Phylogenetic Analyses and Plastome Comparison to Confirm the Taxonomic Position of Ligusticum multivittatum (Apiaceae, Apioideae)
Abstract
1. Introduction
2. Materials and Methods
2.1. Phylogenetic Analysis
2.2. Plastome Comparison Analyses
2.3. Simple Sequence Repeats and Codon Bias Analyses
2.4. Identification of Divergence Hotspots
3. Results
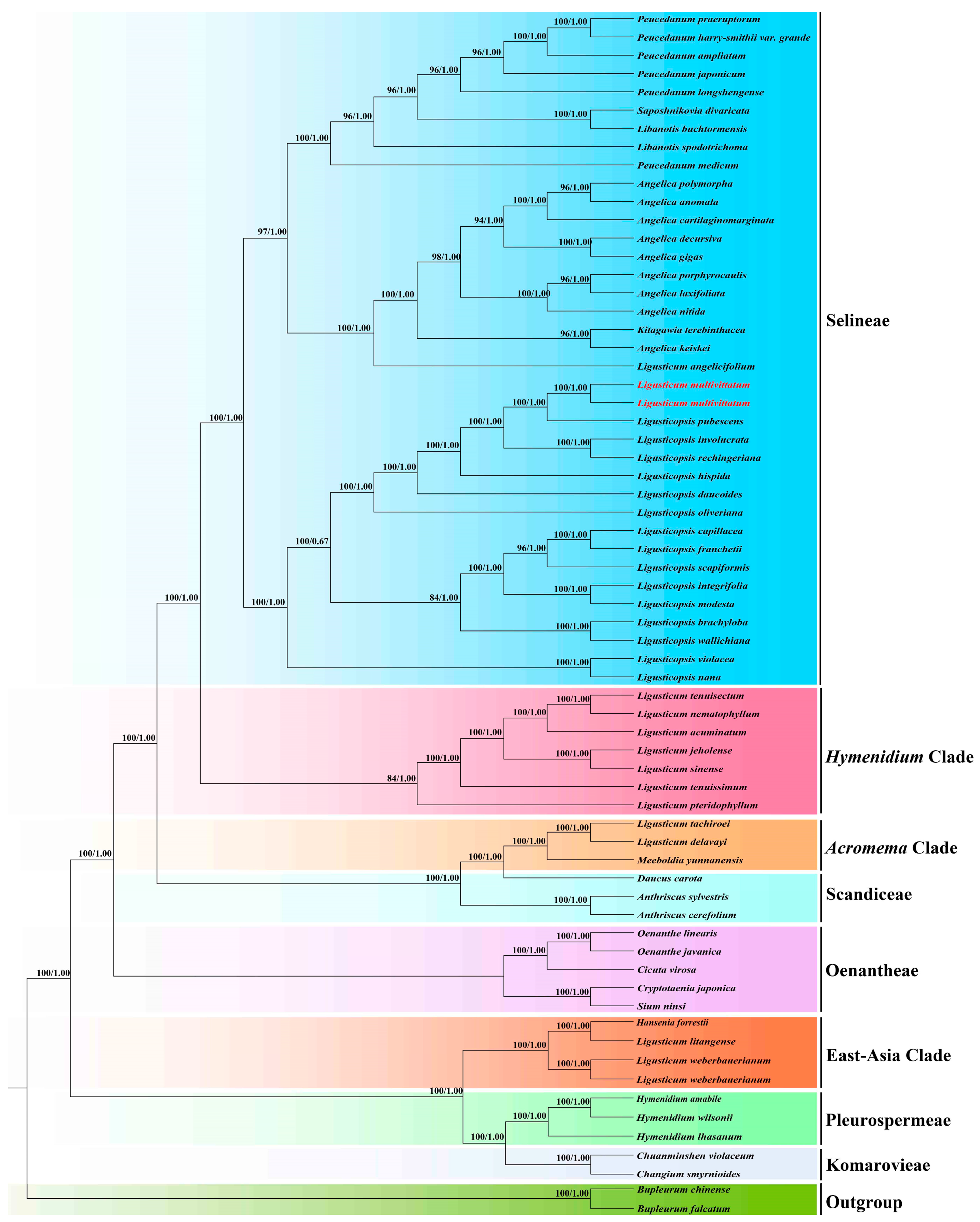
3.1. Phylogenetic Analyses
3.2. Characteristics of Plastome
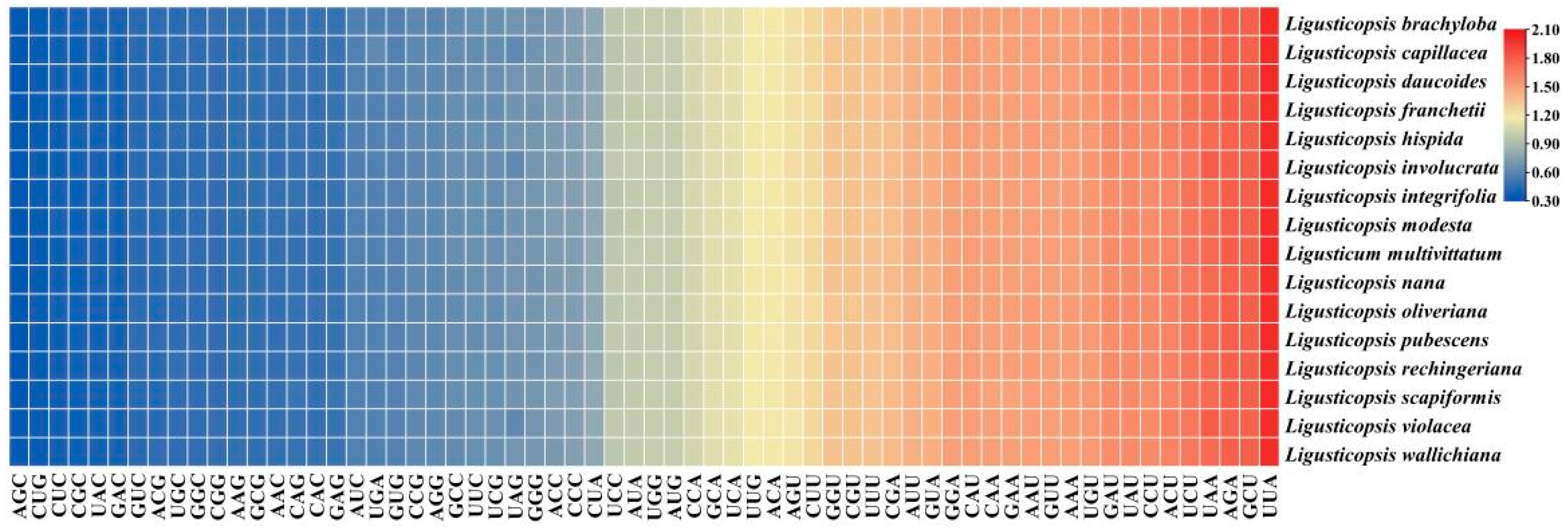
3.3. Simple Sequence Repeats and Codon Bias
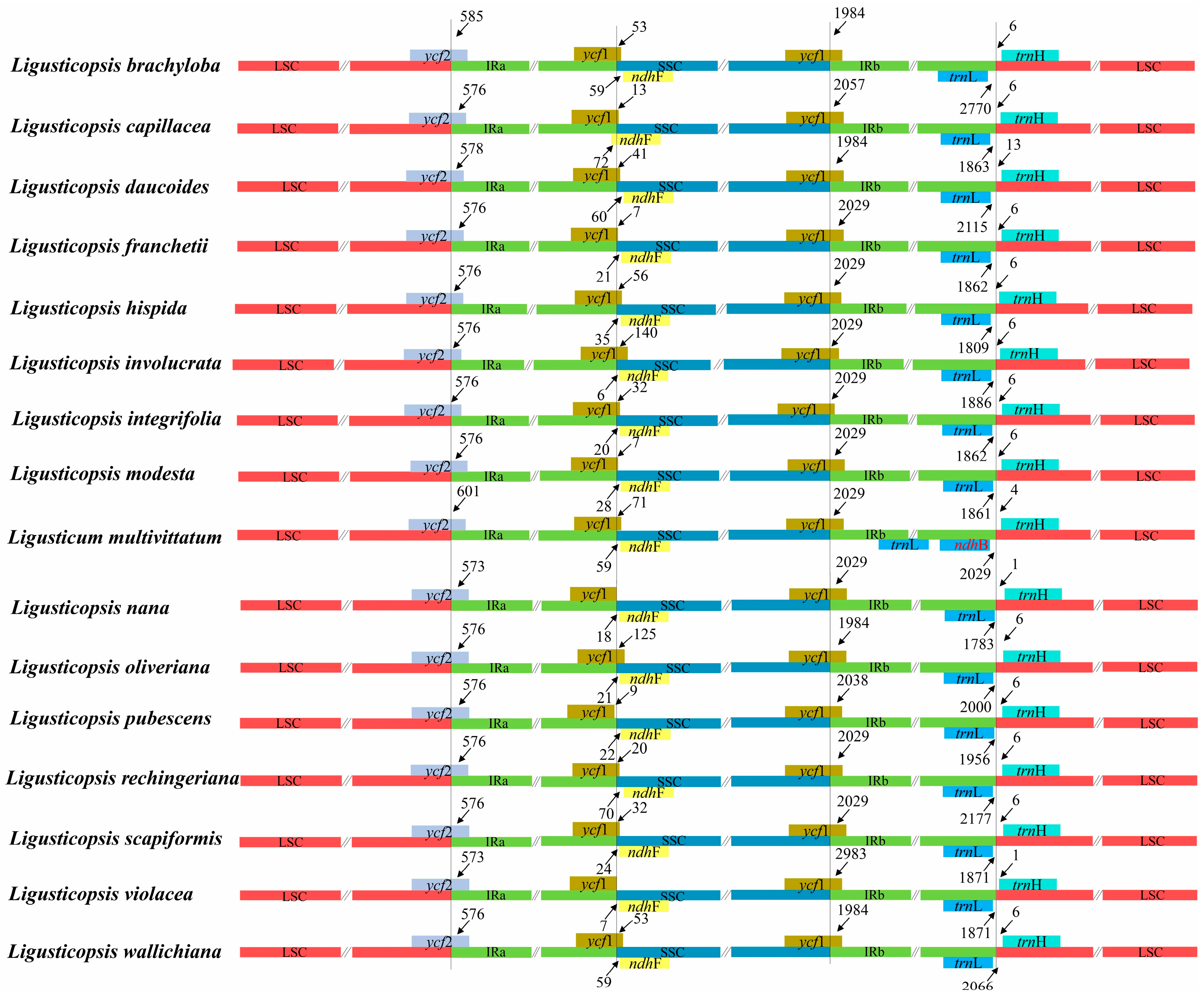
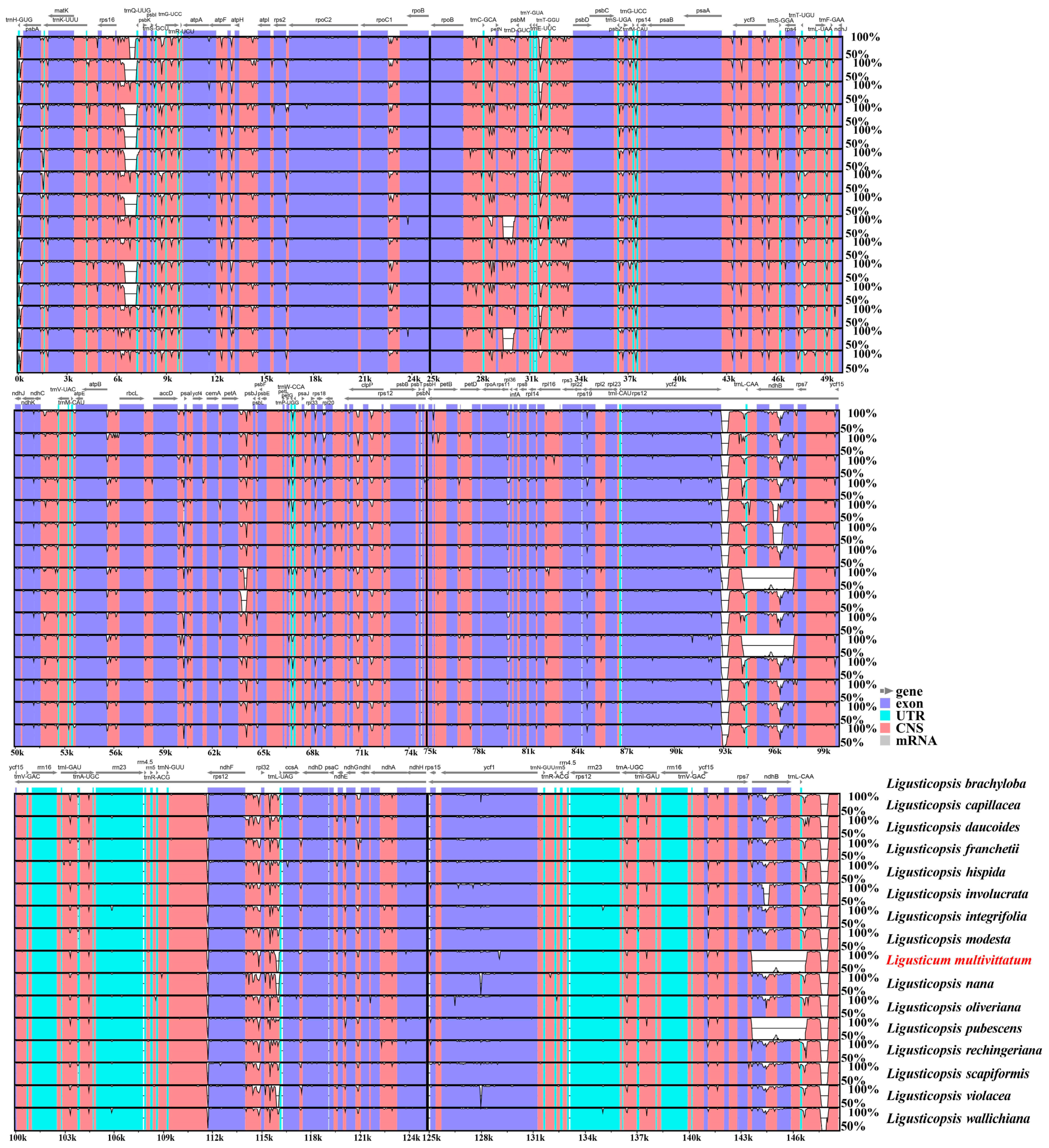
3.4. Plastome Comparison
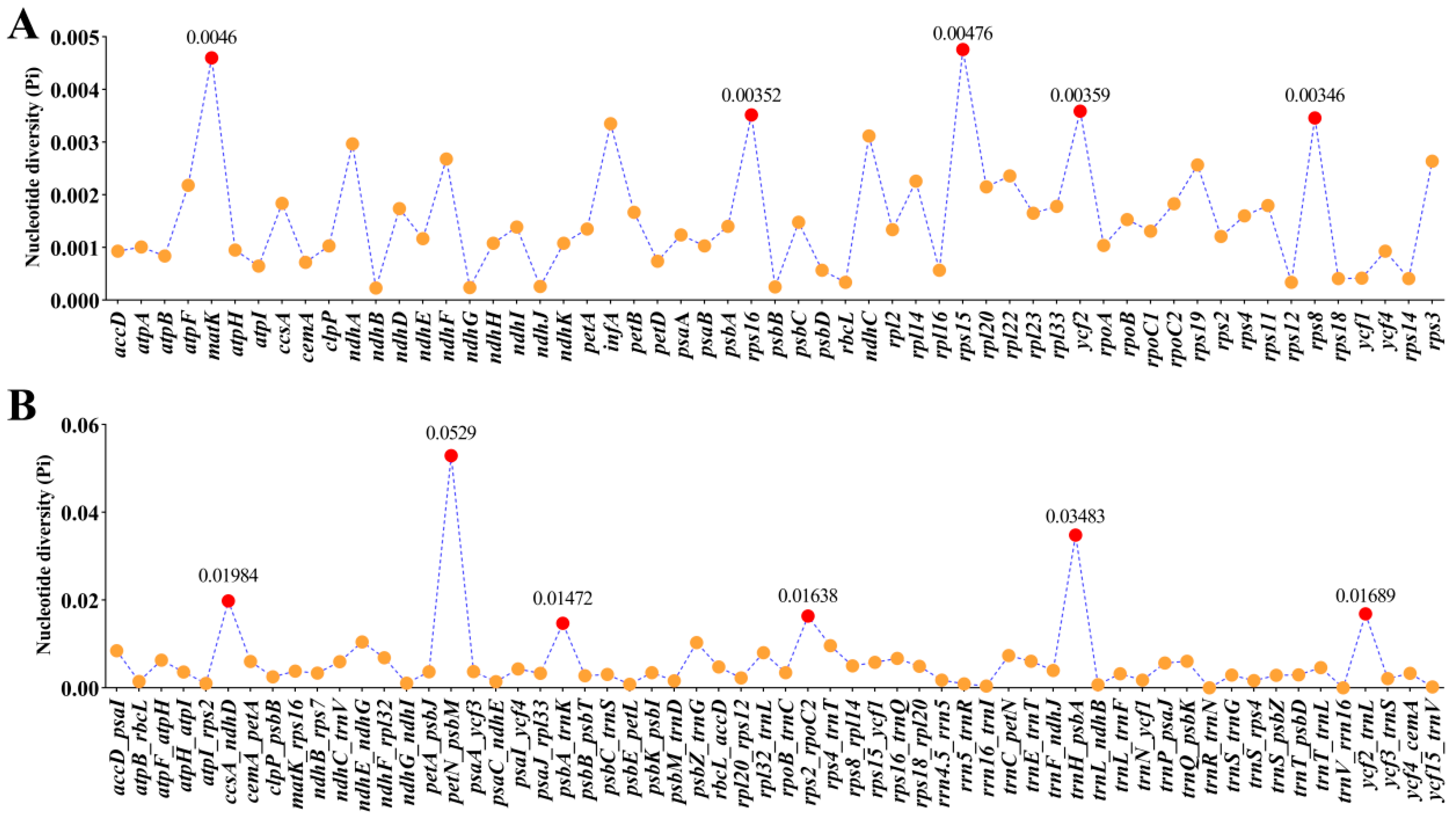
3.5. Nucleotide Diversity (Pi) Analysis
4. Discussion
4.1. Plastome Comparative Analyses
4.2. Phylogenetic Inference and Taxonomic Implication of Ligusticum Multivittatum
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- von Linnaeus, C. Species Plantarum; Laurentii Salvii: Stockholm, Sweden, 1753. [Google Scholar]
- Koch, W.D.J. Generum tribuumque plantarum Umbelliferarum nova disposition. Nova Acta Acad. Caesareae Leopold. Carol. Ger. Nat. Curiosorum 1824, 12, 55–156. [Google Scholar]
- Pu, F.D.; Watson, M.F. Ligusticum L. Flora of China; Science Press: Beijing, China, 2005; Volume 14, pp. 140–150. [Google Scholar]
- Pu, F.D. A revision of the genus Ligusticum L. (Umbelliferae) in China. Acta Phytotax Sin. 1991, 29, 385–393. [Google Scholar]
- Sun, N.; He, X.J.; Zhou, S.D. Morphological cladistic analysis of Ligusticum (Umbelliferae) in China. Nord. J. Bot. 2008, 26, 118–128. [Google Scholar] [CrossRef]
- Zhang, H.C. Ligusticum L. In Flora Reipublicae Popularis Sinicae; Shan, R.H., She, M.L., Eds.; Science Press: Beijing, China, 1985; Volume 55, pp. 234–257. [Google Scholar]
- Leute, G.H. Untersuchungen über den Verwandtschaftskreis der Gattung Ligusticum L. (Umbelliferae). Teil I. Ann Naturhist Mus Wien. 1969, 73, 55–98. [Google Scholar]
- Pimenov, M.G. Updated checklist of Chinese Umbelliferae: Nomenclature, synonymy, typification, distribution. Turczaninowia 2017, 20, 106–239. [Google Scholar] [CrossRef]
- Li, Z.X.; Guo, X.L.; Price, M.; Zhou, S.D.; He, X.J. Phylogenetic position of Ligusticopsis (Apiaceae, Apioideae): Evidence from molecular data and carpological characters. AoB Plants 2022, 14, plac008. [Google Scholar] [CrossRef]
- Ren, T.; Xie, D.F.; Peng, C.; Gui, L.J.; Price, M.; Zhou, S.D.; He, X.J. Molecular evolution and phylogenetic relationships of Ligusticum (Apiaceae) inferred from the whole plastome sequences. BMC Ecol. Evol. 2022, 22, 55. [Google Scholar] [CrossRef]
- Weng, L.; Jiang, Y.H.; Wang, Y.; Zhang, X.M.; Zhou, P.; Wu, M.; Li, H.Z.; Sun, H.; Chen, S.T. Chloroplast genome characteristics and phylogeny of the sinodielsia clade (Apiaceae: Apioideae). BMC Plant Biol. 2023, 23, 284. [Google Scholar] [CrossRef]
- Song, B.N.; Liu, C.K.; Xie, D.F.; Xiao, Y.L.; Tian, R.M.; Li, Z.X.; Zhou, S.D.; He, X.J. Plastid phylogenomic analyses reveal the taxonomic position of Peucedanum franchetii. Plants 2023, 12, 97. [Google Scholar] [CrossRef]
- Liu, C.K.; Deng, J.J.; Zhou, R.X.; Song, B.N.; Zhou, S.D.; He, X.J. Plastid Phylogenomics Provide Evidence to Accept Two New Members of Ligusticopsis (Apiaceae, Angiosperms). Int. J. Mol. Sci. 2023, 24, 382. [Google Scholar] [CrossRef]
- Xie, D.F.; Tan, J.B.; Yu, Y.; Gui, L.J.; Su, D.M.; Zhou, S.D.; He, X.J. Insights into phylogeny, age, and evolution of Allium (Amaryllidaceae) based on the whole plastome sequences. Ann. Bot. 2020, 125, 1039–1055. [Google Scholar] [CrossRef]
- Li, J.; Cai, J.; Qin, H.H.; Price, M.; Zhang, Z.; Yu, Y.; Xie, D.F.; He, X.J.; Zhou, S.D.; Gao, X.F. Phylogeny, Age, and Evolution of Tribe Lilieae (Liliaceae) Based on Whole Plastid Genomes. Front. Plant Sci. 2022, 12, 699226. [Google Scholar] [CrossRef]
- Wang, M.L.; Wang, X.; Sun, J.H.; Wang, Y.H.; Ge, Y.; Dong, W.P.; Yuan, Q.J.; Huang, L.Q. Phylogenomic and evolutionary dynamics of inverted repeats across Angelica plastomes. BMC Plant Biol. 2021, 21, 26. [Google Scholar] [CrossRef]
- Metzker, M.L. Sequencing technologies—The next generation. Nature reviews. Genetics 2010, 11, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Qin, H.H.; Lei, J.Q.; Liu, C.K.; He, X.J.; Zhou, S.D. The phylogeny of Seseli (Apiaceae, Apioideae): Insights from molecular and morphological data. BMC Plant Biol. 2022, 22, 534. [Google Scholar] [CrossRef]
- Song, B.N.; Liu, C.K.; Ren, T.; Xiao, Y.L.; Chen, L.; Xie, D.F.; He, A.G.; Xu, P.; Fan, X.; Zhou, S.D.; et al. Plastid phylogenomics contributes to the taxonomic revision of taxa within the genus Sanicula L. and acceptance of two new members of the genus. Front. Plant Sci. 2024, 15, 1351023. [Google Scholar] [CrossRef]
- Tseng, Y.H.; Chien, H.C.; Zhu, G.X. Comparative plastome analyses and phylogenetic insights of Elatostema. BMC Plant Biol. 2025, 25, 537. [Google Scholar] [CrossRef]
- Chen, K.Y.; Wang, J.D.; Xiang, R.Q.; Yang, X.D.; Yun, Q.Z.; Huang, Y.; Sun, H.; Chen, J.H. Backbone phylogeny of Salix based on genome skimming data. Plant Divers. 2025, 47, 178–188. [Google Scholar] [CrossRef]
- Chen, L.; Song, B.N.; Yang, L.; Wang, Y.; Wang, Y.Y.; Aou, X.Y.M.; He, X.J.; Zhou, S.D. Phylogeny, adaptive evolution, and taxonomy of Acronema (Apiaceae): Evidence from plastid phylogenomics and morphological data. Front. Plant Sci. 2024, 15, 1425158. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.; Huelsenbeck, J.P. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Posada, D.; Crandall, K.A. MODELTEST: Testing the model of DNA substitution. Bioinformatics 1998, 14, 817–818. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A. FigTree, Version 1.4.2. 2015. Available online: http://tree.bio.ed.ac.uk/software/fgtree/ (accessed on 1 May 2025).
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef]
- Thiel, T.; Michalek, W.; Varshney, R.K.; Graner, A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Angew. Genet. 2003, 106, 411–422. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlic, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Peden, J.F. Analysis of Codon Usage. Ph.D. Thesis, Nottingham University: Department of Genetics, Nottingham, UK, 1999. [Google Scholar]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Gitzendanner, M.A.; Soltis, P.S.; Yi, T.S.; Li, D.Z.; Soltis, D.E. Plastome phylogenetics: 30 years of inferences into plant evolution. In Advances in Botanical Research; Academic Press: Cambridge, MA, USA, 2018; pp. 293–313. [Google Scholar]
- Yao, G.; Jin, J.J.; Li, H.T.; Yang, J.B.; Mandala, V.S.; Croley, M.; Mostow, R.; Douglas, A.N.; Chase, W.M.; Christenhusz, J.M.; et al. Plastid phylogenomic insights into the evolution of Caryophyllales. Mol. Phylogenet. Evol. 2019, 134, 74–86. [Google Scholar] [CrossRef]
- Song, B.N.; Liu, C.K.; Zhao, A.Q.; Tian, R.M.; Xie, D.F.; Xiao, Y.L.; Chen, H.; Zhou, S.D.; He, X.J. Phylogeny and diversification of genus Sanicula L. (Apiaceae): Novel insights from plastid phylogenomic analyses. BMC Plant Biol. 2024, 24, 70. [Google Scholar] [CrossRef]
- Liu, L.J.; Liu, C.K.; Cai, J.; Deng, J.J.; He, X.J.; Zhou, S.D. The complete plastomes of thirteen Libanotis (Apiaceae, Apioideae) plants: Comparative and phylogenetic analyses provide insights into the plastome evolution and taxonomy of Libanotis. BMC Plant Biol. 2024, 24, 106. [Google Scholar] [CrossRef]
- Liu, C.K.; Lei, J.Q.; Jiang, Q.P.; Zhou, S.D.; He, X.J. The complete plastomes of seven Peucedanum plants: Comparative and phylogenetic analyses for the Peucedanum genus. BMC Plant Biol. 2022, 22, 101. [Google Scholar] [CrossRef]
- Lei, J.Q.; Liu, C.K.; Cai, J.; Price, M.; Zhou, S.D.; He, X.J. Evidence from phylogenomics and morphology provide insights into the phylogeny, plastome evolution, and taxonomy of Kitagawia. Plants 2022, 11, 3275. [Google Scholar] [CrossRef]
- Hollingsworth, P.M.; Li, D.Z.; van der Bank, M.; Twyford, A.D. Telling plant species apart with DNA: From barcodes to genomes. Philosophical transactions of the Royal Society of London. Ser. B Biol. Sci. 2016, 371, 20150338. [Google Scholar] [CrossRef]
- Zhou, J.; Gao, Y.Z.; Wei, J.; Liu, Z.W.; Downie, S.R. Molecular phylogenetics of Ligusticum (Apiaceae) based on nrDNA ITS sequences: Rampant polyphyly, placement of the Chinese endemic species, and a much-reduced circumscription of the genus. Int. J. Plant Sci. 2020, 181, 306–323. [Google Scholar] [CrossRef]
- Jugal, V.; Shrishail, H.C.; Singh, P.S.; Shah, K.; Chauhan, N.S. Molecular phylogeny of genus Nothopegia Blume nom. cons.(Anacardiaceae) in Central Western Ghats Karnataka, based on the ITS, rbcL and trnLF markers. Ecol. Genet. Genom. 2025, 36, 100379. [Google Scholar] [CrossRef]
- Chen, Z.; Gao, L.; Wang, H.; Feng, S. Molecular identification and phylogenetic analysis of Cymbidium species (Orchidaceae) based on the potential DNA barcodes matK, rbcL, psbA-trnH, and internal transcribed spacer. Agronomy 2024, 14, 933. [Google Scholar] [CrossRef]
- Gou, W.; Guo, X.L.; Zhou, S.D.; He, X.J. Phylogeny and taxonomy of Meeboldia, Sinodielsia and their relatives (Apiaceae: Apioideae) inferred from nrDNA ITS, plastid DNA intron (rpl16 and rps16) sequences and morphological characters. Phytotaxa 2021, 482, 121–142. [Google Scholar] [CrossRef]
- Peng, C.; Guo, X.L.; Zhou, S.D.; He, X.J. Backbone phylogeny and adaptive evolution of Pleurospermum sl.: New insights from phylogenomic analyses of complete plastome data. Front. Plant Sci. 2023, 14, 1148303. [Google Scholar] [CrossRef]
- Gui, L.J.; Xie, D.F.; Peng, C.; Ren, T.; Yu, L.Y.; Zhou, S.D.; He, X.J. Chloroplast genomes and ribosomal DNA provide insights into divergence and morphological evolution of alpine Tongoloa. J. Syst. Evol. 2023, 63, 72–84. [Google Scholar] [CrossRef]
- Guo, X.L.; Gou, W.; Price, M.; Jiang, Q.P.; Peng, C.; Zhou, S.D.; He, X.J. Reinterpreting the phylogenetic position and taxonomic revision of the genus Pterocyclus (Apiaceae, Apioideae) based on nrITS, complete plastid genome, and morphological evidence. J. Syst. Evol. 2023, 62, 384–402. [Google Scholar] [CrossRef]
- Liu, C.K.; Deng, J.J.; Song, B.N.; Qin, H.H.; Zhou, S.D.; He, X.J. Plastid phylogenomics provide evidence to accept a new genus Pseudopeucedanum (Apiaceae) separated from Peucedanum sl. Bot. J. Linn. Soc. 2024, 205, 243–252. [Google Scholar] [CrossRef]
- Song, B.N.; Aou, X.Y.M.; Tian, R.M.; Cai, J.; Tan, W.Y.; Liu, C.K.; He, X.J.; Zhou, S.D. Morphology, phylogeography, phylogeny, and taxonomy of Cyclorhiza (Apiaceae). Front. Plant Sci. 2025, 15, 1504734. [Google Scholar] [CrossRef]



| Taxon | Length (bp) | Number of Genes (Unique) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Genome | LSC | SSC | IR | Total | CDS | rRNA | tRNA | GC (%) | |
| Ligusticopsis brachyloba | 148,633 | 92,265 | 17,588 | 19,390 | 113 | 79 | 4 | 30 | 37.40% |
| Ligusticopsis capillacea | 147,808 | 91,907 | 17,503 | 19,199 | 113 | 79 | 4 | 30 | 37.50% |
| Ligusticopsis daucoides | 148,078 | 91,666 | 17,582 | 19,415 | 113 | 79 | 4 | 30 | 37.40% |
| Ligusticopsis franchetii | 148,281 | 92,298 | 17,691 | 19,146 | 113 | 79 | 4 | 30 | 37.50% |
| Ligusticopsis hispida | 147,797 | 91,846 | 17,627 | 19,162 | 113 | 79 | 4 | 30 | 37.40% |
| Ligusticopsis involucrata | 147,752 | 91,782 | 17,560 | 19,205 | 113 | 79 | 4 | 30 | 37.40% |
| Ligusticopsis intergrifolia | 148,196 | 92,305 | 17,575 | 19,158 | 113 | 79 | 4 | 30 | 37.50% |
| Ligusticopsis modesta | 148,133 | 92,247 | 17,568 | 19,159 | 113 | 79 | 4 | 30 | 37.50% |
| Ligusticopsis nana | 146,900 | 91,480 | 17,308 | 19,056 | 113 | 79 | 4 | 30 | 37.50% |
| Ligusticopsis oliveriana | 148,175 | 92,273 | 17,534 | 19,184 | 113 | 79 | 4 | 30 | 37.50% |
| Ligusticopsis pubescens | 148,260 | 91,819 | 17,619 | 19,411 | 113 | 79 | 4 | 30 | 37.4% |
| Ligusticopsis rechingeriana | 148,525 | 91,813 | 17,654 | 19,529 | 113 | 79 | 4 | 30 | 37.30% |
| Ligusticopsis scapiformis | 148,107 | 92,214 | 17,581 | 19,156 | 113 | 79 | 4 | 30 | 37.50% |
| Ligusticopsis violacea | 148,190 | 91,811 | 16,335 | 20,022 | 113 | 79 | 4 | 30 | 37.50% |
| Ligusticopsis wallichiana | 148,594 | 92,281 | 17,567 | 19,373 | 113 | 79 | 4 | 30 | 37.40% |
| Ligusticum multivittatum | 148,262 | 91,571 | 17,631 | 19,530 | 113 | 79 | 4 | 30 | 37.40% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Song, B.; Yong, F.; Xu, C.; Dong, Q.; Wang, X.; Sun, C.; Wang, Z. Phylogenetic Analyses and Plastome Comparison to Confirm the Taxonomic Position of Ligusticum multivittatum (Apiaceae, Apioideae). Genes 2025, 16, 823. https://doi.org/10.3390/genes16070823
Liu C, Song B, Yong F, Xu C, Dong Q, Wang X, Sun C, Wang Z. Phylogenetic Analyses and Plastome Comparison to Confirm the Taxonomic Position of Ligusticum multivittatum (Apiaceae, Apioideae). Genes. 2025; 16(7):823. https://doi.org/10.3390/genes16070823
Chicago/Turabian StyleLiu, Changkun, Boni Song, Feng Yong, Chengdong Xu, Quanying Dong, Xiaoyi Wang, Chao Sun, and Zhenji Wang. 2025. "Phylogenetic Analyses and Plastome Comparison to Confirm the Taxonomic Position of Ligusticum multivittatum (Apiaceae, Apioideae)" Genes 16, no. 7: 823. https://doi.org/10.3390/genes16070823
APA StyleLiu, C., Song, B., Yong, F., Xu, C., Dong, Q., Wang, X., Sun, C., & Wang, Z. (2025). Phylogenetic Analyses and Plastome Comparison to Confirm the Taxonomic Position of Ligusticum multivittatum (Apiaceae, Apioideae). Genes, 16(7), 823. https://doi.org/10.3390/genes16070823





