Comparison of Chloroplast Genome Sequences of Saxifraga umbellulata var. pectinata in Qinghai–Xizang Plateau
Abstract
1. Introduction
2. Materials and Methods
2.1. Test Materials
2.2. DNA Extraction and Sequencing
2.3. Chloroplast Genome Assembly and Annotation
2.4. Analysis of Codon Usage Frequency and Simple Repeat Sequence (SSR) of Chloroplast Genome
2.5. Comparative Analysis of the Whole Chloroplast Genome of Saxifraga
2.6. Phylogenetic Analysis
3. Results
3.1. Basic Characteristics of Chloroplast Genome of S. umbellulata var. pectinata
3.2. Relative Synonymous Codon Usage Frequency (RSCU), Repetitive Sequence, and SSR Analysis of S. umbellulata var. pectinata
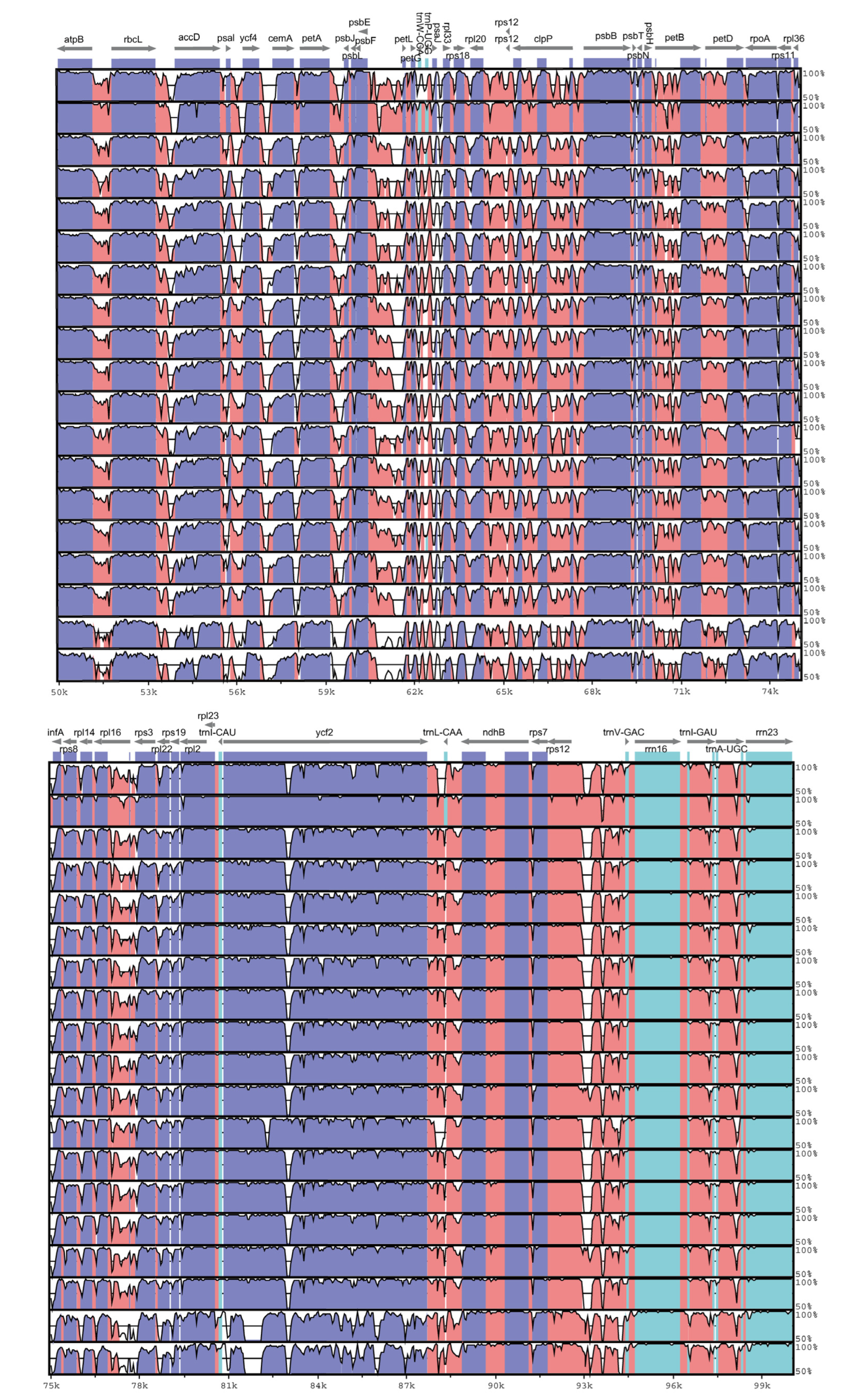
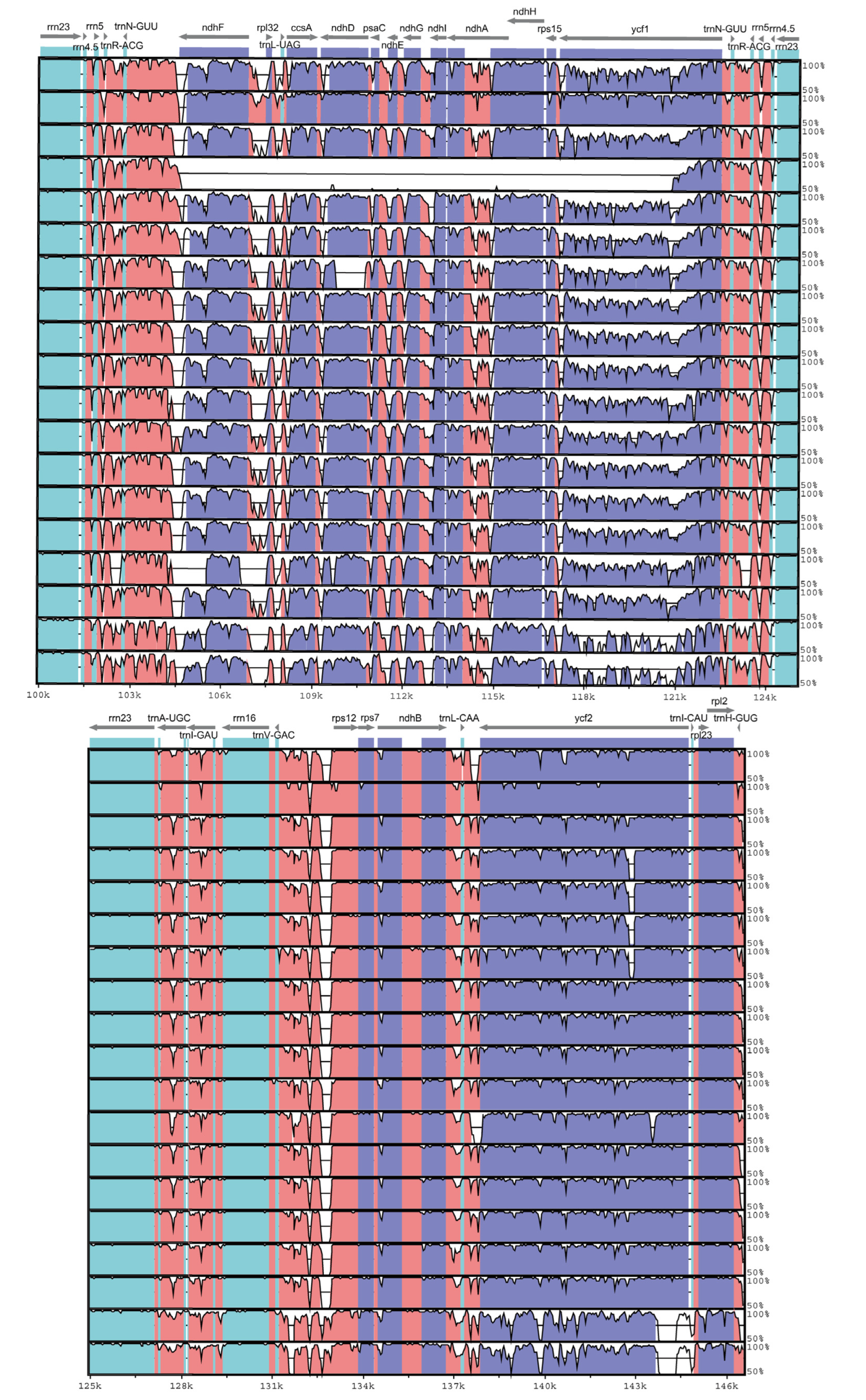
3.3. Sequence Analysis of Chloroplast Genome of S. umbellulata var. pectinata and Its Related Taxa
3.4. Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, X.J.; Jiang, K.Z.; Kuai, X.Y.; Chen, J.T.; Luo, P.R.; Sun, H.; Deng, T. Taxonomic resurrection of Saxifraga lancangensis (Saxifragaceae). Bot. Stud. 2024, 65, 12. [Google Scholar] [CrossRef] [PubMed]
- Thach, N.; Roser, M.; Miehe, G. Molecular phylogenetics, morphology and a revised classification of the complex genus Saxifra-ga (Saxifragaceae). Taxon 2015, 64, 1159–1187. [Google Scholar] [CrossRef]
- Ebersbach, J.; Muellner-Riehl, A.N.; Michalak, I. In and out of the Qinghai-Tibet Plateau: Divergence time estimation and historical biogeography of the large arctic-alpine genus Saxifraga L. J. Biogeogr. 2017, 44, 900–910. [Google Scholar] [CrossRef]
- Ebersbach, J.; Schnitzler, J.; Favre, A. Evolutionary radiations in the species-rich mountain genus Saxifraga L. BMC Evol. Biol. 2017, 17, 119. [Google Scholar] [CrossRef]
- Zhao, W.Y.; Meng, K.K.; Fan, Q.; Jin, J.H.; Liao, W.B. Saxifraga damingshanensis (S. sect. Irregulares, Saxifragaceae), a new species from Guangxi, China. PhytoKeys 2019, 133, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.M.; Zhong, G.Y.; Jiang, W.; Ren, G. Analysis of varieties and standards of Saxifragaceae medicinal plants used in Tibetan medicine. China J. Chin. Mater. Medica 2021, 46, 488–493. (In Chinese) [Google Scholar] [CrossRef]
- Jiang, Y.; Zhong, M.; Zhan, H.; Tao, X.; Zhang, Y.; Mao, J.; Geng, Z.; Gao, B. Integrated strategy of network pharmacology, molecular docking, HPLC-DAD and mice model for exploring active ingredients and pharmacological mechanisms of Penthorum chinense Pursh against alcoholic liver injury. J. Ethnopharmacol. 2022, 298, 115589. [Google Scholar] [CrossRef]
- Ksouda, G.; Sellimi, S.; Merlier, F.; Falcimaigne-Cordin, A.; Thomasset, B.; Nasri, M.; Hajji, M. Composition, antibacterial and antioxidant activities of Pimpinella saxifraga essential oil and application to cheese preservation as coating additive. Food Chem. 2019, 288, 47–56. [Google Scholar] [CrossRef]
- Wu, X.D.; Chen, H.G.; Zhou, X.; Huang, Y.; Hu, E.M.; Jiang, Z.M.; Zhao, C.; Gong, X.J.; Deng, Q.F. Studies on Chromatographic Fingerprint and Fingerprinting Profile-Efficacy Relationship of Saxifraga stolonifera Meerb. Molecules 2015, 20, 22781–22798. [Google Scholar] [CrossRef]
- Parmar, R.; Cattonaro, F.; Phillips, C.; Vassiliev, S.; Morgante, M.; Rajora, O.P. Assembly and Annotation of Red Spruce (Picea rubens) Chloroplast Genome, Identification of Simple Sequence Repeats, and Phylogenetic Analysis in Picea. Int. J. Mol. Sci. 2022, 23, 15243. [Google Scholar] [CrossRef]
- Chen, J.; Huang, T.; Fan, H.; Lin, F.; Ma, H.; Cao, J.; Chai, T.; Ma, L.; Wang, H. A Comparative Phylogenetic Analysis on the Chloroplast Genome in Different Reynoutria japonica Populations. Genes 2022, 13, 1979. [Google Scholar] [CrossRef] [PubMed]
- Twyford, A.D.; Ness, R.W. Strategies for complete plastid genome sequencing. Mol. Ecol. Resour. 2017, 17, 858–868. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Yu, X.; Yang, Y.; Wei, P.; Zhang, W.; Li, X.; Liu, C.; Zhao, S.; Li, X.; Liu, X. Comparative Analysis of Chloroplast Genomes within Saxifraga (Saxifragaceae) Takes Insights into Their Genomic Evolution and Adaption to the High-Elevation Environment. Genes 2022, 13, 1673. [Google Scholar] [CrossRef]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Cock, P.J.; Fields, C.J.; Goto, N.; Heuer, M.L.; Rice, P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010, 38, 1767–1771. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Brudno, M.; Malde, S.; Poliakov, A.; Do, C.B.; Couronne, O.; Dubchak, I.; Batzoglou, S. Glocal alignment: Finding rearrangements during alignment. Bioinformatics 2003, 19 (Suppl. S1), i54–i62. [Google Scholar] [CrossRef]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 2018, 34, 3030–3031. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Gerschwitz-Eidt, M.A.; Dillenberger, M.S.; Kadereit, J.W. Phylogeny of Saxifraga section Saxifraga subsection Arachnoideae (Saxifragaceae) and the origin of low elevation shade-dwelling species. Ecol. Evol. 2023, 13, e9728. [Google Scholar] [CrossRef]
- Zhang, Y.; Tian, L.; Lu, C. Chloroplast gene expression: Recent advances and perspectives. Plant Commun. 2023, 4, 100611. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.F.; Zhang, Y.X.; Guo, Z.H.; Li, D.Z. Evidence for horizontal transfer of mitochondrial DNA to the plastid genome in a bamboo genus. Sci. Rep. 2015, 5, 11608. [Google Scholar] [CrossRef]
- Guisinger, M.M.; Kuehl, J.V.; Boore, J.L.; Jansen, R.K. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: Rearrangements, repeats, and codon usage. Mol. Biol. Evol. 2011, 28, 583–600. [Google Scholar] [CrossRef] [PubMed]
- Yuan, R.; Ma, X.; Zhang, Z.; Gornall, R.J.; Wang, Y.; Chen, S.; Gao, Q. Chloroplast phylogenomics and the taxonomy of Saxifraga section Ciliatae (Saxifragaceae). Ecol. Evol. 2023, 13, e9694. [Google Scholar] [CrossRef]
- Huang, Y.; Li, J.; Yang, Z.; An, W.; Xie, C.; Liu, S.; Zheng, X. Comprehensive analysis of complete chloroplast genome and phylogenetic aspects of ten Ficus species. BMC Plant Biol. 2022, 22, 253. [Google Scholar] [CrossRef] [PubMed]







| Category | Gene Group | Gene Name |
|---|---|---|
| Subunits of photosystem I | PsaA, psaB, psaC, psaI, psaJ | |
| Subunits of photosystem II | PsbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Subunits of NADH dehydrogenase | ndhA*, ndhB*(2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Photosynthesis | Subunits of cytochrome b/f complex | petA, petB*, petD*, petG, petL, petN |
| Subunits of ATP synthase | atpA, atpB, atpE, atpF*, atpH, atpI | |
| Large subunit of rubisco | rbcL | |
| Subunits photochlorophyllide reductase | - | |
| Proteins of large ribosomal subunit | rpl14, rpl16*, rpl2(2), rpl20, rpl22, rpl23(2), rpl32, rpl33, rpl36 | |
| Proteins of small ribosomal subunit | rps11, rps12**(2), rps14, rps15, rps16*, rps18, rps19, rps2, rps3, rps4, rps7(2), rps8 | |
| Expression of related genes | Subunits of RNA polymerase | rpoA, rpoB, rpoC1*, rpoC2 |
| Ribosomal RNAs | rrn16(2), rrn23(2), rrn4.5(2), rrn5(2) | |
| Transfer RNAs | trnA-UGC*(2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC*, trnH-GUG, trnI-CAU(2), trnI-GAU*(2), trnK-UUU*, trnL-CAA(2), trnL-UAA*, trnL-UAG, trnM-CAU, trnN-GUU(2), trnP-UGG, trnQ-UUG, trnR-ACG(2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(2), trnV-UAC*, trnW-CCA, trnY-GUA, trnfM-CAU | |
| Other genes | Envelope membrane protein | cemA |
| Acetyl-CoA carboxylase | accD | |
| c-type cytochrome synthesis gene | ccsA | |
| Translation initiation factor | infA | |
| Maturase | matK | |
| Protease | clpP** | |
| Conserved hypothetical chloroplast ORF | ycf1(2), ycf2(2), ycf3**, ycf4 |
| Repeat Types | Repeat Sequence | Repeat Time | Totality | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | |||
| Mononucleotide | A/T | 37 | 32 | 17 | 11 | 6 | 4 | 2 | 2 | 1 | 112 | |||||
| C/G | 1 | 3 | 1 | 5 | ||||||||||||
| Dinucleotide | AG/CT | 17 | 1 | 18 | ||||||||||||
| AT/AT | 16 | 9 | 4 | 29 | ||||||||||||
| Trinucleotide | AAT/ATT | 1 | 1 | 2 | ||||||||||||
| ATC/ATG | 1 | 1 | ||||||||||||||
| Tetranucleotide | AAAG/CTTT | 2 | 2 | |||||||||||||
| AACC/GGTT | 1 | 1 | ||||||||||||||
| ACAT/ATGT | 1 | 1 | ||||||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Su, K.; Li, Q.; Sun, R.; Liu, H.; Du, J.; Li, J.; Liu, L. Comparison of Chloroplast Genome Sequences of Saxifraga umbellulata var. pectinata in Qinghai–Xizang Plateau. Genes 2025, 16, 789. https://doi.org/10.3390/genes16070789
Wang C, Su K, Li Q, Sun R, Liu H, Du J, Li J, Liu L. Comparison of Chloroplast Genome Sequences of Saxifraga umbellulata var. pectinata in Qinghai–Xizang Plateau. Genes. 2025; 16(7):789. https://doi.org/10.3390/genes16070789
Chicago/Turabian StyleWang, Cui, Kaidi Su, Qiwen Li, Rui Sun, Haoyu Liu, Jingxuan Du, Jinping Li, and Likuan Liu. 2025. "Comparison of Chloroplast Genome Sequences of Saxifraga umbellulata var. pectinata in Qinghai–Xizang Plateau" Genes 16, no. 7: 789. https://doi.org/10.3390/genes16070789
APA StyleWang, C., Su, K., Li, Q., Sun, R., Liu, H., Du, J., Li, J., & Liu, L. (2025). Comparison of Chloroplast Genome Sequences of Saxifraga umbellulata var. pectinata in Qinghai–Xizang Plateau. Genes, 16(7), 789. https://doi.org/10.3390/genes16070789





