Mitochondrial Genome of Scutiger ningshanensis (Anura, Megophryidae, Scutiger): Insights into the Characteristics of the Mitogenome and the Phylogenetic Relationships of Megophryidae Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimen Collection and DNA Extraction
2.2. Primer Design and PCR Amplification
2.3. Sequence Assembly, Analysis, and Annotation
2.4. Phylogenetic Analyses
2.5. Divergence Time Estimates
2.6. Ka and Ks Analysis
3. Results
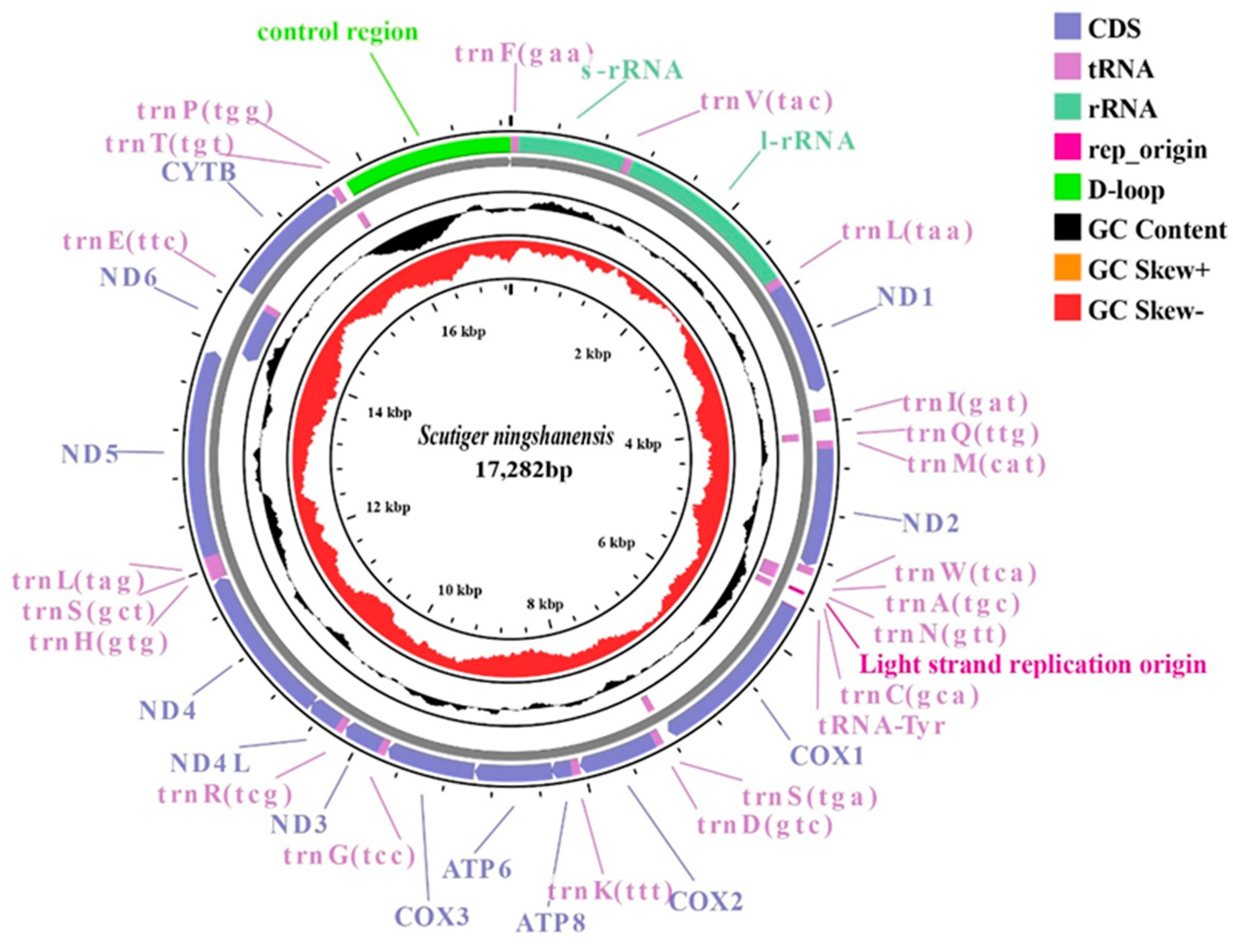
3.1. Characterization of the Mitogenome Structure
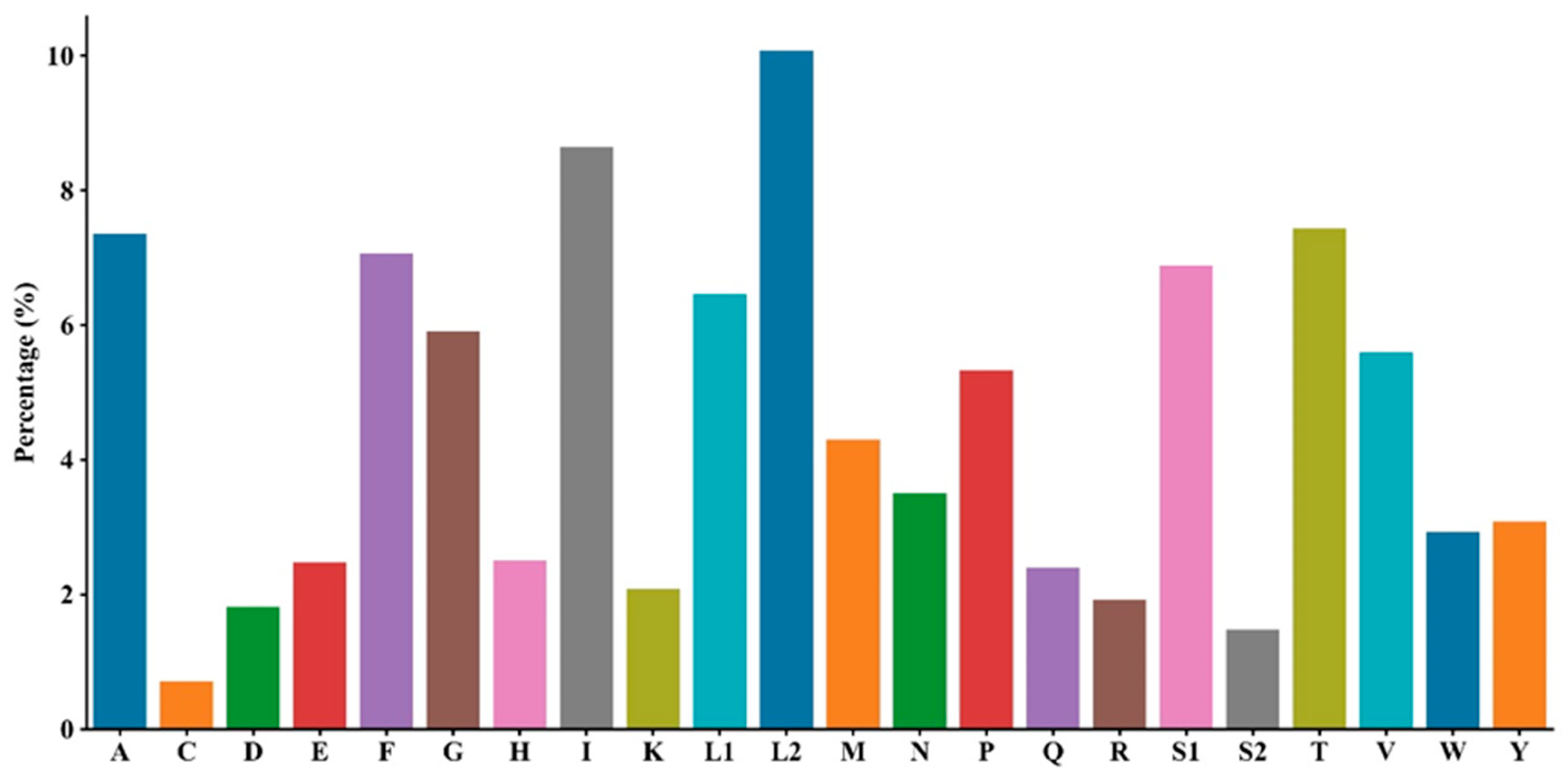
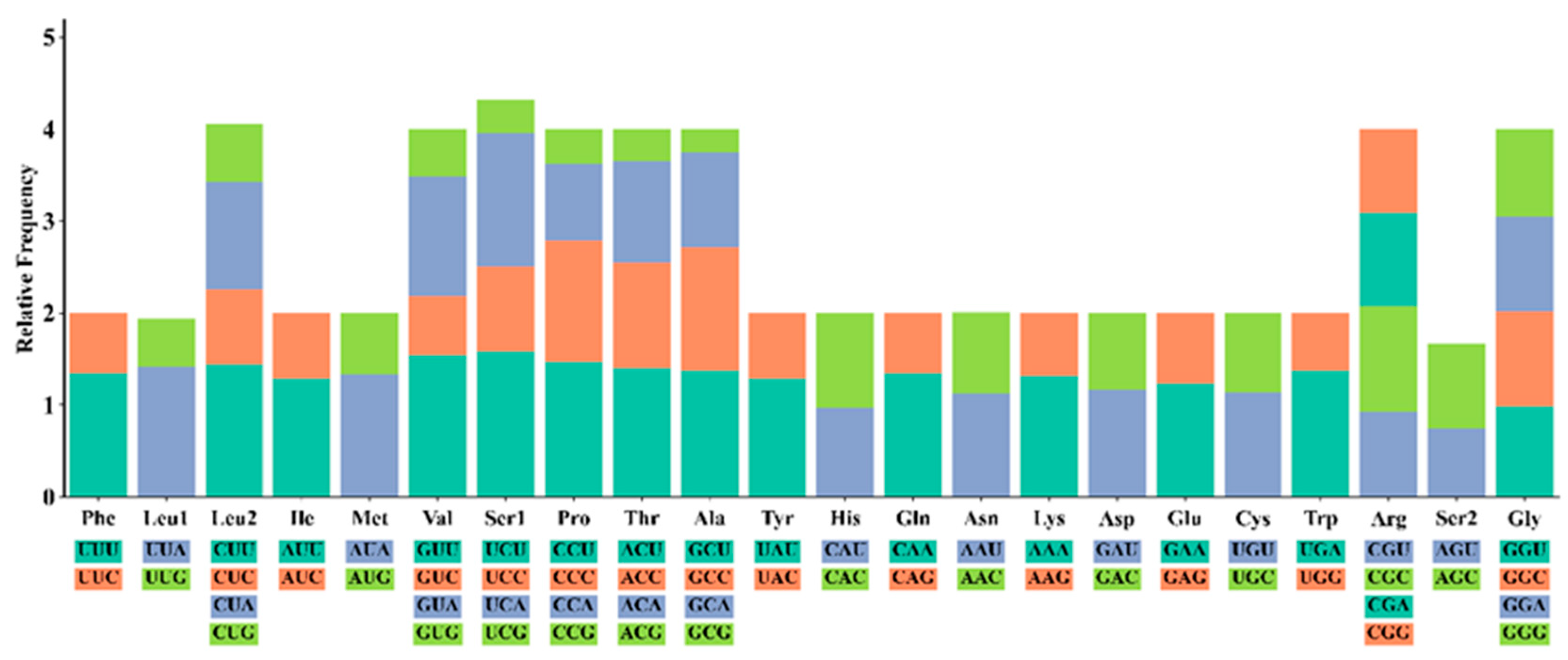
3.2. Protein-Coding Genes and Codon Usage
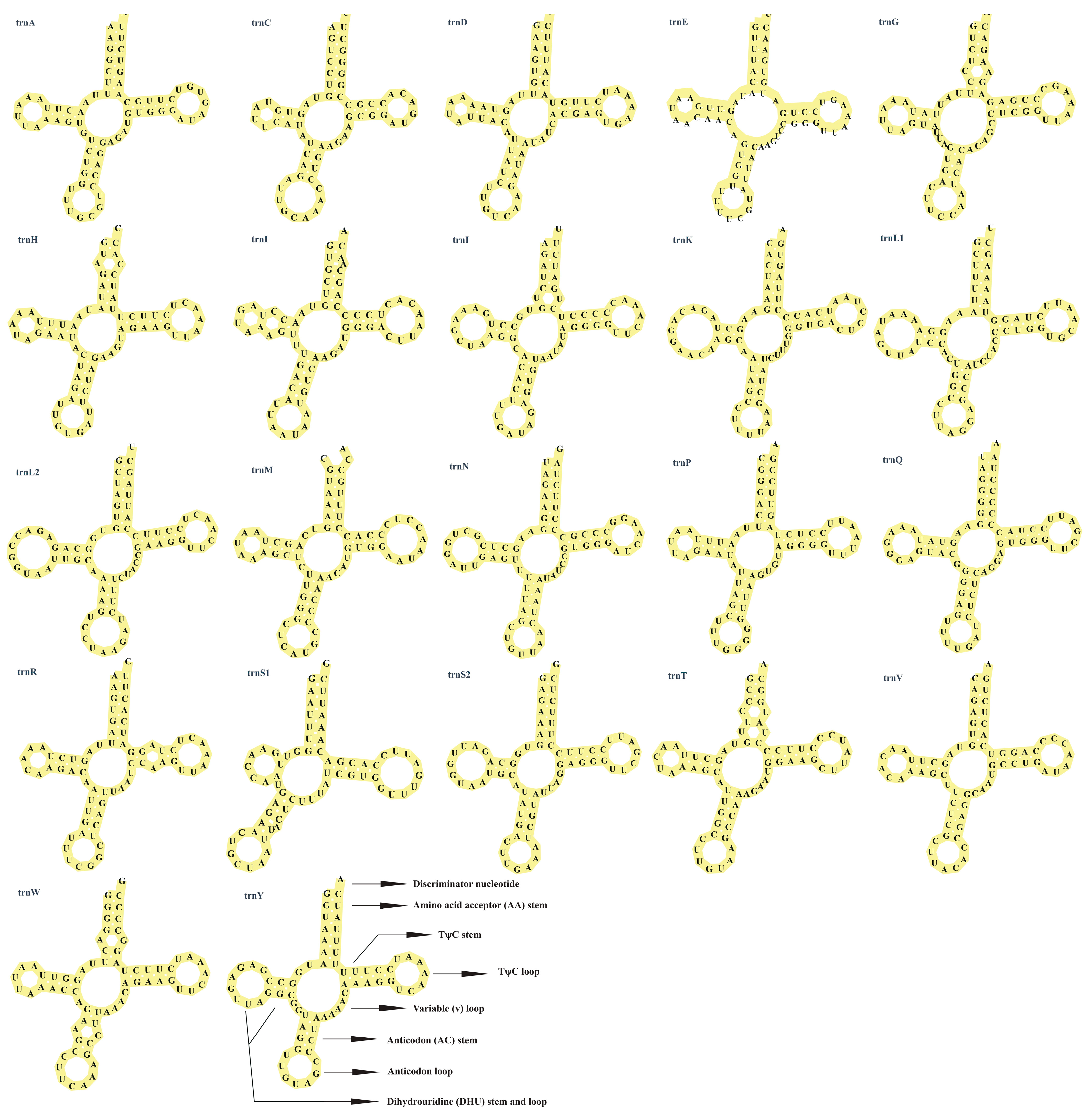
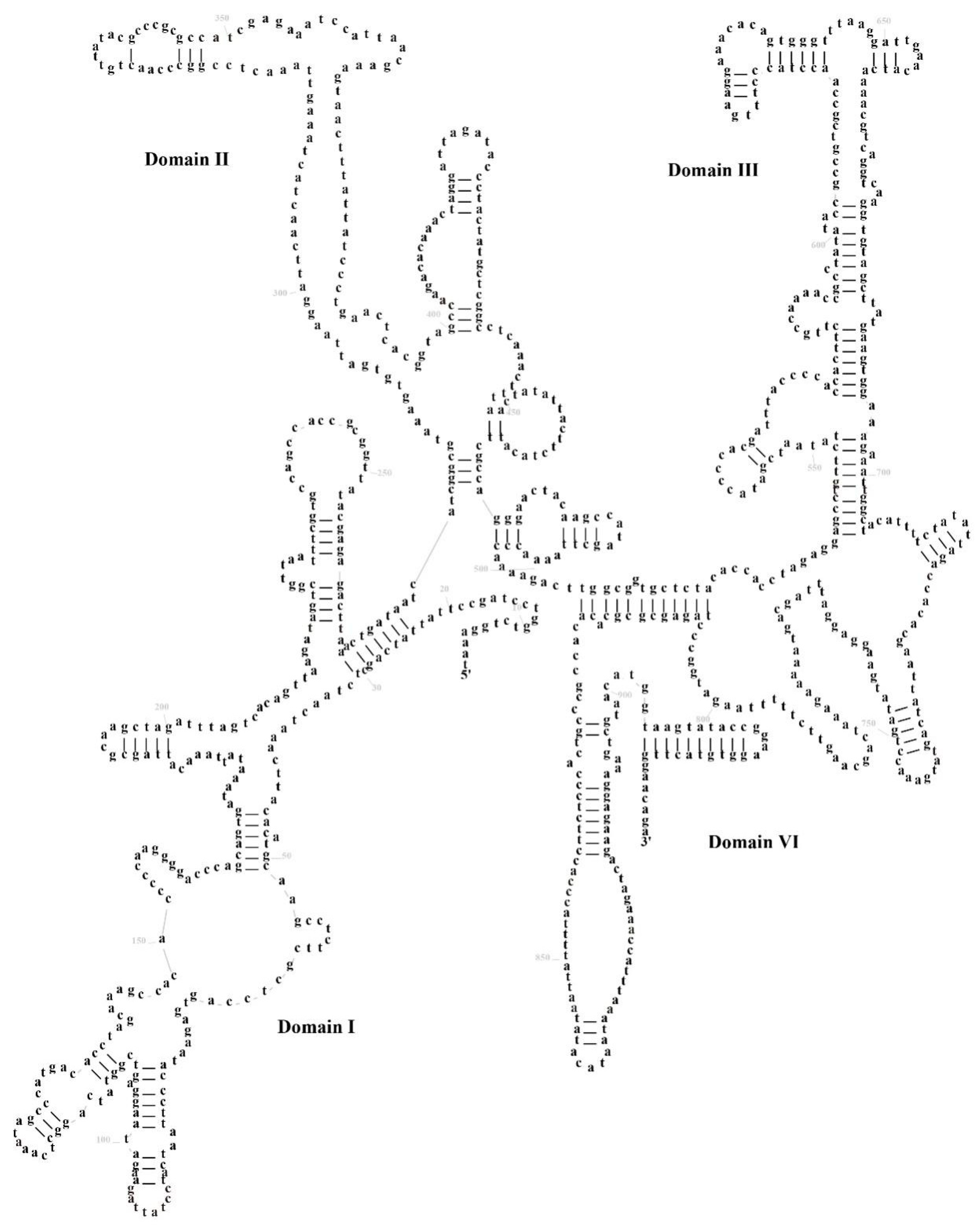
3.3. Transfer RNA and Ribosomal RNA Genes and Control Region
3.4. Phylogenetic Analysis
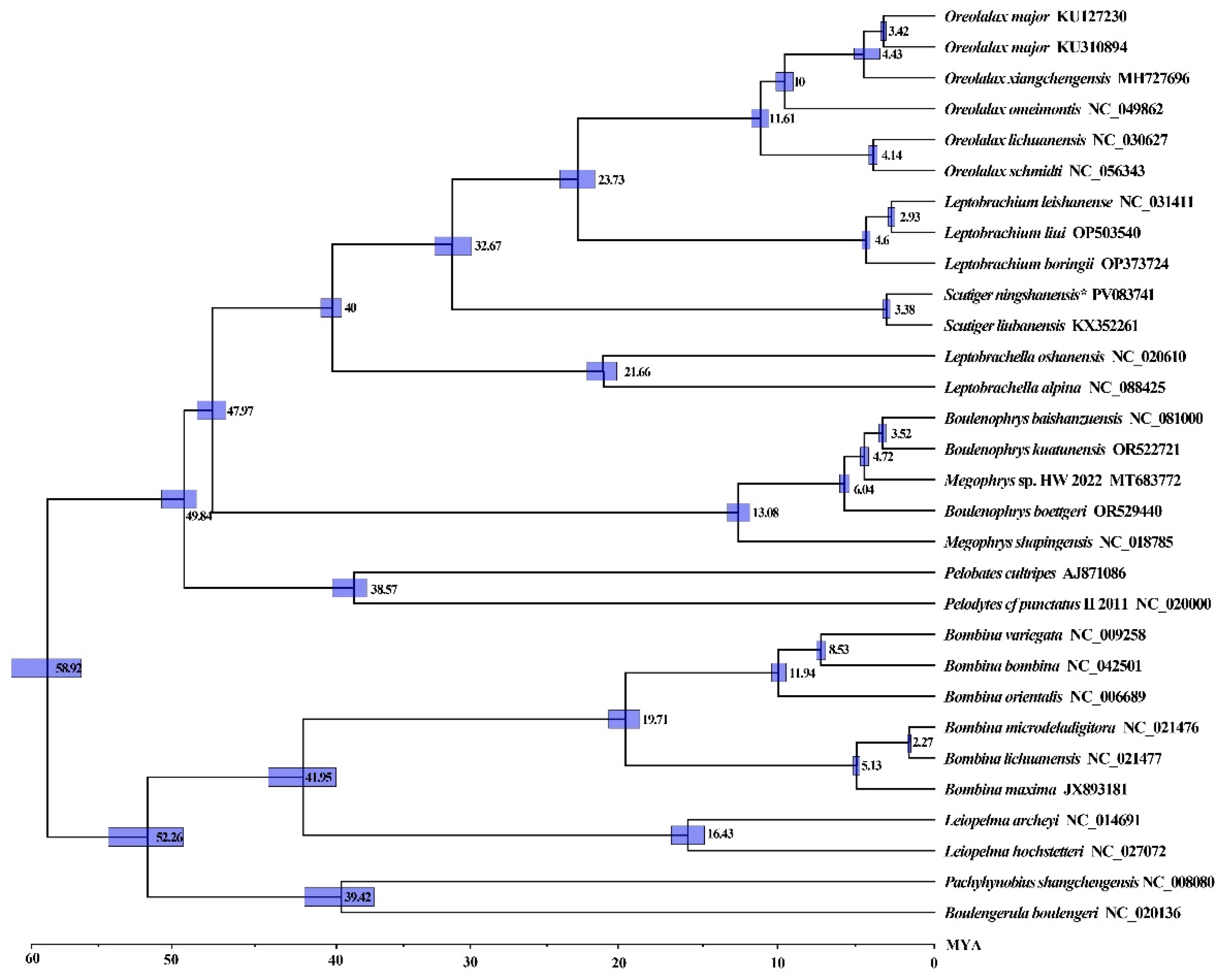
3.5. Divergence Time of the Megophryidae Species
3.6. Selective Pressure Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cataldo, C.S. The gastropod family Aporrhaidae in the Lower Cretaceous of the Neuquén Basin, west-central Argentina. J. Paleontol. 2014, 88, 1222–1239. [Google Scholar] [CrossRef]
- Munir, M.; Nishikawa, K.; Hamidy, A.; Smith, E.N. Two new species of Megophrys Kuhl and Van Hasselt (Amphibia: Megophryidae) from Sumatra, Indonesia. Zootaxa 2021, 5057, 503–529. [Google Scholar] [CrossRef]
- Frost, D.R.; Grant, T.; Faivovich, J.; Bain, R.H.; Haas, A.; Haddad, C.F.B.; Desa, R.O.; Channing, A.; Wilkinson, M.; Donnellan, S.C.; et al. The amphibian tree of life. Bull. Am. Mus. Nat. Hist. 2006, 297, 1–291. [Google Scholar] [CrossRef]
- Pyron, R.A.; Wiens, J.J. A large-scale phylogeny of Amphibia including over 2800 species, and a revised classification of extant frogs, salamanders, and caecilians. Mol. Phylogenet. Evol. 2011, 61, 543–583. [Google Scholar] [CrossRef]
- Jiang, K.; Wang, K.; Zou, D.H.; Yan, F.; Li, P.P.; Che, J. A new species of the genus Scutiger (Anura: Megophryidae) from Medog of southeastern Tibet, China. Zool. Res. 2016, 37, 21–30. [Google Scholar] [CrossRef]
- Song, J.; Tian, Y.; Guan, D.L. Characterization of the complete mitochondrial genome of an endangered alpine toad, Scutiger ningshanensis (Amphibia: Anura: Megophryidae). Conserv. Genet. Resour. 2017, 9, 35–38. [Google Scholar] [CrossRef]
- Dufresnes, C.; Litvinchuk, S.N. Diversity, distribution and molecular species delimitation in frogs and toads from the Eastern Palaearctic. Zool. J. Linn. Soc. 2022, 195, 695–760. [Google Scholar] [CrossRef]
- Primmer, C.R. From conservation genetics to conservation genomics. Ann. N. Y. Acad. Sci. 2009, 1162, 357–368. [Google Scholar] [CrossRef]
- Fu, J.; Weadick, C.J.; Bi, K. A phylogeny of the high-elevation Tibetan megophryid frogs and evidence for the multiple origins of reversed sexual size dimorphism. J. Zool. 2007, 273, 315–325. [Google Scholar] [CrossRef]
- Chen, W.; Bi, K.; Fu, J. Frequent mitochondrial gene introgression among high elevation Tibetan megophryid frogs revealed by conflicting gene genealogies. Mol. Ecol. 2010, 18, 2856–2876. [Google Scholar] [CrossRef]
- Hofmann, S.; Stöck, M.; Zheng, Y.; Ficetola, F.G.; Li, J.T.; Scheidt, U.; Schmidt, J. Molecular phylogenies indicate a Paleo-Tibetan origin of Himalayan Lazy Toads (Scutiger). Sci. Rep. 2017, 7, 3308. [Google Scholar] [CrossRef]
- Barr, C.M.; Neiman, M.; Taylor, D. Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol. 2005, 168, 39–50. [Google Scholar] [CrossRef]
- Guo, X.; Bao, P.J.; Pei, J.; Ding, X.Z.; Liang, C.N.; Yan, P.; Lu, D.X. Complete mitochondrial genome of Qingyang donkey (Equus asinus). Conserv. Genet. Resour. 2017, 9, 269–271. [Google Scholar] [CrossRef]
- Moritz, C.; Dowling, T.E.; Brown, W.M. Evolution of animal mitochondrial DNA: Relevance for population biology and systematics. Annu. Rev. Ecol. Syst. 1987, 18, 269–292. [Google Scholar] [CrossRef]
- Xiang, Z.Y.; Zhang, C.Y.; Zheng, W.C.; Yu, S.S.; Zhang, L.; Ding, G.H. Characterization of the complete mitochondrial genome of the Chong’an Moustache Toad, Leptobrachium liui (Pope, 1947) with a phylogenetic analysis of Megophryidae. Mitochondrial DNA Part B 2021, 6, 1061–1063. [Google Scholar] [CrossRef]
- Qiu, Z.Y.; Chang, H.H.; Yuan, H.; Huang, Y.; Lu, H.M.; Li, X.; Gou, X.C. Comparative mitochondrial genomes of four species of Sinopodisma and phylogenetic implications (Orthoptera, Melanoplinae). ZooKeys 2020, 969, 23–42. [Google Scholar] [CrossRef]
- Bao, X.L.; Cui, L.; Li, D.Y.; Fan, X.L.; Yang, M.Y.; Yang, D.Y.; Ni, Q.Y.; Yao, Y.F.; Xu, H.L.; Zeng, B.; et al. The near complete mitochondrial genome of Oreolalax schmidti (Anura: Megophryidae). Mitochondrial DNA Part B 2020, 5, 3536–3537. [Google Scholar] [CrossRef]
- Shu, G.C.; Yu, M.; He, Z.P.; Xie, F.; Liang, X.X. Complete mitochondrial genome of the Alpine Metacarpal-tubercled Toad Leptobrachella alpina (Amphibia, Anura, Megophryidae). Mitochondrial DNA Part B 2021, 6, 3242–3243. [Google Scholar] [CrossRef]
- Zhang, J.F.; Miao, G.P.; Hu, S.J.; Sun, Q.; Ding, H.W.; Ji, Z.C.; Guo, P.; Yan, S.B.; Wang, C.R.; Kan, X.Z.; et al. Quantification and evolution of mitochondrial genome rearrangement in Amphibians. BMC Ecol. Evol. 2021, 21, 19. [Google Scholar] [CrossRef]
- Kurabayashi, A.; Sumida, M. PCR primers for the Neobatrachian mitochondrial genome. Curr. Herpetol. 2009, 28, 1–11. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Lohse, M.; Drechsel, O.; Kahlau, S.; Bock, R. Organellar Genome DRAW—A Suite of Tools for Generating Physical Maps of Plastid and Mitochondrial Genomes and Visualizing Expression Data Sets. Nucleic Acids Res. 2013, 41, W575–W581. [Google Scholar] [CrossRef]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved Detection and Functional Classification of Transfer RNAo Genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Li, W.X.; Jakovli’c, I.; Zou, H.; Zhang, J.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N.; et al. BEAST 2.5: An Advanced Software Platform for Bayesian Evolutionary Analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Zhang, Z. KaKs_Calculator 3.0: Calculating Selective Pressure on Coding and Non-Coding Sequences. Genom. Proteom. Bioinform. 2022, 20, 536–540. [Google Scholar] [CrossRef]
- Zhang, D.-X.; Hewitt, G.M. Insect Mitochondrial Control Region: A Review of Its Structure, Evolution and Usefulness in Evolutionary Studies. Biochem. Syst. Ecol. 1997, 25, 99–120. [Google Scholar] [CrossRef]
- Ingman, M.; Kaessmann, H.; Pääbo, S.; Gyllensten, U. Mitochondrial Genome Variation and the Origin of Modern Humans. Nature 2000, 408, 708–713. [Google Scholar] [CrossRef]
- Taanman, J.W. The mitochondrial genome: Structure, transcription, translation and replication. Biochim. Biophys. Acta Bioenerg. 1999, 1410, 103–123. [Google Scholar] [CrossRef]
- Meiklejohn, C.D.; Montooth, K.L.; Rand, D.M. Positive and negative selection on the mitochondrial genome. Trends Genet. 2007, 23, 259–263. [Google Scholar] [CrossRef]
- Montaña-Lozano, P.; Moreno-Carmona, M.; Ochoa-Capera, M.; Medina, N.S.; Boore, J.L.; Prada, C.F. Comparative genomic analysis of vertebrate mitochondrial reveals a differential of rearrangements rate between taxonomic class. Sci. Rep. 2022, 12, 5479. [Google Scholar] [CrossRef]
- Zhou, Q.; Xiang, H.M.; Zhang, M.Y.; Liu, Y.; Gu, Z.R.; Lan, X.Y.; Wang, J.X.; Jiang, W.S. Two Complete Mitochondrial Genomes of Leptobrachium (Anura: Megophryidae: Leptobrachiinae): Characteristics, Population Divergences, and Phylogenetic Implications. Genes 2023, 14, 768. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Zhang, L.P.; Yu, D.N.; Storey, K.B.; Zheng, R.Q. Complete mitochondrial genomes of Nanorana taihangnica and N. yunnanensis (Anura: Dicroglossidae) with novel gene arrangements and phylogenetic relationship of Dicroglossidae. BMC Evol. Biol. 2018, 18, 26. [Google Scholar] [CrossRef]
- Zhang, P.; Liang, D.; Mao, R.; Hillis, D.M.; Wake, D.B.; Cannatella, D.C. Efficient sequencing of Anuran mtDNAs and a mitogenomic exploration of the phylogeny and evolution of frogs. Mol. Biol. Evol. 2013, 30, 1899–1915. [Google Scholar] [CrossRef]
- Yu, X.; Du, Y.; Fang, M.; Li, H.; Lin, L. The mitochondrial genome of Pseudocalotes microlepis (Squamata: Agamidae) and its Phylogenetic position in agamids. Asian Herpetol. Res. 2018, 9, 24–34. [Google Scholar]
- Yuan, S.; Xia, Y.; Zheng, Y.; Zeng, X. Next-generation sequencing of mixed genomic DNA allows efficient assembly of rearranged mitochondrial genomes in Amolops chunganensis and Quasipaa boulengeri. PeerJ 2016, 4, e2786. [Google Scholar] [CrossRef][Green Version]
- Bloom, J.D.; Labthavikul, S.T.; Otey, C.R.; Arnold, F.H. Protein stability promotes evolvability. Proc. Natl. Acad. Sci. USA 2006, 103, 5869–5874. [Google Scholar] [CrossRef]
- Yang, Z.H.; Nielsen, R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 2000, 17, 32–43. [Google Scholar] [CrossRef]
- Zhang, L.N.; Ma, P.F.; Zhang, Y.X.; Zeng, C.X.; Zhao, L. Using nuclear loci and allelic variation to disentangle the phylogeny of Phyllostachys (Poaceae, Bambusoideae). Mol. Phylogenet. Evol. 2019, 137, 222–235. [Google Scholar] [CrossRef]
- Lyu, Z.T.; Qi, S.; Wang, J.; Zhang, S.Z.; Zhao, J.; Zeng, Z.C.; Wan, H.; Yang, J.H.; Mo, Y.M.; Wang, Y.Y. Generic classification of Asian horned toads (Anura: Megophryidae: Megophryinae) and monograph of Chinese species. Zool. Res. 2023, 44, 380–450. [Google Scholar] [CrossRef]
- Shi, S.C.; Zhang, M.H.; Xie, F.; Jiang, J.P.; Liu, W.L.; Ding, L.; Li, L.; Wang, B. Multiple data revealed two new species of the Asian horned toad Megophrys Kuhl & Van Hasselt, 1822 (Anura, Megophryidae) from the eastern corner of the Himalayas. ZooKeys 2020, 977, 101–161. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, S.; Tian, Z.; Wang, Y.; Xu, S.; Wang, D. Comprehensive analysis of phylogenetic relationship and optimal codons in mitochondrial genomes of the genus Pseudogastromyzon. Animals 2024, 14, 495. [Google Scholar] [CrossRef]



| No. | Primer Name | Sequence 5′-3′ | Primer Length (bp) | References |
|---|---|---|---|---|
| 1 | CTF1 | ATTAAGATAAAGCCCTTCTAGAA | 23 | This study |
| CTR1 | AATACCATTGGTGTCCCACG | 20 | ||
| 2 | CTF2 | CAAGAYGCRRYHTCHCCNATYATAGAAGA | 29 | [20] |
| CTR2 | CCTTCWCGRAYNAYRTCTCGYCAYCAYTG | 29 | ||
| 3 | CTF3 | GCMCACCAAGCWCAYGCHTWYCAYATRGT | 29 | [20] |
| CTR3 | GADCCDGCRATDGGDGCYTCDACRTG | 26 | ||
| 4 | CTF4 | CACTACGCAGCAGACACCTC | 20 | This study |
| CTR4 | CAAGGGAAGGTCCTATCAAGT | 21 | ||
| 5 | CTF5 | ATGGTGGTATAATAGTATGGTGT | 23 | This study |
| CTR5 | GTTGTTGGGAATAAGGGTGT | 20 | ||
| 6 | CTF6 | ACCTCATACGCAAACTCAGC | 20 | This study |
| CTR6 | TACCATCATTTTAATAGGTGGA | 22 | ||
| 7 | CTF7 | CCCACATGTATAATTAACAGATT | 23 | This study |
| CTR7 | GGAGTAATCTTTCGTTTTGTAT | 22 | ||
| 8 | CTF8 | TTCGCAAAGCAAATACCCACA | 21 | This study |
| CTR8 | CGCCGACTAATATCAATTTG | 20 | ||
| 9 | CTF9 | CCACACCYHCAAGGGHAYTCAGCAGT | 26 | [20] |
| CTR9 | CTTYGCACGGTYAGRRTACCGCGGCCGT | 28 | ||
| 10 | CTF10 | CCCGCCTGTTTACCAAAAACAT | 22 | [20] |
| CTR10 | ACRTTRAANCCNGANACHAGTTCWGAYTC | 29 | ||
| 11 | CTF11 | CGRGCHGTHGCHCAAACNATYTCHTAYGA | 29 | [20] |
| CTR11 | AAGCTCKCTGGAWWGAGYGTTTAGCTGTTAA | 31 | ||
| 12 | CTF12 | GTCGCCCAAACAATCTCATATGA | 23 | [20] |
| CTR12 | AGGAGGGCTTTATCTTAAT | 19 |
| Gene | Direction | Position | Size (bp) | IGS (bp) | Codon | ||
|---|---|---|---|---|---|---|---|
| From | To | Start | Stop | ||||
| tRNA-Phe | H | 2 | 66 | 65 | 0 | ||
| 12S rRNA | H | 67 | 1001 | 935 | −4 | ||
| tRNA-Val | H | 998 | 1067 | 68 | 1 | ||
| 16S rRNA | H | 1069 | 2680 | 1610 | −3 | ||
| tRNA-Leu | H | 2678 | 2752 | 75 | 0 | ||
| ND1 | H | 2753 | 3730 | 978 | 154 | ATG | TAA |
| tRNA-Ile | H | 3885 | 3995 | 71 | 87 | ||
| tRNA-Gln | L | 4083 | 4153 | 71 | 7 | ||
| tRNA-Met | H | 4161 | 4229 | 68 | 0 | ||
| ND2 | H | 4230 | 5273 | 1044 | 0 | ATG | TAA |
| tRNA-Trp | H | 5274 | 5342 | 69 | 4 | ||
| tRNA-Ala | L | 5347 | 5416 | 70 | 0 | ||
| tRNA-Asn | L | 5417 | 5489 | 73 | −1 | ||
| rep_origin | H | 5489 | 5520 | 32 | −3 | ||
| tRNA-Cys | L | 5518 | 5580 | 63 | 0 | ||
| tRNA-Tyr | L | 5581 | 5650 | 70 | 1 | ||
| COX1 | H | 5652 | 7203 | 1552 | 1 | GTG | T−− |
| tRNA-Ser | L | 7205 | 7275 | 71 | 4 | ||
| tRNA-Asp | H | 7280 | 7347 | 68 | 1 | ||
| COX2 | H | 7349 | 8036 | 688 | 0 | ATG | T−− |
| tRNA-Lys | H | 8037 | 8110 | 74 | 0 | ||
| ATP8 | H | 8111 | 8290 | 180 | −10 | GTG | TAA |
| ATP6 | H | 8281 | 8962 | 682 | 0 | ATG | T−− |
| COX3 | H | 8963 | 9746 | 784 | 0 | ATG | T−− |
| tRNA-Gly | H | 9747 | 9816 | 70 | 0 | ||
| ND3 | H | 9817 | 10,159 | 343 | 0 | ATG | T−− |
| tRNA-Arg | H | 10,160 | 10,228 | 69 | 0 | ||
| ND4L | H | 10,229 | 10,525 | 297 | −7 | ATG | TAA |
| ND4 | H | 10,519 | 11,896 | 1378 | 0 | ATG | T−− |
| tRNA-His | H | 11,897 | 11,965 | 69 | 0 | ||
| tRNA-Ser | H | 11,966 | 12,032 | 67 | 0 | ||
| tRNA-Leu | H | 12,033 | 12,103 | 71 | 0 | ||
| ND5 | H | 12,104 | 13,924 | 1821 | 15 | ATG | TAG |
| ND6 | L | 13,940 | 14,449 | 510 | 0 | ATG | AGG |
| tRNA-Glu | L | 14,450 | 14,518 | 69 | 2 | ||
| CYTB | H | 14,521 | 15,661 | 1141 | 0 | ATG | T−− |
| tRNA-Thr | H | 15,662 | 15,731 | 70 | 2 | ||
| tRNA-Pro | L | 15,734 | 15,802 | 69 | 0 | ||
| D-loop | H | 15,803 | 17,282 | 1480 | 0 | ||
| Gens | Size(bp) | A% | T% | G% | C% | A + T% | G + C% | AT-Skew | GC-Skew |
|---|---|---|---|---|---|---|---|---|---|
| Mitogenome | 17,282 | 28.54 | 32.51 | 14.3 | 24.64 | 61.05 | 38.94 | −0.065 | −0.265 |
| PCGs | 11,398 | 26 | 34.87 | 14.67 | 24.46 | 60.87 | 39.13 | −0.145 | −0.250 |
| PCGs(1st) | 3797 | 27.44 | 27.23 | 23.12 | 22.2 | 54.67 | 45.33 | 0.0039 | 0.0203 |
| PCGs(2nd) | 3797 | 17.99 | 42.09 | 12.96 | 26.97 | 60.07 | 39.93 | −0.4011 | −0.3509 |
| PCGs(3rd) | 3797 | 32.6 | 35.19 | 7.95 | 24.26 | 67.79 | 32.21 | −0.0381 | −0.5061 |
| tRNAs | 1504 | 29.79 | 29.26 | 20.88 | 20.08 | 59.05 | 40.96 | 0.009 | 0.0195 |
| rRNAs | 2547 | 32.08 | 27.21 | 17.71 | 23.01 | 59.29 | 40.71 | 0.082 | −0.13 |
| D-loop | 1480 | 32.23 | 36.22 | 10.27 | 21.28 | 68.45 | 31.55 | −0.058 | −0.349 |
| ND1 | 978 | 26.07 | 33.33 | 26.28 | 14.31 | 59.4 | 40.59 | −0.122 | −0.294 |
| ND2 | 1044 | 25.96 | 37.36 | 25.48 | 11.21 | 63.32 | 36.69 | −0.18 | −0.389 |
| COX1 | 1552 | 25.97 | 33.31 | 23.71 | 17.01 | 59.28 | 40.72 | −0.123 | −0.164 |
| COX2 | 688 | 29.36 | 31.1 | 24.85 | 14.68 | 60.46 | 39.53 | −0.028 | −0.257 |
| ATP8 | 180 | 29.44 | 38.89 | 23.33 | 8.33 | 68.33 | 31.67 | −0.138 | −0.473 |
| ATP6 | 682 | 25.51 | 36.36 | 26.39 | 11.73 | 61.87 | 38.12 | −0.175 | −0.384 |
| COX3 | 784 | 23.72 | 33.42 | 25.89 | 16.96 | 62.1 | 42.86 | −0.169 | −0.208 |
| ND3 | 343 | 23.32 | 38.78 | 23.62 | 14.29 | 62.1 | 37.9 | −0.248 | −0.246 |
| ND4L | 297 | 24.92 | 33 | 28.96 | 13.13 | 57.92 | 42.09 | −0.139 | −0.376 |
| ND4 | 1378 | 27.07 | 34.83 | 25.33 | 12.77 | 61.9 | 38.1 | −0.125 | −0.329 |
| ND5 | 1821 | 27.4 | 35.15 | 24.66 | 12.8 | 62.55 | 37.45 | −0.123 | −0.316 |
| ND6 | 510 | 18.82 | 38.43 | 11.18 | 31.57 | 57.25 | 42.75 | −0.342 | 0.477 |
| CYTB | 1141 | 26.03 | 35.14 | 24.45 | 14.37 | 61.17 | 38.83 | −0.149 | −0.259 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shan, S.; Chen, S.; Li, C.; Peng, L.; Zhao, D.; Liao, Y.; Liu, P.; Jiang, L. Mitochondrial Genome of Scutiger ningshanensis (Anura, Megophryidae, Scutiger): Insights into the Characteristics of the Mitogenome and the Phylogenetic Relationships of Megophryidae Species. Genes 2025, 16, 879. https://doi.org/10.3390/genes16080879
Shan S, Chen S, Li C, Peng L, Zhao D, Liao Y, Liu P, Jiang L. Mitochondrial Genome of Scutiger ningshanensis (Anura, Megophryidae, Scutiger): Insights into the Characteristics of the Mitogenome and the Phylogenetic Relationships of Megophryidae Species. Genes. 2025; 16(8):879. https://doi.org/10.3390/genes16080879
Chicago/Turabian StyleShan, Siqi, Simin Chen, Chengmin Li, Lingyu Peng, Dongmei Zhao, Yaqing Liao, Peng Liu, and Lichun Jiang. 2025. "Mitochondrial Genome of Scutiger ningshanensis (Anura, Megophryidae, Scutiger): Insights into the Characteristics of the Mitogenome and the Phylogenetic Relationships of Megophryidae Species" Genes 16, no. 8: 879. https://doi.org/10.3390/genes16080879
APA StyleShan, S., Chen, S., Li, C., Peng, L., Zhao, D., Liao, Y., Liu, P., & Jiang, L. (2025). Mitochondrial Genome of Scutiger ningshanensis (Anura, Megophryidae, Scutiger): Insights into the Characteristics of the Mitogenome and the Phylogenetic Relationships of Megophryidae Species. Genes, 16(8), 879. https://doi.org/10.3390/genes16080879





