Long-Term Noninvasive Genetic Monitoring Guides Recovery of the Endangered Columbia Basin Pygmy Rabbits (Brachylagus idahoensis)
Abstract
1. Introduction
2. Materials and Methods
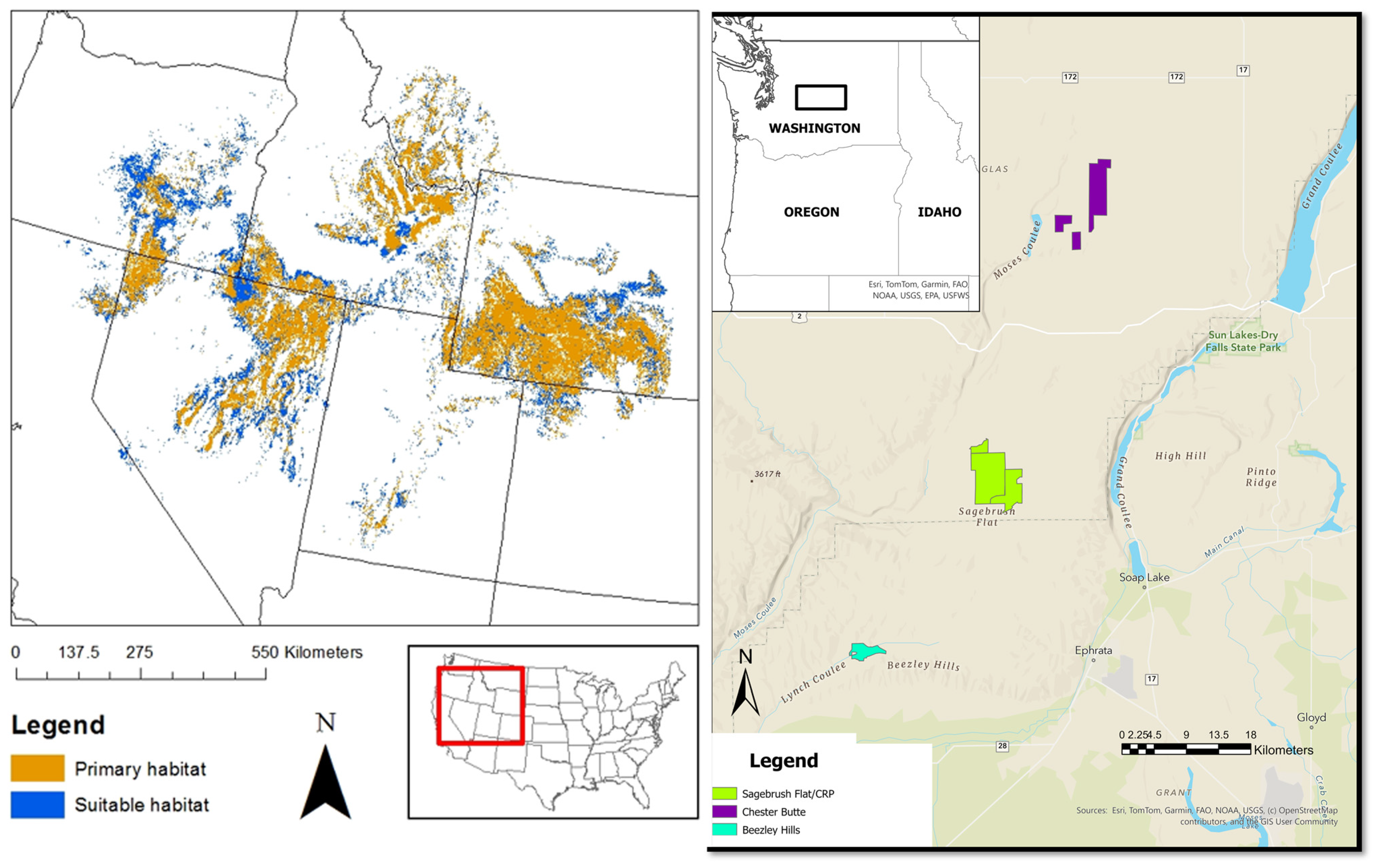
2.1. Study Area
2.2. Release Efforts
2.3. Winter Surveys
2.4. Summer Surveys
2.5. Laboratory Methods
2.6. Analytical Methods
2.7. Apparent Survival Models
3. Results
3.1. Genetic Monitoring at Sagebrush Flat
3.2. Genetic Diversity and CB Ancestry at Sagebrush Flat
3.3. Genetic Monitoring at Beezley Hills
3.4. Genetic Diversity and CB Ancestry at Beezley Hills
3.5. Genetic Monitoring at Chester Butte
3.6. Genetic Diversity and CB Ancestry at Chester Butte
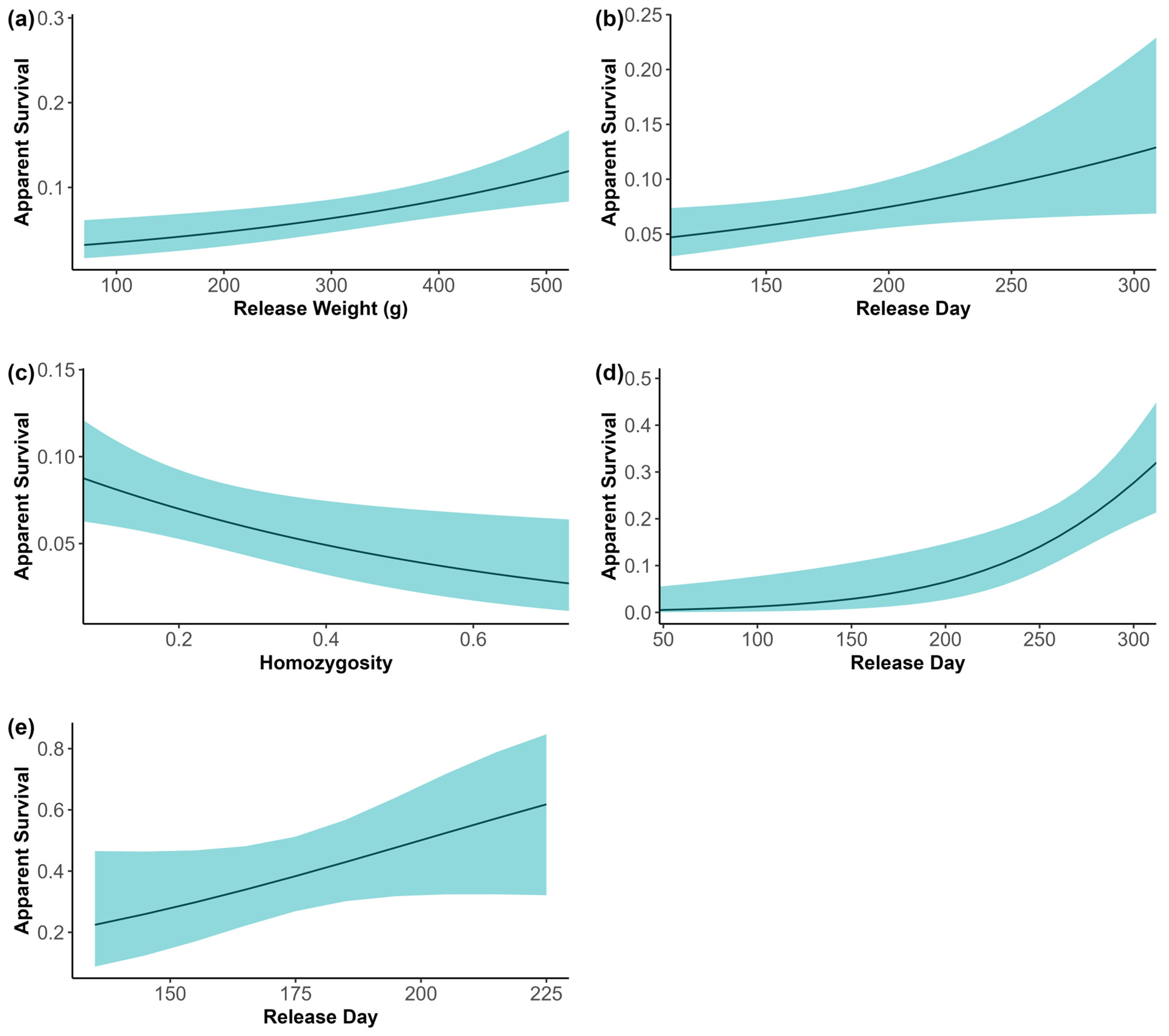
3.7. Apparent Survival
4. Discussion
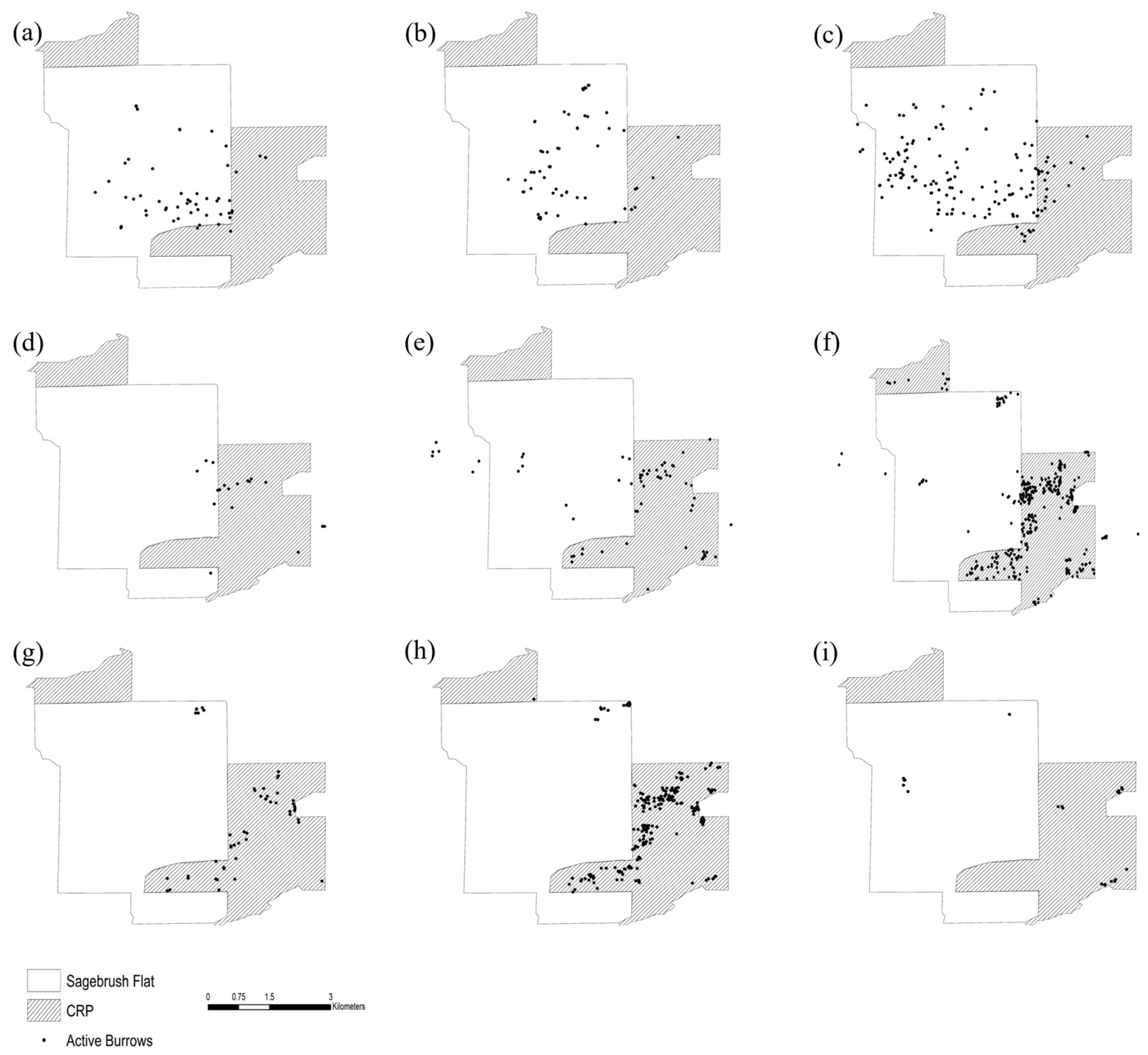
4.1. Habitat Use and Population Density
4.2. Survival and Inbreeding Depression
4.3. Prospects for Population Persistence
4.4. CB Ancestry and Genetic Diversity
4.5. Management and Conservation Implications
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pimm, S.L.; Jenkins, C.N.; Abell, R.; Brooks, T.M.; Gittleman, J.L.; Joppa, L.N.; Raven, P.H.; Roberts, C.M.; Sexton, J.O. The Biodiversity of Species and Their Rates of Extinction, Distribution, and Protection. Science 2014, 344, 1246752. [Google Scholar] [CrossRef]
- Hedrick, P.W.; Peterson, R.O.; Vucetich, L.M.; Adams, J.R.; Vucetich, J.A. Genetic Rescue in Isle Royale Wolves: Genetic Analysis and the Collapse of the Population. Conserv. Genet. 2014, 15, 1111–1121. [Google Scholar] [CrossRef]
- Schwartz, M.K.; Luikart, G.; Waples, R.S. Genetic Monitoring as a Promising Tool for Conservation and Management. Trends Ecol. Evol. 2007, 22, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Stetz, J.B.; Kendall, K.C.; Vojta, C.D.; Genetic Monitoring (GeM) Working Group. Genetic Monitoring for Managers: A New Online Resource. J. Fish Wildl. Manag. 2011, 2, 216–219. [Google Scholar] [CrossRef][Green Version]
- Hoban, S.; Bruford, M.W.; Funk, W.C.; Galbusera, P.; Griffith, M.P.; Grueber, C.E.; Heuertz, M.; Hunter, M.E.; Hvilsom, C.; Stroil, B.K.; et al. Global Commitments to Conserving and Monitoring Genetic Diversity Are Now Necessary and Feasible. BioScience 2021, 71, 964–976. [Google Scholar] [CrossRef]
- Hunter, M.E.; Hoban, S.M.; Bruford, M.W.; Segelbacher, G.; Bernatchez, L. Next-Generation Conservation Genetics and Biodiversity Monitoring. Evol. Appl. 2018, 11, 1029–1034. [Google Scholar] [CrossRef]
- Blouin, M.S.; Parsons, M.; Lacaille, V.; Lotz, S. Use of Microsatellite Loci to Classify Individuals by Relatedness. Mol. Ecol. 1996, 5, 393–401. [Google Scholar] [CrossRef]
- Koskinen, M.T. Individual Assignment Using Microsatellite DNA Reveals Unambiguous Breed Identification in the Domestic Dog. Anim. Genet. 2003, 34, 297–301. [Google Scholar] [CrossRef]
- Maes, G.; Pujolar, J.M.; Raeymaekers, J.; Dannewitz, J.; Volckaert, F. Microsatellite Conservation and Bayesian Individual Assignment in Four Anguilla Species. Mar. Ecol. Prog. Ser. 2006, 319, 251–261. [Google Scholar] [CrossRef][Green Version]
- Arandjelovic, M.; Head, J.; Kühl, H.; Boesch, C.; Robbins, M.M.; Maisels, F.; Vigilant, L. Effective Non-Invasive Genetic Monitoring of Multiple Wild Western Gorilla Groups. Biol. Conserv. 2010, 143, 1780–1791. [Google Scholar] [CrossRef]
- Cooper, A.M.; Miller, L.M.; Kapuscinski, A.R. Conservation of Population Structure and Genetic Diversity under Captive Breeding of Remnant Coaster Brook Trout (Salvelinus fontinalis) Populations. Conserv. Genet. 2010, 11, 1087–1093. [Google Scholar] [CrossRef]
- Khrustaleva, A.M.; Klovach, N.V.; Seeb, J.E. Genetic Variability and Population Structure of Sockeye Salmon from the Asian Coast of Pacific Ocean. Russ. J. Genet. 2017, 53, 1126–1136. [Google Scholar] [CrossRef]
- Cardoso, M.J.; Mooney, N.; Eldridge, M.D.B.; Firestone, K.B.; Sherwin, W.B. Genetic Monitoring Reveals Significant Population Structure in Eastern Quolls: Implications for the Conservation of a Threatened Carnivorous Marsupial. Aust. Mammal. 2014, 36, 169–177. [Google Scholar] [CrossRef]
- Younger, J.L.; Clucas, G.V.; Kao, D.; Rogers, A.D.; Gharbi, K.; Hart, T.; Miller, K.J. The Challenges of Detecting Subtle Population Structure and Its Importance for the Conservation of Emperor Penguins. Mol. Ecol. 2017, 26, 3883–3897. [Google Scholar] [CrossRef] [PubMed]
- Rudnick, J.A.; Katzner, T.E.; Bragin, E.A.; Rhodes, O.E., Jr.; Dewoody, J.A. Using Naturally Shed Feathers for Individual Identification, Genetic Parentage Analyses, and Population Monitoring in an Endangered Eastern Imperial Eagle (Aquila heliaca) Population from Kazakhstan. Mol. Ecol. 2005, 14, 2959–2967. [Google Scholar] [CrossRef]
- Roques, S.; Chancerel, E.; Boury, C.; Pierre, M.; Acolas, M.-L. From Microsatellites to Single Nucleotide Polymorphisms for the Genetic Monitoring of a Critically Endangered Sturgeon. Ecol. Evol. 2019, 9, 7017–7029. [Google Scholar] [CrossRef]
- Weeks, A.R.; Stoklosa, J.; Hoffmann, A.A. Conservation of Genetic Uniqueness of Populations May Increase Extinction Likelihood of Endangered Species: The Case of Australian Mammals. Front Zool 2016, 13, 31. [Google Scholar] [CrossRef]
- Albert, E.M.; García-Navas, V. Population Structure and Genetic Diversity of the Threatened Pygmy Newt Triturus Pygmaeus in a Network of Natural and Artificial Ponds. Conserv. Genet. 2022, 23, 575–588. [Google Scholar] [CrossRef]
- Kotzé, A.; Smith, R.M.; Moodley, Y.; Luikart, G.; Birss, C.; Wyk, A.M.V.; Grobler, J.P.; Dalton, D.L. Lessons for Conservation Management: Monitoring Temporal Changes in Genetic Diversity of Cape Mountain Zebra (Equus zebra zebra). PLoS ONE 2019, 14, e0220331. [Google Scholar] [CrossRef]
- Ramey II, R.R.; Luikart, G.; Singer, F.J. Genetic Bottlenecks Resulting from Restoration Efforts: The Case of Bighorn Sheep in Badlands National Park. Restor. Ecol. 2000, 8, 85–90. [Google Scholar] [CrossRef]
- Blanco, G.; Morinha, F. Genetic Signatures of Population Bottlenecks, Relatedness, and Inbreeding Highlight Recent and Novel Conservation Concerns in the Egyptian Vulture. PeerJ 2021, 9, e11139. [Google Scholar] [CrossRef] [PubMed]
- Corti, P.; Shafer, A.B.A.; Coltman, D.W.; Festa-Bianchet, M. Past Bottlenecks and Current Population Fragmentation of Endangered Huemul Deer (Hippocamelus bisulcus): Implications for Preservation of Genetic Diversity. Conserv. Genet. 2011, 12, 119–128. [Google Scholar] [CrossRef]
- Taberlet, P.; Waits, L.P.; Luikart, G. Noninvasive Genetic Sampling: Look before You Leap. Trends Ecol. Evol. 1999, 14, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Beja-Pereira, A.; Oliveira, R.; Alves, P.C.; Schwartz, M.K.; Luikart, G. Advancing Ecological Understandings through Technological Transformations in Noninvasive Genetics. Mol. Ecol. Resour. 2009, 9, 1279–1301. [Google Scholar] [CrossRef]
- Dufresnes, C.; Remollino, N.; Stoffel, C.; Manz, R.; Weber, J.-M.; Fumagalli, L. Two Decades of Non-Invasive Genetic Monitoring of the Grey Wolves Recolonizing the Alps Support Very Limited Dog Introgression. Sci. Rep. 2019, 9, 148. [Google Scholar] [CrossRef]
- Schultz, A.J.; Strickland, K.; Cristescu, R.H.; Hanger, J.; de Villiers, D.; Frère, C.H. Testing the Effectiveness of Genetic Monitoring Using Genetic Non-Invasive Sampling. Ecol. Evol. 2022, 12, e8459. [Google Scholar] [CrossRef]
- Tallmon, D.A.; Luikart, G.; Waples, R.S. The Alluring Simplicity and Complex Reality of Genetic Rescue. Trends Ecol. Evol. 2004, 19, 489–496. [Google Scholar] [CrossRef]
- Mills, L.S.; Allendorf, F.W. The One-Migrant-per-Generation Rule in Conservation and Management. Conserv. Biol. 1996, 10, 1509–1518. [Google Scholar] [CrossRef]
- Edmands, S. Between a Rock and a Hard Place: Evaluating the Relative Risks of Inbreeding and Outbreeding for Conservation and Management. Mol. Ecol. 2007, 16, 463–475. [Google Scholar] [CrossRef]
- Frankham, R.; Ballou, J.D.; Eldridge, M.D.B.; Lacy, R.C.; Ralls, K.; Dudash, M.R.; Fenster, C.B. Predicting the Probability of Outbreeding Depression. Conserv. Biol. 2011, 25, 465–475. [Google Scholar] [CrossRef]
- Whiteley, A.R.; Fitzpatrick, S.W.; Funk, W.C.; Tallmon, D.A. Genetic Rescue to the Rescue. Trends Ecol. Evol. 2015, 30, 42–49. [Google Scholar] [CrossRef]
- Fredrickson, R.J.; Siminski, P.; Woolf, M.; Hedrick, P.W. Genetic Rescue and Inbreeding Depression in Mexican Wolves. Proc. R. Soc. B Biol. Sci. 2007, 274, 2365–2371. [Google Scholar] [CrossRef] [PubMed]
- Pimm, S.L.; Dollar, L.; Bass, O.L. The Genetic Rescue of the Florida Panther. Anim. Conserv. 2006, 9, 115–122. [Google Scholar] [CrossRef]
- Mussmann, S.M.; Douglas, M.R.; Anthonysamy, W.J.B.; Davis, M.A.; Simpson, S.A.; Louis, W.; Douglas, M.E. Genetic Rescue, the Greater Prairie Chicken and the Problem of Conservation Reliance in the Anthropocene. R. Soc. Open Sci. 2017, 4, 160736. [Google Scholar] [CrossRef]
- Hasselgren, M.; Angerbjörn, A.; Eide, N.E.; Erlandsson, R.; Flagstad, Ø.; Landa, A.; Wallén, J.; Norén, K. Genetic Rescue in an Inbred Arctic Fox (Vulpes lagopus) Population. Proc. R. Soc. B Biol. Sci. 2018, 285, 20172814. [Google Scholar] [CrossRef]
- Miller, J.M.; Poissant, J.; Hogg, J.T.; Coltman, D.W. Genomic Consequences of Genetic Rescue in an Insular Population of Bighorn Sheep (Ovis canadensis). Mol. Ecol. 2012, 21, 1583–1596. [Google Scholar] [CrossRef]
- Robinson, Z.L.; Bell, D.A.; Dhendup, T.; Luikart, G.; Whiteley, A.R.; Kardos, M. Evaluating the Outcomes of Genetic Rescue Attempts. Conserv. Biol. 2020, 35, 666–677. [Google Scholar] [CrossRef]
- Lyman, R.L. Late Quaternary Biogeography of the Pygmy Rabbit (Brachylagus idahoensis) in Eastern Washington. J. Mammal. 1991, 72, 110–117. [Google Scholar] [CrossRef]
- Warheit, K. Genetic Diversity and Population Differentiation of Pygmy Rabbits (Brachylagus idahoensis); Washington Department of Fish and Wildlife: Olympia, WA, USA, 2001. [Google Scholar]
- WDFW. Washington State Recovery Plan for the Pygmy Rabbit; Wildlife Management Program; Washington Department of Fish and Wildlife: Olympia, WA, USA, 1995; p. 84. [Google Scholar]
- WDFW. Washington Pygmy Rabbit 2003 Recovery Plan Update; Washington Department of Fish and Wildlife: Olympia, WA, USA, 2003. [Google Scholar]
- U.S. Fish and Wildlife Service, Department of the Interior: Fish and Wildlife Service. Rules and Regulations: Endangered and Threatened Wildlife and Plants; Final Rule to List the Columbia Basin Distinct Population Segment of the Pygmy Rabbit (Brachylagus idahoensis) Final Rule. Fed. Regist. 2003, 75, 10388–10409. [Google Scholar]
- Becker, P.A.; Hays, D.W.; Sayler, R.D. Columbia Basin Pygmy Rabbit Reintroduction and Genetic Management Plan; Washington Department of Fish and Wildlife: Olympia, WA, USA, 2011; p. 31. [Google Scholar]
- Elias, B.A.; Shipley, L.A.; McCusker, S.; Sayler, R.D.; Johnson, T.R. Effects of Genetic Management on Reproduction, Growth, and Survival in Captive Endangered Pygmy Rabbits (Brachylagus idahoensis). J. Mammal. 2013, 94, 1282–1292. [Google Scholar] [CrossRef]
- U.S. Fish and Wildlife Service. Recovery Plan for the Columbia Basin Distinct Population Segment of the Pygmy Rabbit (Brachylagus idahoensis); U.S. Fish and Wildlife Service: Portland, OR, USA, 2012; p. ix+109. [Google Scholar]
- Hayes, G.E. Periodic Status Review for the Pygmy Rabbit in Washington; Washington Department of Fish and Wildlife: Olympia, WA, USA, 2018; pp. 1–26. [Google Scholar]
- DeMay, S.M.; Becker, P.A.; Eidson, C.A.; Rachlow, J.L.; Johnson, T.R.; Waits, L.P. Evaluating DNA Degradation Rates in Faecal Pellets of the Endangered Pygmy Rabbit. Mol. Ecol. Resour. 2013, 13, 654–662. [Google Scholar] [CrossRef] [PubMed]
- DeMay, S.M.; Becker, P.A.; Rachlow, J.L.; Waits, L.P. Genetic Monitoring of an Endangered Species Recovery: Demographic and Genetic Trends for Reintroduced Pygmy Rabbits (Brachylagus idahoensis). J. Mammal. 2017, 98, 350–364. [Google Scholar] [CrossRef]
- DeMay, S.M.; Becker, P.A.; Waits, L.P.; Johnson, T.R.; Rachlow, J.L. Consequences for Conservation: Population Density and Genetic Effects on Reproduction of an Endangered Lagomorph. Ecol. Appl. 2016, 26, 784–795. [Google Scholar] [CrossRef] [PubMed]
- Scott, P.A.; Allison, L.J.; Field, K.J.; Averill-Murray, R.C.; Shaffer, H.B. Individual Heterozygosity Predicts Translocation Success in Threatened Desert Tortoises. Science 2020, 370, 1086–1089. [Google Scholar] [CrossRef]
- Rödel, H.G.; Bora, A.; Kaetzke, P.; Khaschei, M.; Hutzelmeyer, H.; von Holst, D. Over-Winter Survival in Subadult European Rabbits: Weather Effects, Density Dependence, and the Impact of Individual Characteristics. Oecologia 2004, 140, 566–576. [Google Scholar] [CrossRef]
- WDFW. Sagebrush Flat Wildlife Area Management Plan; Wildlife Management Program; Washington Department of Fish and Wildlife: Olympia, WA, USA, 2006; p. 66. [Google Scholar]
- Tullis, J.A. Characteristics and Origin of Earth-Mounds on the Eastern Snake River Plain, Idaho; Idaho State University: Pocatello, ID, USA, 1995. [Google Scholar]
- Western Regional Climate Center Western Regional Climate Center. Available online: https://wrcc.dri.edu (accessed on 1 November 2020).
- DeMay, S.M.; Rachlow, J.L.; Waits, L.P.; Becker, P.A. Comparing Telemetry and Fecal Dna Sampling Methods to Quantify Survival and Dispersal of Juvenile Pygmy Rabbits. Wildl. Soc. Bull. 2015, 39, 413–421. [Google Scholar] [CrossRef]
- Gallie, J.A.; Zinke, B.M. 2017 Annual Report for the Columbia Basin Pygmy Rabbit Recovery Project; Washington Department of Fish and Wildlife: Olympia, WA, USA, 2018; p. 38. [Google Scholar]
- Sikes, R.S. 2016 Guidelines of the American Society of Mammalogists for the Use of Wild Mammals in Research and Education. J. Mammal. 2016, 97, 663–688. [Google Scholar] [CrossRef]
- Adams, J.R.; Goldberg, C.S.; Bosworth, W.R.; Rachlow, J.L.; Waits, L.P. Rapid Species Identification of Pygmy Rabbits (Brachylagus idahoensis) from Faecal Pellet DNA. Mol. Ecol. Resour. 2011, 11, 808–812. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research—An Update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Marshall, T.C.; Slate, J.; Kruuk, L.E.B.; Pemberton, J.M. Statistical Confidence for Likelihood-Based Paternity Inference in Natural Populations. Mol. Ecol. 1998, 7, 639–655. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising How the Computer Program Cervus Accommodates Genotyping Error Increases Success in Paternity Assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Nerkowski, S.; Waits, L.; Warheit, K.I.; Hohenlohe, P. Range-Wide Genomic Analysis of Pygmy Rabbits (Brachylagus idahoensis) Reveals Genetic Distinctiveness of the Endangered Columbia Basin Population. Authorea 2024. [Google Scholar] [CrossRef]
- Goudet, J. Hierfstat, a Package for r to Compute and Test Hierarchical F-Statistics. Mol. Ecol. Notes 2005, 5, 184–186. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Do, C.; Waples, R.S.; Peel, D.; Macbeth, G.M.; Tillett, B.J.; Ovenden, J.R. NeEstimator v2: Re-Implementation of Software for the Estimation of Contemporary Effective Population Size (Ne) from Genetic Data. Mol. Mol. Mol. Ecol. Resour. 2014, 14, 209–214. [Google Scholar] [CrossRef]
- Nomura, T. Estimation of Effective Number of Breeders from Molecular Coancestry of Single Cohort Sample. Evol. Appl. 2008, 1, 462–474. [Google Scholar] [CrossRef]
- Arnold, T.W. Uninformative Parameters and Model Selection Using Akaike’s Information Criterion. J. Wildl. Manag. 2010, 74, 1175–1178. [Google Scholar] [CrossRef]
- Mazerolle, M.J. Model Selection and Multimodel Inference Using the AICcmodavg Package. 2020. Available online: https://cran.r-project.org/web/packages/AICcmodavg/vignettes/AICcmodavg.pdf (accessed on 1 July 2025).
- Coulon, A. Genhet: An Easy-to-Use R Function to Estimate Individual Heterozygosity. Mol. Ecol. Resour. 2010, 10, 167–169. [Google Scholar] [CrossRef]
- Gallie, J.A. Columbia Basin Pygmy Rabbit Project—2016 Annual Report; Washington Department of Fish and Wildlife: Olympia, WA, USA, 2016. [Google Scholar]
- Anderson, D.R. Model Based Inference in the Life Sciences: A Primer on Evidence; Anderson, D.R., Ed.; Springer: New York, NY, USA, 2008; pp. 51–82. ISBN 978-0-387-74075-1. [Google Scholar]
- Pierce, J.E.; Larsen, R.T.; Flinders, J.T.; Whiting, J.C. Fragmentation of Sagebrush Communities: Does an Increase in Habitat Edge Impact Pygmy Rabbits? Anim. Conserv. 2011, 14, 314–321. [Google Scholar] [CrossRef]
- Gallie, J.A.; Zinke, B.M. Annual Report for the Columbia Basin Pygmy Rabbit Recovery Project: 2018; Washington Department of Fish and Wildlife: Olympia, WA, USA, 2019; p. 38. [Google Scholar]
- Schroeder, M.A.; Vander Haegen, M.W. Response of Greater Sage-Grouse to the Conservation Reserve Program in Washington State. In Greater Sage-Grouse Ecology and Conservation of a Landscape Species and Its Habitats; University of California Press: Oakland, CA, USA, 2011; ISBN 978-0-520-26711-4. [Google Scholar]
- Schroeder, M.; Vander Haegen, M. Use of CRP Fields by Greater Sage-Grouse and Other Shrubsteppe Associated Wildlife in Washington; Technical Report Prepared for US Department of Agriculture Farm Service Agency; Washington Department of Fish and Wildlife: Olympia, WA, USA, 2006. [Google Scholar]
- Vander Haegen, W.M.; Dobler, F.C.; Pierce, D.J. Shrubsteppe Bird Response to Habitat and Landscape Variables in Eastern Washington, U.S.A. Conserv. Biol. 2000, 14, 1145–1160. [Google Scholar] [CrossRef]
- Lupis, S.G.; Messmer, T.A.; Black, T. Gunnison Sage-Grouse Use of Conservation Reserve Program Fields in Utah and Response to Emergency Grazing: A Preliminary Evaluation. Wildl. Soc. Bull. 2006, 34, 957–962. [Google Scholar] [CrossRef]
- Sanchez, D.M.; Rachlow, J.L. Spatio-Temporal Factors Shaping Diurnal Space Use by Pygmy Rabbits. J. Wildl. Manag. 2008, 72, 1304–1310. [Google Scholar] [CrossRef]
- Wilde, D.B. A Population Analysis of the Pygmy Rabbit (Brachylagus idahoensis) on the Idaho National Engineering Laboratory. Ph.D. Thesis, Idaho State University, Pocatello, ID, USA, 1978. [Google Scholar]
- Sanchez, D.M. Survivorship of Pygmy Rabbits in East Central Idaho. Ph.D. Thesis, University of Idaho, Moscow, ID, USA, 2007. [Google Scholar]
- Price, A.J.; Rachlow, J.L. Development of an Index of Abundance for Pygmy Rabbit Populations. J. Wildl. Manag. 2011, 75, 929–937. [Google Scholar] [CrossRef]
- Crawford, J.A.; Anthony, R.G.; Forbes, J.T.; Lorton, G.A. Survival and Causes of Mortality for Pygmy Rabbits (Brachylagus idahoensis) in Oregon and Nevada. J. Mammal. 2010, 91, 838–847. [Google Scholar] [CrossRef]
- Estes-Zumpf, W.A.; Rachlow, J.L. Natal Dispersal by Pygmy Rabbits (Brachylagus idahoensis). J. Mammal. 2009, 90, 363–372. [Google Scholar] [CrossRef]
- Korpimaki, E.; Norrdahl, K. Numerical and Functional Responses of Kestrels, Short-Eared Owls, and Long-Eared Owls to Vole Densities. Ecology 1991, 72, 814–826. [Google Scholar] [CrossRef]
- Sinclair, A.R.E.; Pech, R.P.; Dickman, C.R.; Hik, D.; Mahon, P.; Newsome, A.E. Predicting Effects of Predation on Conservation of Endangered Prey. Conserv. Biol. 1998, 12, 564–575. [Google Scholar] [CrossRef]
- Gilg, O.; Sittler, B.; Sabard, B.; Hurstel, A.; Sané, R.; Delattre, P.; Hanski, I. Functional and Numerical Responses of Four Lemming Predators in High Arctic Greenland. Oikos 2006, 113, 193–216. [Google Scholar] [CrossRef]
- Goodrich, L.J.; Smith, J.P. Raptor Migration in North America. In State of North America’s Birds of Prey (Studies in Ornithology); Amer Ornithologists Union: Cambridge, MA, USA; Washington, DC, USA, 2008; p. 113. [Google Scholar]
- O’Donoghue, M.; Boutin, S.; Krebs, C.J.; Hofer, E.J. Numerical Responses of Coyotes and Lynx to the Snowshoe Hare Cycle. Oikos 1997, 80, 150–162. [Google Scholar] [CrossRef]
- Gillis, E.A. Survival of Juvenile Hares during a Cyclic Population Increase. Can. J. Zool. 1998. [Google Scholar] [CrossRef]
- Bond, B.T.; Burger, L.W., Jr.; Leopold, B.D.; Godwin, K.D. Survival of Cottontail Rabbits (Sylvilagus floridanus) in Mississippi and an Examination of Latitudinal Variation. Am. Midl. Nat. 2001, 145, 127–136. [Google Scholar] [CrossRef]
- Willi, Y.; Van Buskirk, J.; Hoffmann, A.A. Limits to the Adaptive Potential of Small Populations. Annu. Rev. Ecol. Evol. Syst. 2006, 37, 433–458. [Google Scholar] [CrossRef]
- O’Grady, J.J.; Reed, D.H.; Brook, B.W.; Frankham, R. What Are the Best Correlates of Predicted Extinction Risk? Biol. Conserv. 2004, 118, 513–520. [Google Scholar] [CrossRef]
- Smith, I.T.; Rachlow, J.L.; Svancara, L.K.; McMahon, L.A.; Knetter, S.J. Habitat Specialists as Conservation Umbrellas: Do Areas Managed for Greater Sage-Grouse Also Protect Pygmy Rabbits? Ecosphere 2019, 10, e02827. [Google Scholar] [CrossRef]
- Nerkowski, S.A. A Rabbit’s Tale: Genetic Monitoring, Genomic Diversity, and Habitat Selection in the Endangered Columbia Basin Pygmy Rabbit (Brachylagus idahoensis). Ph.D. Thesis, University of Idaho, Moscow, ID, USA, 2021. [Google Scholar]
- InciWeb Final Update, 9-15-20, Pearl Hill/Apple Acres Fires—InciWeb the Incident Information System. Available online: https://inciweb.nwcg.gov/incident/article/7169/55999/ (accessed on 17 March 2021).
- Lynch, M. The Genetic Interpretation of Inbreeding Depression and Outbreeding Depression. Evolution 1991, 45, 622–629. [Google Scholar] [CrossRef] [PubMed]
- Hedrick, P.W.; Adams, J.R.; Vucetich, J.A. Reevaluating and Broadening the Definition of Genetic Rescue. Conserv. Biol. 2011, 25, 1069–1070. [Google Scholar] [CrossRef] [PubMed]
- Leberg, P.L. Strategies for Population Reintroduction: Effects of Genetic Variability on Population Growth and Size. Conserv. Biol. 1993, 7, 194–199. [Google Scholar] [CrossRef]
- Earnhardt, J.M. Reintroduction Programmes: Genetic Trade-Offs for Populations. Anim. Conserv. 1999, 2, 279–286. [Google Scholar] [CrossRef]
- Miller, K.A.; Nelson, N.J.; Smith, H.G.; Moore, J.A. How Do Reproductive Skew and Founder Group Size Affect Genetic Diversity in Reintroduced Populations? Mol. Ecol. 2009, 18, 3792–3802. [Google Scholar] [CrossRef]
- Estes-Zumpf, W.A.; Rachlow, J.L.; Waits, L.P.; Warheit, K.I. Dispersal, Gene Flow, and Population Genetic Structure in the Pygmy Rabbit (Brachylagus idahoensis). J. Mammal. 2010, 91, 208–219. [Google Scholar] [CrossRef]
- Thimmayya, A.C.; Buskirk, S.W. Genetic Connectivity and Diversity of Pygmy Rabbits (Brachylagus idahoensis) in Southern Wyoming. J. Mammal. 2012, 93, 29–37. [Google Scholar] [CrossRef]
- Frankham, R.; Bradshaw, C.J.A.; Brook, B.W. Genetics in Conservation Management: Revised Recommendations for the 50/500 Rules, Red List Criteria and Population Viability Analyses. Biol. Conserv. 2014, 170, 56–63. [Google Scholar] [CrossRef]
- Newmark, W.D. Extinction of Mammal Populations in Western North American National Parks. Conserv. Biol. 1995, 9, 512–526. [Google Scholar] [CrossRef]
- Fenderson, L.E.; Kovach, A.I.; Litvaitis, J.A.; O’Brien, K.M.; Boland, K.M.; Jakubas, W.J. A Multiscale Analysis of Gene Flow for the New England Cottontail, an Imperiled Habitat Specialist in a Fragmented Landscape. Ecol. Evol. 2014, 4, 1853–1875. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.L. Assessing the Effects of Habitat Restoration on Shrubland Specialists: Case Study on the New England Cottontail and Shrubland Birds. Ph.D. Thesis, University of New Hampshire, Durham, NH, USA, 2018. [Google Scholar]
- La Haye, M.J.J.; Reiners, T.E.; Raedts, R.; Verbist, V.; Koelewijn, H.P. Genetic Monitoring to Evaluate Reintroduction Attempts of a Highly Endangered Rodent. Conserv. Genet. 2017, 18, 877–892. [Google Scholar] [CrossRef]
- Bauer, M.L.; Ferry, B.; Holman, H.; Kovach, A.I. Monitoring a New England Cottontail Reintroduction with Noninvasive Genetic Sampling. Wildl. Soc. Bull. 2020, 44, 110–121. [Google Scholar] [CrossRef]
- Sard, N.M.; Johnson, M.A.; Jacobson, D.P.; Hogansen, M.J.; O’Malley, K.G.; Banks, M.A. Genetic Monitoring Guides Adaptive Management of a Migratory Fish Reintroduction Program. Anim. Conserv. 2016, 19, 570–577. [Google Scholar] [CrossRef]
- Pierson, J.C.; Berry, L.; Alexander, L.; Anson, J.; Birkett, M.; Kemp, L.; Pascoe, B.A.; Farquharson, K.A.; Hogg, C.J. Adaptive Genetic Management of a Reintroduction Program from Captive Breeding to Metapopulation Management of an Arboreal Marsupial. Diversity 2023, 15, 848. [Google Scholar] [CrossRef]
- White, S.L.; Johnson, T.C.; Rash, J.M.; Lubinski, B.A.; Kazyak, D.C. Using Genetic Data to Advance Stream Fish Reintroduction Science: A Case Study in Brook Trout. Restor. Ecol. 2023, 31, e13662. [Google Scholar] [CrossRef]
- Giglio, R.M.; Ivy, J.A.; Jones, L.C.; Latch, E.K. Evaluation of Alternative Management Strategies for Maintenance of Genetic Variation in Wildlife Populations. Anim. Conserv. 2016, 19, 380–390. [Google Scholar] [CrossRef]
- Koelewijn, H.P.; Pérez-Haro, M.; Jansman, H.A.H.; Boerwinkel, M.C.; Bovenschen, J.; Lammertsma, D.R.; Niewold, F.J.J.; Kuiters, A.T. The Reintroduction of the Eurasian Otter (Lutra lutra) into the Netherlands: Hidden Life Revealed by Noninvasive Genetic Monitoring. Conserv. Genet. 2010, 11, 601–614. [Google Scholar] [CrossRef]
- Ali, O.A.; O’Rourke, S.M.; Amish, S.J.; Meek, M.H.; Luikart, G.; Jeffres, C.; Miller, M.R. RAD Capture (Rapture): Flexible and Efficient Sequence-Based Genotyping. Genetics 2016, 202, 389–400. [Google Scholar] [CrossRef]
| Objective | Parameter | Genetic Monitoring Approach |
|---|---|---|
| Wild Apparent Survival | Released Individuals | Compare tissue sample genotypes to winter pellet genotypes |
| Adults After 1st Winter | Compare tissue sample genotypes to winter pellet genotypes | |
| Factors Influencing Survival Rates (Genetic) | Logistic regression models of winter monitoring data | |
| Wild Population Information | Habitat Occupancy and Spatial Distribution | Compare GPS locations of burrows and identified species and individuals from winter monitoring data each year |
| Minimum Population Size | Winter monitoring fecal DNA genotyping | |
| Sex Ratios in Wild Population | Ratio of male/female individuals identified in winter monitoring surveys | |
| Rabbits Per Active Burrow | Ratio of number of rabbits identified to total number of active burrows located during winter monitoring surveys | |
| Genetic Diversity | Estimates of observed and expected heterozygosity, and allelic richness from winter and summer fecal DNA genotypes | |
| CB Ancestry | Genetic estimates based on winter and summer monitoring fecal DNA genotypes | |
| Effective Population Size | Parametric point estimates using linkage disequilibrium method and minor allele frequency 0.05, from winter and summer monitoring fecal DNA genotypes |
| Breeding Season | ENC | Release Period | Released | Survey Period | AS | Fecal Samples | Pygmy Fecal | SPID Success | ID Success | Individuals Detected (Year Released or First Detected) | Contributing Breeders of Wild-Born Juveniles |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2012 | 2 | May–July | 104 JV | Dec 2012– Jan 2013 | 9.71 km2 | 117 | 111 | NA | 78% | 45 | 1 female |
| 0 AD | 41 released (2012) | 1 male | |||||||||
| 4 wild-born (2012) | |||||||||||
| 2013 | 3 | May–August | 265 JV | Jan–Feb 2014 | 10.52 km2 | 296 | 273 | NA | 46% | 44 | 7 females |
| 7 AD | 3 released (2012) | 7 males | |||||||||
| 34 released (2013) | |||||||||||
| 7 wild-born (2013) | |||||||||||
| 2014 | 4 | March– November | 717 JV | Jan–Mar 2015 | 13.76 km2 | 265 | 212 | NA | 76% | 91 | 2 females |
| 113 AD | 1 released (2013) | 3 males | |||||||||
| 87 released (2014) | |||||||||||
| 3 wild-born (2014) | |||||||||||
| 2015 | 4 | February–October | 149 JV | Jan–Feb 2016 | 10.84 km2 | 105 | 105 | NA | 20% | 18 | 11 females (1 unknown) |
| 4 AD | 1 released (2014) | 8 males (5 unknown) | |||||||||
| 1 released (2015) | |||||||||||
| 16 wild-born (2015) | |||||||||||
| 2016 | 4 | May–October | 119 JV | Dec 2016– Mar 2017 | 24.28 km2 | 193 | 124 | 46% | 52% | 60 | 18 females (25 unknown) |
| 1 AD | 1 wild-born (2015) | 17 males (32 unknown) | |||||||||
| 5 released (2016) | |||||||||||
| 54 wild-born (2016) | |||||||||||
| 2017 | 4 | May–October | 0 JV | Dec 2017– Mar 2018 | 14.67 km2 | 357 | 296 | 72% | 56% | 158 | 47 females (98 unknowns) |
| 0 AD | 2 wild-born (2016) | 46 fathers (92 unknowns) | |||||||||
| 156 wild-born (2017) | |||||||||||
| 2018 | 2 | May–August | 0 JV | June–Aug 2018 | 7.40 km2 | 98 | 98 | NA | 56% | 54 | 19 females (33 unknowns) |
| 0 AD | 2 wild-born (2017) | ||||||||||
| 49 wild-born adults (2017) | 17 males (31 unknowns) | ||||||||||
| 3 wild-born juveniles (2018) | |||||||||||
| Dec 2018– Apr 2019 | 11.51 km2 | 447 | 296 | 77% | 73% | 138 | 19 females (88 unknowns) | ||||
| 1 wild-born (2016) | |||||||||||
| 14 wild-born (2017) | 20 males (81 unknowns) | ||||||||||
| 123 wild-born (2018) | |||||||||||
| 2019 | 2 | May–August | 0 JV | Jan–Mar 2020 | 5.89 km2 | 59 | 27 | 97% | 83% | 8 | 2 females (3 unknowns) |
| 0 AD | 3 wild-born (2018) | ||||||||||
| 5 wild-born (2019) | 3 males (2 unknowns) |
| Breeding Season | Release Area | Release Period | Released | Survey Period | AS | Fecal Samples | Pygmy Fecal | SPID Success | ID Success | Individuals Detected (Year Released) | Contributing Breeders to Wild-Born Juveniles |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2015 | BH | February–May | 369 JV | Jan–Feb 2016 | 3.09 km2 | 0 | 0 | - | - | - | - |
| 51 AD | |||||||||||
| 2017 | BH | May–October | 37 JV | Dec 2017–Mar 2018 | 0.21 km2 | 9 | 8 | - | 75% | 5 | 0 females |
| 0 AD | 5 released ENC (2017) | 0 males | |||||||||
| 2018 | BH | May–August | 14 JV | Dec 2018–Apr 2019 | 0.69 km2 | 10 | 8 | 80% | 88% | 3 | 0 females |
| 11 ENC | 2 released ENC (2018) | 0 males | |||||||||
| 3 WLD | 1 released WLD (2018) | ||||||||||
| CHB | May–August | 17 JV | Dec 2018–Apr 2019 | 1.07 km2 | 20 | 19 | 95% | 84% | 6 | 0 females | |
| 8 ENC | 1 released ENC (2018) | 0 males | |||||||||
| 9 ENC | 5 released WLD (2018) | ||||||||||
| 2019 | BH | May–August | 14 JV | June–Sept 2019 | 0.69 km2 | 34 | 27 | 85% | 67% | 7 | 1 female (1 unknown) |
| 10 ENC | 2 released ENC (2019) | 1 male (1 unknown) | |||||||||
| 4 WLD | 3 escaped ENC (2019) | ||||||||||
| 2 wild-born (2019) | |||||||||||
| CHB | May–August | 20 JV | June–Sept 2019 | 1.53 km2 | 20 | 14 | 80% | 93% | 5 | 0 females | |
| 19 ENC | 5 released ENC (2019) | 0 males | |||||||||
| 1 WLD | |||||||||||
| BH | May–August | * | Oct 2019–Feb 2020 | 0.69 km2 | 15 | 13 | 93% | 92% | 5 | 0 females | |
| 4 released ENC (2019) | 0 males | ||||||||||
| 1 released WLD (2019) | |||||||||||
| CHB | May–August | * | Oct 2019–Feb 2020 | 2.43 km2 | 39 | 37 | 97% | 81% | 10 | 0 females | |
| 9 released ENC (2019) | 0 males | ||||||||||
| 1 released WLD (2019) |
| YEAR | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2012 | 2013 | 2014 | 2015 | 2016 | 2017 | 2018 | 2019 | |||
| SURVEY PERIOD | ||||||||||
| Winter | Winter | Winter | Winter | Winter | Winter | Summer | Winter | Winter | ||
| Category | Parameter | |||||||||
| Demographic | Minimum count | 45 | 44 | 91 | 18 | 60 | 158 | 54 | 138 | 8 |
| M/F Sex Ratio (actual numbers) | 1:1.5 (18:27) | 1:1 (22:22) | 1:1.1 (44:47) | 1:1.6 (7:11) | 1.1:1 (32:28) | 1.8:1 (101:57) | 1:1.8 (19:35) | 1.9:1 (91:47) | 1:1 (4:4) | |
| Rabbits/Active Burrow | 0.87 | 0.75 | 0.63 | 1 | 0.92 | 0.96 | 0.56 | 0.65 | 0.33 | |
| Density (rabbits/ha) | 0.03 | 0.02 | 0.05 | 0.01 | 0.03 | 0.09 | 0.03 | 0.08 | 0.004 | |
| Genetic | Average CB ancestry | 19.69% | 19.72% | 19.10% | 21.06% | 21.89% | 15.31% | 18.48% | 17.97% | 16.1% |
| % of identified individuals containing CB ancestry | 48.89% | 88.64% | 71.43% | 100% | 100% | 100% | 100%% | 100% | 100% | |
| % of wild-born individuals containing CB ancestry | 100%% | 100% | 33.33% | 100% | 100% | 100% | 100%% | 100% | 100% | |
| Effective population size (95% Confidence Interval) | 15.4 (13.7–17.3) | 29.6 (25.3–34.9) | 30.4 (27.7–33.5) | 19.3 (14.7–27.0) | 40.7 (35.0–47.9) | 44.3 (40.6–48.5) | 36.9 (23.7–30.8) | 27.6 (25.5–29.9) | 12.3 (7.0–26.8) | |
| Observed Heterozygosity | 0.76 | 0.81 | 0.81 | 0.75 | 0.7 | 0.84 | 0.76 | 0.62 | 0.64 | |
| Expected Heterozygosity | 0.80 | 0.79 | 0.80 | 0.80 | 0.80 | 0.82 | 0.79 | 0.79 | 0.72 | |
| Allelic Richness | 5.22 | 5.15 | 5.13 | 5.29 | 5.16 | 5.35 | 5.00 | 4.95 | 4.67 | |
| SURVEY PERIOD | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Winter 2017–18 | Winter 2018–19 | Summer 2019 | Winter 2019–20 | ||||||
| LOCATION | CHB | BH | CHB | BH | CHB | BH | CHB | BH | |
| Demographic Parameters | |||||||||
| Minimum count | - | 5 | 5 | 3 | 5 | 7 | 10 | 5 | |
| M:F Sex Ratio (actual #s) | - | 1:1.5 (2:3) | 1.5:1 (3:2) | 1:2 (1:2) | 1:1.5 (2:3) | 6:1 (6:1) | 1:1.5 (4:6) | 1:1.5 (2:3) | |
| Rabbits/Active Burrow | - | 0.42 | 0.45 | 0.43 | 0.5 | 0.7 | 0.35 | 0.5 | |
| Density (rabbits/ha) | - | 0.06 | 0.01 | 0.04 | 0.01 | 0.09 | 0.01 | 0.04 | |
| Genetic Parameters | |||||||||
| Average CB ancestry | - | 23.98% | 14.85% | 23.97% | 20.89% | 22.87% | 19.04% | 27.46% | |
| % of identified individuals containing CB ancestry | - | 100% | 100% | 100% | 100% | 100% | 100% | 100% | |
| Effective population size (95% Confidence Interval) | - | - | - | - | - | 9.3 (3.3–32.0) | 12.5 (7.1–26.6) | - | |
| Observed Heterozygosity | - | 0.78 | 0.77 | 0.75 | 0.73 | 0.8 | 0.74 | 0.59 | |
| Expected Heterozygosity | - | 0.71 | 0.71 | - | 0.66 | 0.66 | 0.64 | 0.64 | |
| Allelic Richness | - | 5.41 | 4.65 | - | 4.59 | 3.82 | 3.69 | 3.71 | |
| Variable | Juvenile Estimate (SBF) | 95% CI | Adult Estimate (SBF) | 95% CI | Juvenile Estimate (Release Pens) | 95% CI | |||
|---|---|---|---|---|---|---|---|---|---|
| Lower | Upper | Lower | Upper | Lower | Upper | ||||
| Release Day | 0.010 | 0.005 | 0.014 | 0.017 | 0.007 | 0.027 | 0.020 | −0.005 | 0.046 |
| Release Weight | 0.003 | 0.001 | 0.005 | 0.000 | −0.005 | 0.005 | 0.000 | −0.007 | 0.007 |
| Sex (female) | 0.056 | −0.318 | 0.430 | 0.109 | −0.768 | 0.986 | 0.282 | −0.786 | 1.350 |
| HL | −1.905 | −3.465 | −0.346 | −2.442 | −6.065 | 1.180 | 1.537 | −3.588 | 6.661 |
| CB | 0.007 | −0.004 | 0.018 | −0.012 | −0.039 | 0.016 | −0.038 | −0.184 | 0.109 |
| Year 2012 | 2.227 | 1.717 | 2.737 | NA | - | - | NA | - | - |
| Year 2013 | 0.544 | 0.066 | 1.021 | NA | - | - | NA | - | - |
| Year 2015 | −3.982 | −5.964 | −2.000 | NA | - | - | NA | - | - |
| Year 2016 | −0.586 | −1.638 | 0.465 | NA | - | - | NA | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nerkowski, S.A.; Hohenlohe, P.A.; Rachlow, J.L.; Warheit, K.I.; Gallie, J.A.; Waits, L.P. Long-Term Noninvasive Genetic Monitoring Guides Recovery of the Endangered Columbia Basin Pygmy Rabbits (Brachylagus idahoensis). Genes 2025, 16, 956. https://doi.org/10.3390/genes16080956
Nerkowski SA, Hohenlohe PA, Rachlow JL, Warheit KI, Gallie JA, Waits LP. Long-Term Noninvasive Genetic Monitoring Guides Recovery of the Endangered Columbia Basin Pygmy Rabbits (Brachylagus idahoensis). Genes. 2025; 16(8):956. https://doi.org/10.3390/genes16080956
Chicago/Turabian StyleNerkowski, Stacey A., Paul A. Hohenlohe, Janet L. Rachlow, Kenneth I. Warheit, Jonathan A. Gallie, and Lisette P. Waits. 2025. "Long-Term Noninvasive Genetic Monitoring Guides Recovery of the Endangered Columbia Basin Pygmy Rabbits (Brachylagus idahoensis)" Genes 16, no. 8: 956. https://doi.org/10.3390/genes16080956
APA StyleNerkowski, S. A., Hohenlohe, P. A., Rachlow, J. L., Warheit, K. I., Gallie, J. A., & Waits, L. P. (2025). Long-Term Noninvasive Genetic Monitoring Guides Recovery of the Endangered Columbia Basin Pygmy Rabbits (Brachylagus idahoensis). Genes, 16(8), 956. https://doi.org/10.3390/genes16080956






