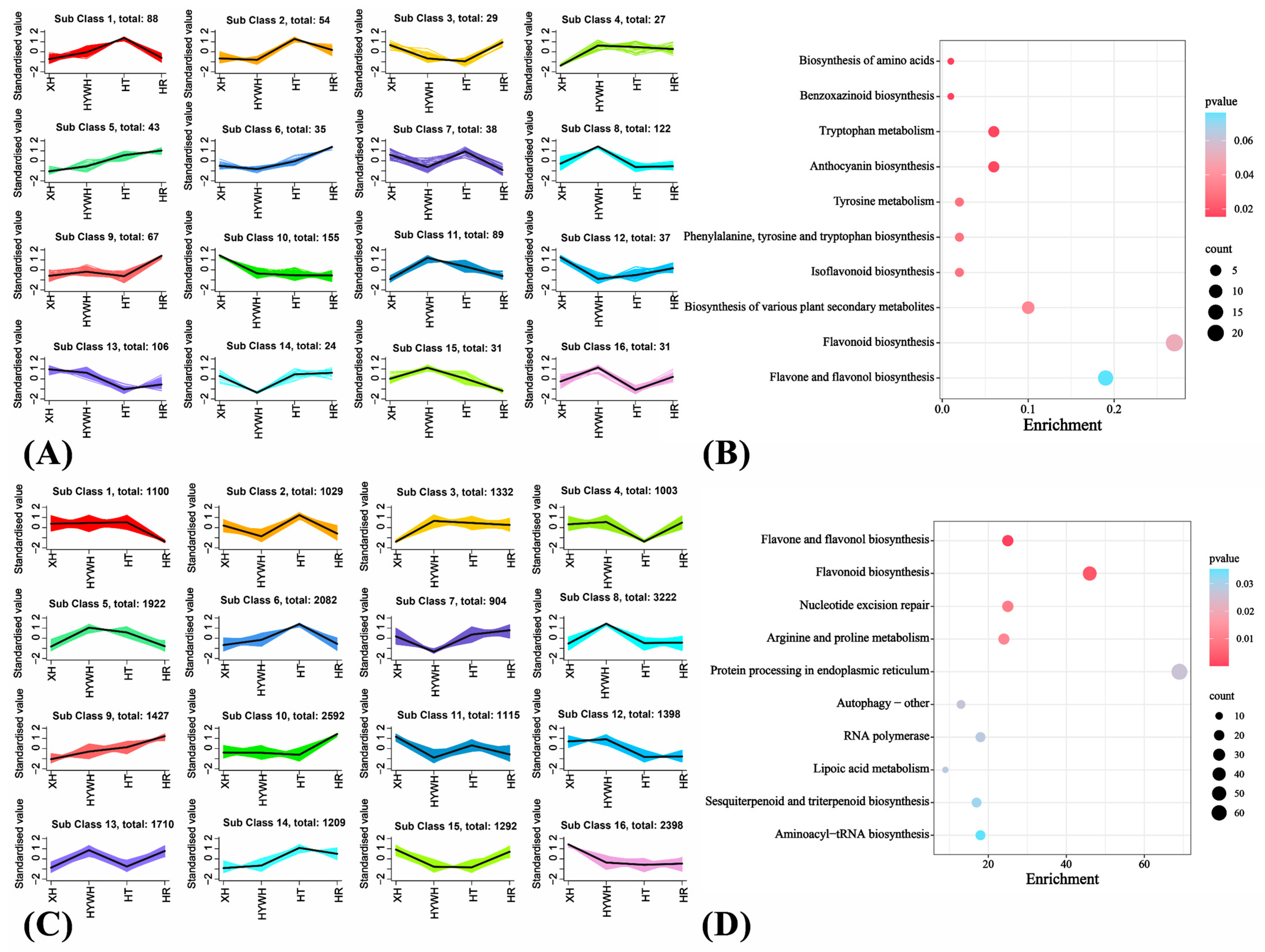
2.3.2. Transcript Assembly, Functional Annotation, and Differential Gene Expression
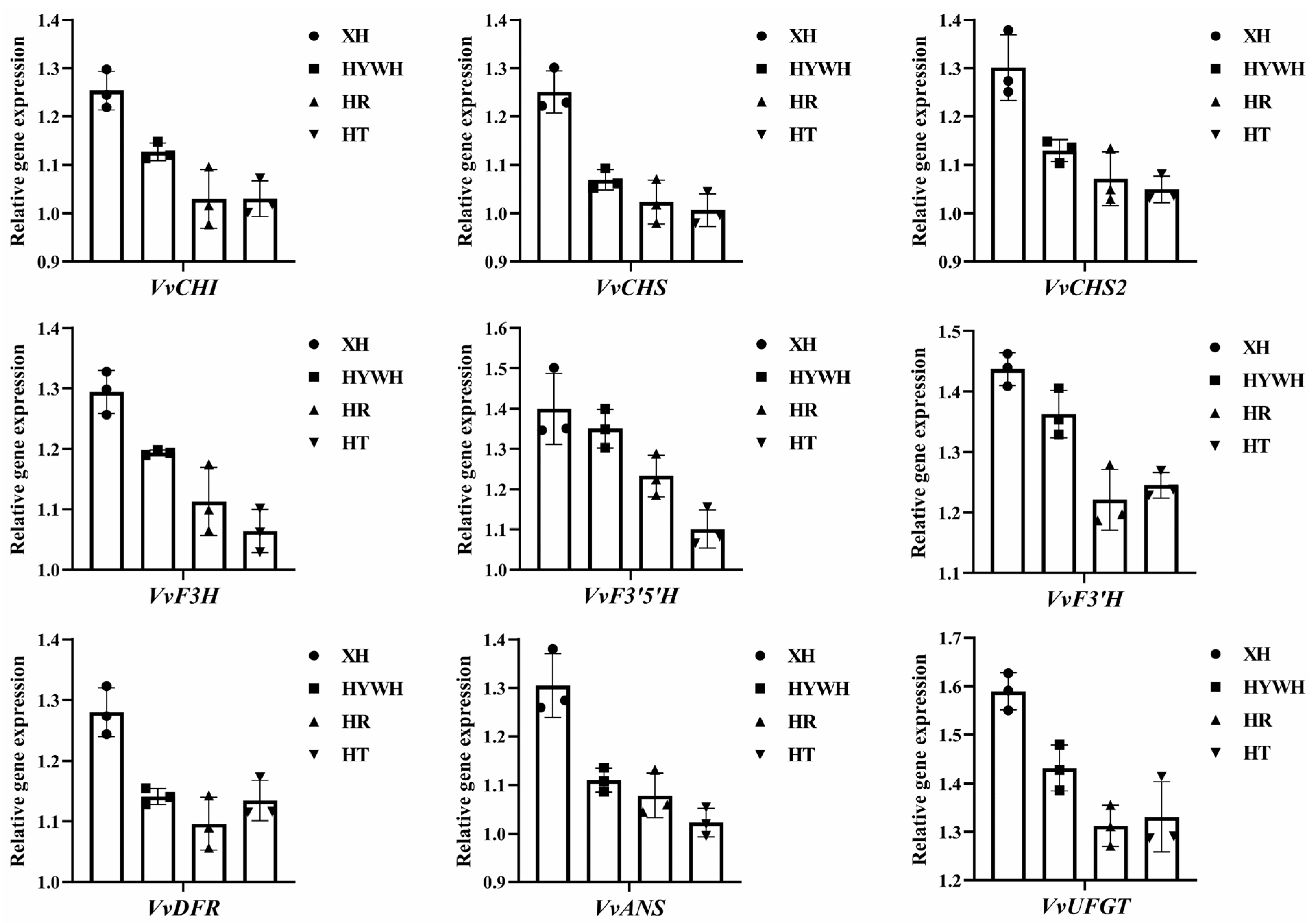
2.4. Verification of RNA Sequences via RT-qPCR
3. Results
3.1. Multivariate Analysis of the Metabolome
3.1.1. Widely Targeted Metabolome Profiles
3.1.2. Qualitative and Quantitative Analysis of Carbohydrates
3.2. Metabolome-Based Comparison of the Four Grape Varieties
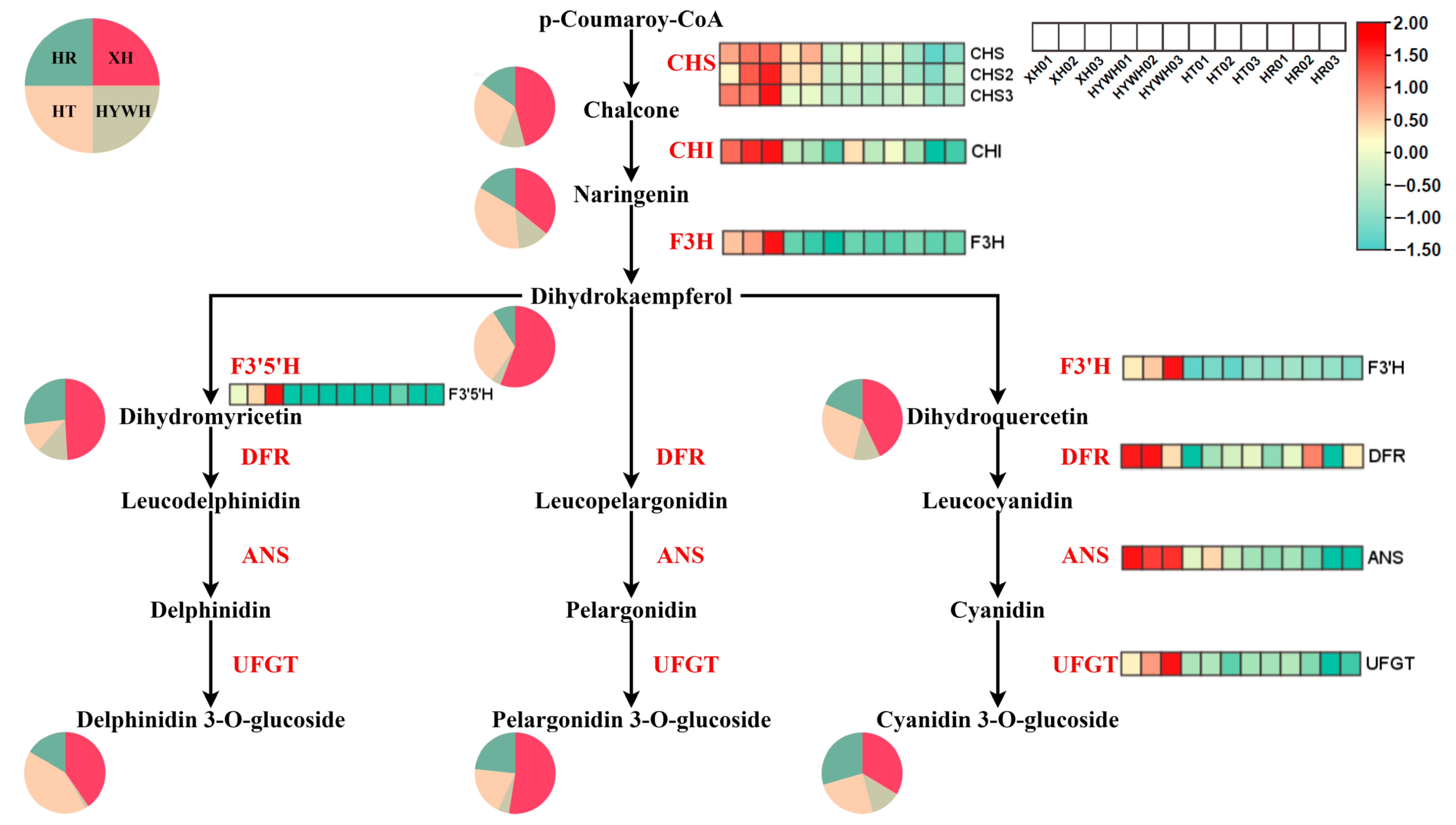
3.3. Beery Coloring and Anthocyanin Accumulation in the Grape Varieties
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviation
| LC-MS/MS | Liquid chromatography–tandem mass spectrometry |
References
- Zhou, D.-D.; Li, J.; Xiong, R.-G.; Saimaiti, A.; Huang, S.-Y.; Wu, S.-X.; Yang, Z.-J.; Shang, A.; Zhao, C.-N.; Gan, R.-Y. Bioactive compounds, health benefits and food applications of grape. Foods 2022, 11, 2755. [Google Scholar] [CrossRef]
- Restani, P.; Fradera, U.; Ruf, J.-C.; Stockley, C.; Teissedre, P.-L.; Biella, S.; Colombo, F.; Lorenzo, C.D. Grapes and their derivatives in modulation of cognitive decline: A critical review of epidemiological and randomized-controlled trials in humans. Crit. Rev. Food Sci. Nutr. 2021, 61, 566–576. [Google Scholar] [CrossRef]
- Yang, J.; Xiao, Y.-Y. Grape phytochemicals and associated health benefits. Crit. Rev. Food Sci. Nutr. 2013, 53, 1202–1225. [Google Scholar] [CrossRef]
- Xia, E.-Q.; Deng, G.-F.; Guo, Y.-J.; Li, H.-B. Biological activities of polyphenols from grapes. Int. J. Mol. Sci. 2010, 11, 622–646. [Google Scholar] [CrossRef]
- Yan, L.; Xin-Ming, L.; Ting, C.; Chao-Yue, G.; Wei-Xiong, H.; Sheng-Hua, C. Early-maturing Cultivation of Xiahei Grapes. Fujian J. Agric. Sci. 2014, 29, 966–969. [Google Scholar]
- Singh, B.; Singh, J.P.; Kaur, A.; Singh, N. Phenolic composition and antioxidant potential of grain legume seeds: A review. Food Res. Int. 2017, 101, 1–16. [Google Scholar] [CrossRef]
- Duenas, M.; Hernandez, T.; Estrella, I. Changes in the content of bioactive polyphenolic compounds of lentils by the action of exogenous enzymes. Effect on their antioxidant activity. Food Chem. 2007, 101, 90–97. [Google Scholar] [CrossRef]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.M.; Visser, R.G.F.; Bovy, A. Anthocyanin Biosynthesis and Degradation Mechanisms in Solanaceous Vegetables: A Review. Front. Chem. 2018, 6, 52. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Zhang, Z.; Zhang, L. Anthocyanin Accumulation, Antioxidant Ability and Stability, and a Transcriptional Analysis of Anthocyanin Biosynthesis in Purple Heading Chinese Cabbage (Brassica rapa L. ssp pekinensis). J. Agric. Food Chem. 2016, 64, 132–145. [Google Scholar] [CrossRef] [PubMed]
- Smeriglio, A.; Barreca, D.; Bellocco, E.; Trombetta, D. Chemistry, pharmacology and health benefits of anthocyanins. Phytother. Res. 2016, 30, 1265–1286. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, S.M.; Soares, M.S.P.; Gutierres, J.M.; Gerzson, M.F.B.; Carvalho, F.B.; Azambuja, J.H.; Schetinger, M.R.C.; Stefanello, F.M.; Spanevello, R.M. Anthocyanins as a potential pharmacological agent to manage memory deficit, oxidative stress and alterations in ion pump activity induced by experimental sporadic dementia of Alzheimer’s type. J. Nutr. Biochem. 2018, 56, 193–204. [Google Scholar] [CrossRef]
- Liobikas, J.; Skemiene, K.; Trumbeckaite, S.; Borutaite, V. Anthocyanins in cardioprotection: A path through mitochondria. Pharmacol. Res. 2016, 113, 808–815. [Google Scholar] [CrossRef]
- Veggi, P.C.; Santos, D.T.; Meireles, M.A.A. Anthocyanin extraction from Jabuticaba (Myrciaria cauliflora) skins by different techniques: Economic evaluation. Procedia Food Sci. 2011, 1, 1725–1731. [Google Scholar] [CrossRef]
- Zeng, T.; Sun, X.; Miao, Y.; Gu, S.; Tian, L.; Zheng, Y.; Jiang, Y.; Zhang, X.; Feng, Z.; Pei, J. Integrating bioclimatic factors and secondary metabolism to predict the suitable producing area of plants with high specific metabolite content in a real-world environment-a case of Carthamus tinctorius L. Ind. Crops Prod. 2022, 177, 114545. [Google Scholar] [CrossRef]
- Thevenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef]
- Shi, S.; Wang, W.; Liu, L.; Shu, B.; Wei, Y.; Jue, D.; Fu, J.; Xie, J.; Liu, C. Physico-chemical properties of longan fruit during development and ripening. Sci. Hortic. 2016, 207, 160–167. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36 (Suppl. 1), D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Botstein, D.; Cherry, J.M.; Ashburner, M.; Ball, C.A.; Blake, J.A.; Butler, H.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Profile hidden Markov models. Bioinformatics 1998, 14, 755–763. [Google Scholar] [CrossRef]
- Li, Y.-M.; Zhang, H.-X.; Tang, X.-S.; Wang, Y.; Cai, Z.-H.; Li, B.; Xie, Z.-S. Abscisic Acid Induces DNA Methylation Alteration in Genes Related to Berry Ripening and Stress Response in Grape (Vitis vinifera L). J. Agric. Food Chem. 2024, 72, 15027–15039. [Google Scholar] [CrossRef] [PubMed]
- Abdi, H.; Williams, L.J. Principal component analysis. Wiley Interdiscip. Rev. Comput. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Anguraj Vadivel, A.K.; Krysiak, K.; Tian, G.; Dhaubhadel, S. Genome-wide identification and localization of chalcone synthase family in soybean (Glycine max [L] Merr). BMC Plant Biol. 2018, 18, 325. [Google Scholar] [CrossRef]
- Zabala, G.; Vodkin, L.O. The wp mutation of Glycine max carries a gene-fragment-rich transposon of the CACTA superfamily. Plant Cell 2005, 17, 2619–2632. [Google Scholar] [CrossRef]
- Toda, K.; Kuroiwa, H.; Senthil, K.; Shimada, N.; Aoki, T.; Ayabe, S.-i.; Shimada, S.; Sakuta, M.; Miyazaki, Y.; Takahashi, R. The soybean F3′ H protein is localized to the tonoplast in the seed coat hilum. Planta 2012, 236, 79–89. [Google Scholar] [CrossRef]
- Takahashi, R.; Dubouzet, J.G.; Matsumura, H.; Yasuda, K.; Iwashina, T. A new allele of flower color gene W1 encoding flavonoid 3’5’-hydroxylase is responsible for light purple flowers in wild soybean Glycine soja. BMC Plant Biol. 2010, 10, 155. [Google Scholar] [CrossRef]
- Park, G.T.; Sundaramoorthy, J.; Lee, J.-D.; Kim, J.H.; Seo, H.S.; Song, J.T. Elucidation of molecular identity of the W3 locus and its implication in determination of flower colors in soybean. PLoS ONE 2015, 10, e0142643. [Google Scholar] [CrossRef][Green Version]
- Kovinich, N.; Saleem, A.; Arnason, J.T.; Miki, B. Functional characterization of a UDP-glucose: Flavonoid 3-O-glucosyltransferase from the seed coat of black soybean (Glycine max (L.) Merr.). Phytochemistry 2010, 71, 1253–1263. [Google Scholar] [CrossRef]
- Hellström, J.; Mattila, P.; Karjalainen, R. Stability of anthocyanins in berry juices stored at different temperatures. J. Food Compos. Anal. 2013, 31, 12–19. [Google Scholar] [CrossRef]
- He, F.; Liang, N.-N.; Mu, L.; Pan, Q.-H.; Wang, J.; Reeves, M.J.; Duan, C.-Q. Anthocyanins and their variation in red wines I. Monomeric anthocyanins and their color expression. Molecules 2012, 17, 1571–1601. [Google Scholar] [CrossRef] [PubMed]
- Naing, A.H.; Kim, C.K. Abiotic stress-induced anthocyanins in plants: Their role in tolerance to abiotic stresses. Physiol. Plant. 2021, 172, 1711–1723. [Google Scholar] [CrossRef]
- Gutterson, N. Anthocyanin biosynthetic genes and their application to flower color modification through sense suppression. HortScience 1995, 30, 964–966. [Google Scholar] [CrossRef]
- Wu, X.; Beecher, G.R.; Holden, J.M.; Haytowitz, D.B.; Gebhardt, S.E.; Prior, R.L. Concentrations of anthocyanins in common foods in the United States and estimation of normal consumption. J. Agric. Food Chem. 2006, 54, 4069–4075. [Google Scholar] [CrossRef]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef]
- Sun, Q.; Jiang, S.; Zhang, T.; Xu, H.; Fang, H.; Zhang, J.; Su, M.; Wang, Y.; Zhang, Z.; Wang, N. Apple NAC transcription factor MdNAC52 regulates biosynthesis of anthocyanin and proanthocyanidin through MdMYB9 and MdMYB11. Plant Sci. 2019, 289, 110286. [Google Scholar] [CrossRef]
- Park, N.I.; Xu, H.; Li, X.; Jang, I.H.; Park, S.; Ahn, G.H.; Lim, Y.P.; Kim, S.J.; Park, S.U. Anthocyanin accumulation and expression of anthocyanin biosynthetic genes in radish (Raphanus sativus). J. Agric. Food Chem. 2011, 59, 6034–6039. [Google Scholar] [CrossRef]
- Ollat, N.; Carde, J.-P.; Gaudillère, J.-P.; Barrieu, F.; Diakou-Verdin, P.; Moing, A. Grape berry development: A review. J. Int. Sci. Vigne Vin 2002, 36, 109–131. [Google Scholar]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007, 49, 414–427. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Tung, C.-W.; Eizenga, G.C.; Wright, M.H.; Ali, M.L.; Price, A.H.; Norton, G.J.; Islam, M.R.; Reynolds, A.; Mezey, J. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2011, 2, 467. [Google Scholar] [CrossRef] [PubMed]


| Time (min) | Solvent A (%) | Solvent B (%) | Description |
|---|---|---|---|
| 0 | 95 | 5 | Initial condition |
| 0.00–9.00 | From 95% to 5% | From 5% to 95% | Linear gradient change |
| 9.00–10.00 | 5 | 95 | Maintained at high organic phase |
| 10.00–11.10 | From 5% to 95% | From 95% to 5% | Linear return to initial composition |
| 11.10–14.00 | 95 | 5 | Column re-equilibration |
| Compound | Class | RT | Equation |
|---|---|---|---|
| 2-Deoxy-d-ribose (2-Deo-ribose) | monosaccharide | 3.504 | y = 0.338298 x + 7.618096 × 10−5 |
| d-Xylose (Xylose) | monosaccharide | 4.239 | y = 0.694375 x + 5.218014 × 10−5 |
| d-Arabinose (d-Ara) | monosaccharide | 4.311 | y = 0.901696 x + 1.438163 × 10−4 |
| d-Ribose (Ribose) | monosaccharide | 4.439 | y = 0.864945 x + 5.076758 × 10−4 |
| d-Xylulose (Xylulose) | monosaccharide | 4.44 | y = 1.107687 x + 0.003011 |
| d-Ribono-1,4-lactone (Ribono-1-4-lactone) | monosaccharide | 4.571 | y = 0.286528 x + 5.477765 × 10−5 |
| Xylitol | monosaccharide | 4.761 | y = 1.311257 x + 4.677106 × 10−4 |
| l-Levoglucosan (Lev) | monosaccharide | 4.871 | y = 0.222450 x − 3.409670 × 10−5 |
| l-Rhamnose (Rha) | monosaccharide | 4.921 | y = 0.590455 x + 2.054933 × 10−4 |
| Arabinitol | monosaccharide | 4.922 | y = 1.200010 x + 6.877416 × 10−4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, K.; Zhao, L.; Li, Y. Comparative Omics Analysis of Four Grape Varieties and Exploration of Their Anthocyanin Synthesis Mechanisms. Genes 2025, 16, 955. https://doi.org/10.3390/genes16080955
Zhang K, Zhao L, Li Y. Comparative Omics Analysis of Four Grape Varieties and Exploration of Their Anthocyanin Synthesis Mechanisms. Genes. 2025; 16(8):955. https://doi.org/10.3390/genes16080955
Chicago/Turabian StyleZhang, Kai, Liyang Zhao, and Yanfeng Li. 2025. "Comparative Omics Analysis of Four Grape Varieties and Exploration of Their Anthocyanin Synthesis Mechanisms" Genes 16, no. 8: 955. https://doi.org/10.3390/genes16080955
APA StyleZhang, K., Zhao, L., & Li, Y. (2025). Comparative Omics Analysis of Four Grape Varieties and Exploration of Their Anthocyanin Synthesis Mechanisms. Genes, 16(8), 955. https://doi.org/10.3390/genes16080955





