Genome-Wide Analysis of the Typical Thioredoxin Gene Family in Hexaploid Oil-Camellia: Identification, Phylogenetic Analysis, and Gene Expression Patterns
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome-Wide Identification of TRXs Genes in the Camellia oleifera Genome
2.2. Analysis of Conserved Motifs and Structural Domains of Typical TRX Protein in Camellia oleifera
2.3. Protein Sequence Analysis and cis-Regulatory Elements Analysis of Typical CoTRX Promoters
2.4. Phylogenetic and Molecular Evolution Analyses
2.5. Gene Expression and Protein–Protein Interaction Network Analyses
2.6. Gene Cloning and Transgenic Plant Construction
3. Results
3.1. Genome-Wide Identification and Physicochemical Properties of Typical TRX in Hexaploid C. oleifera Genome
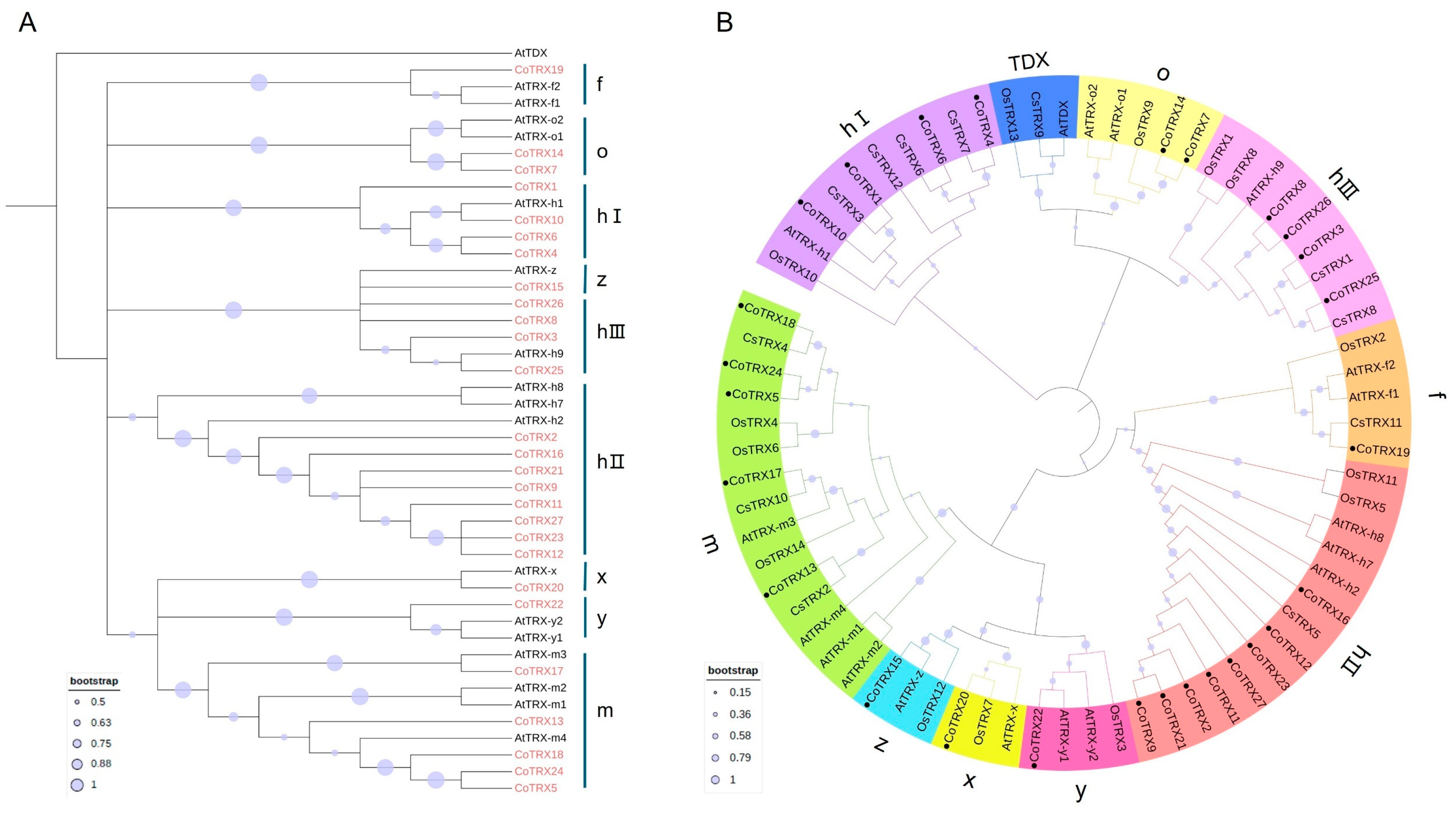
3.2. Evolutionary Analysis of Typical TRX Gene Family in Camellia oleifera
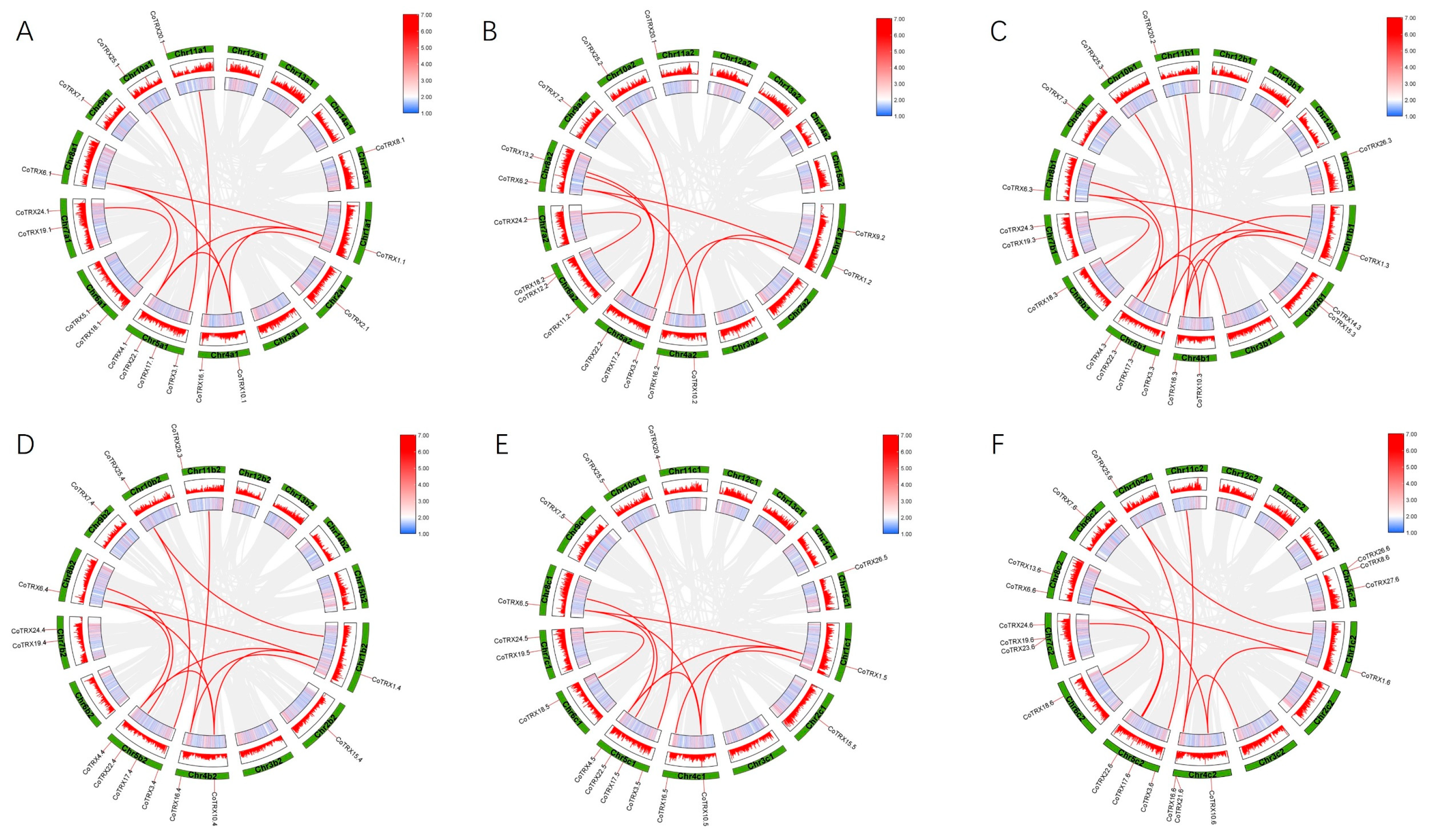
3.3. Collinearity and Selective Pressure Analysis
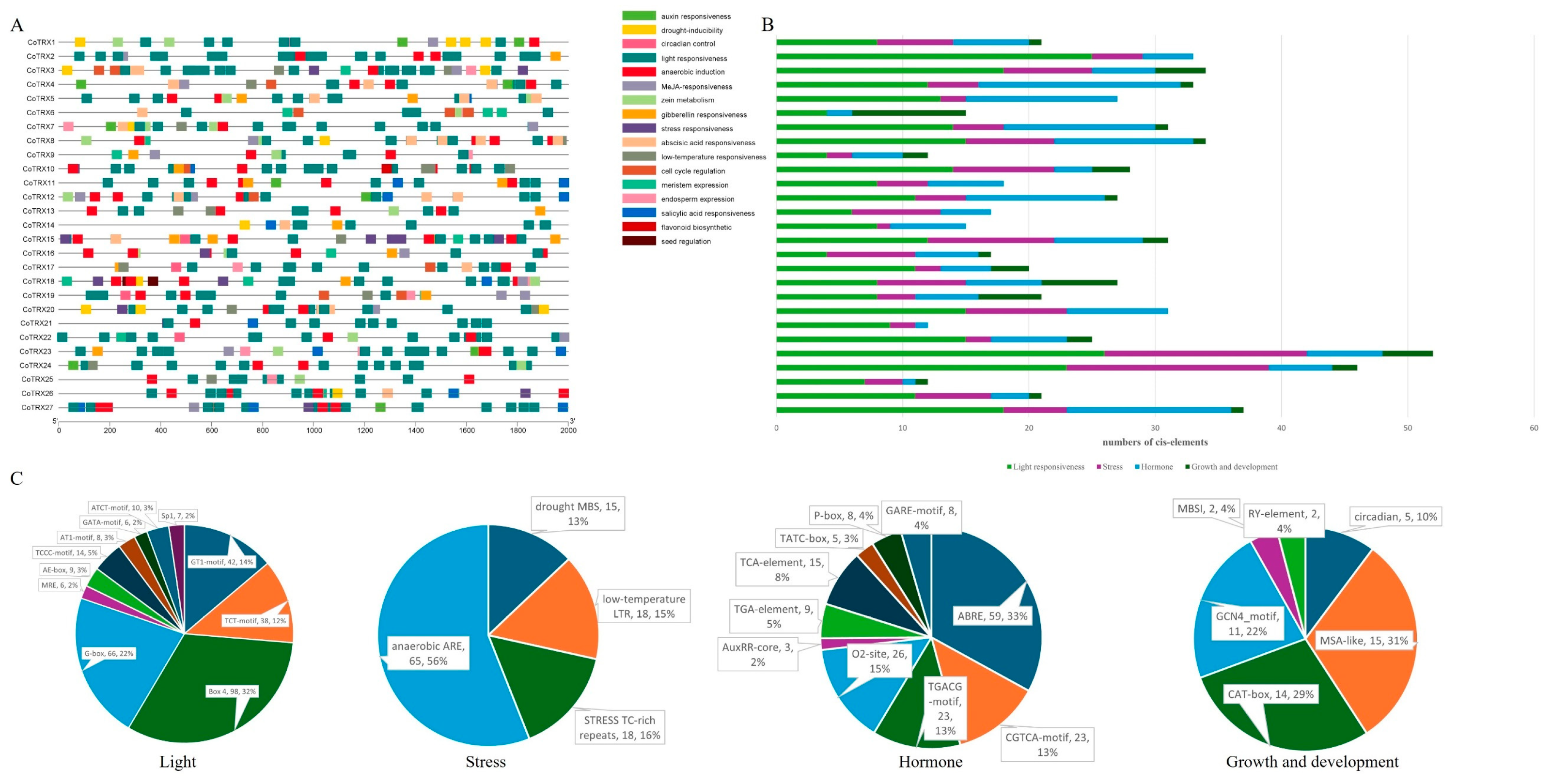
3.4. cis-Regulatory Element Analysis of Typical CoTRX Promoters
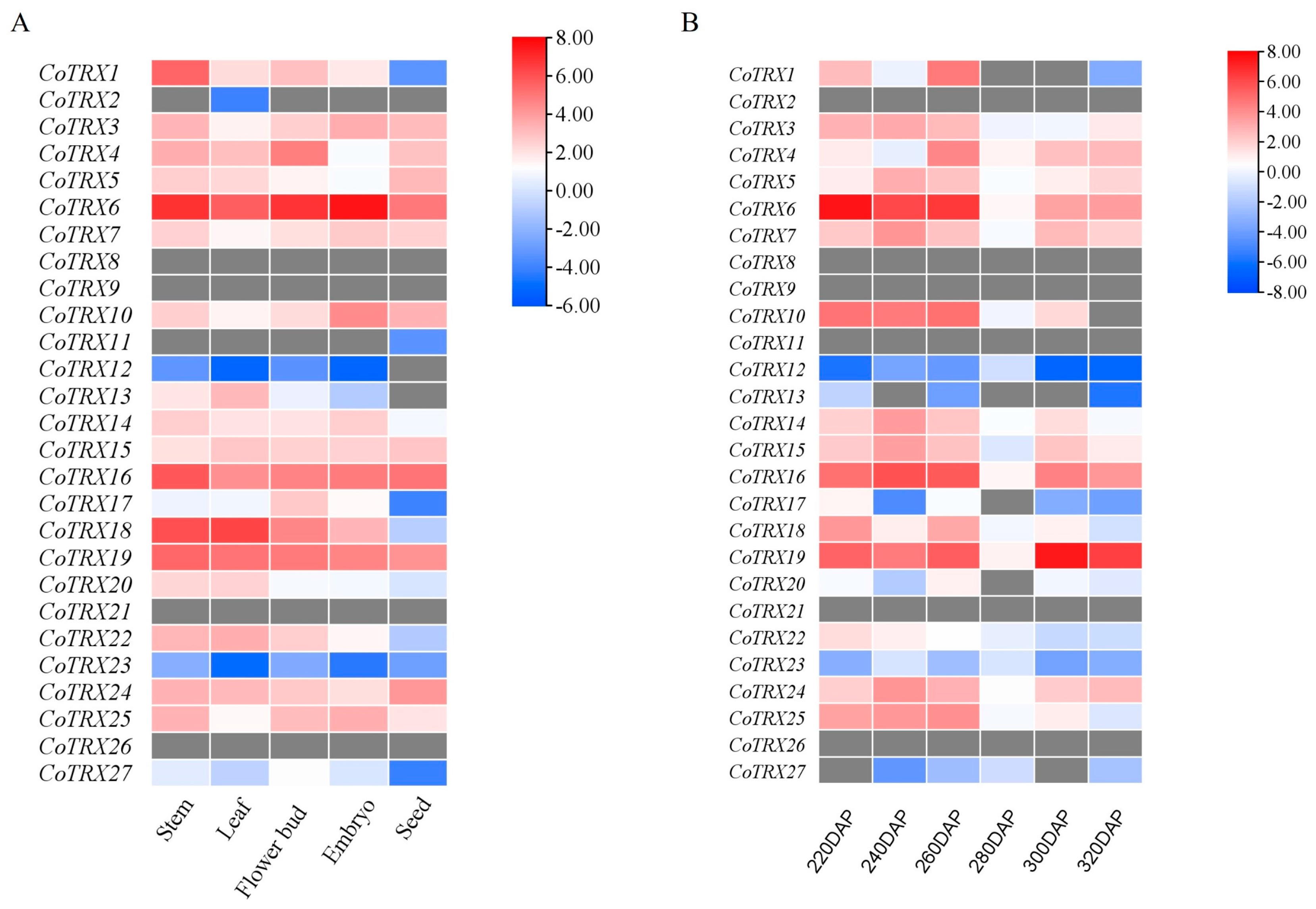
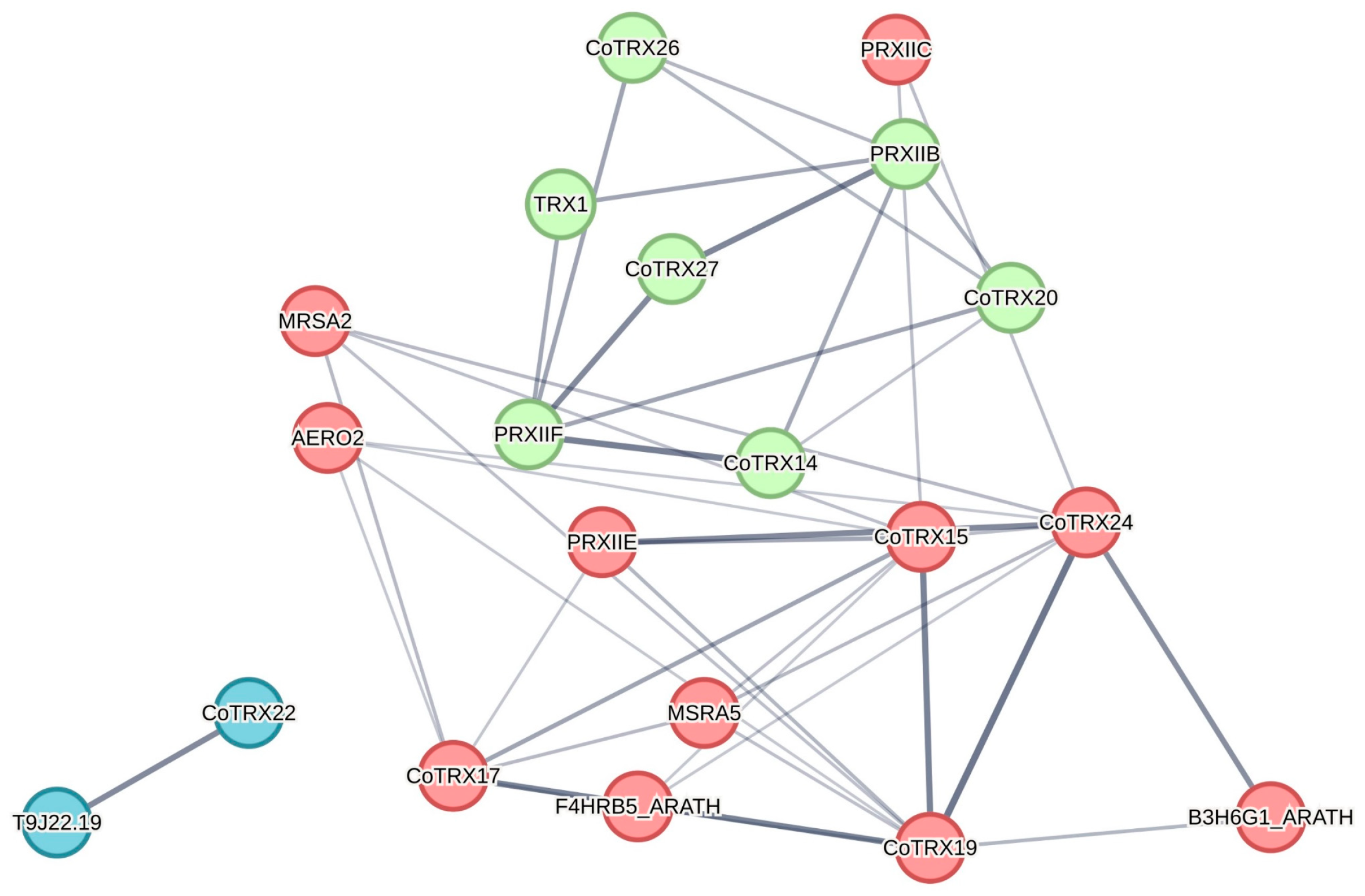
3.5. Gene Expression and PPI Network Analysis of Typical CoTRXs
3.6. Overexpression of CoTRX25 Leaded to Late Flowering of Transgenic Arabidopsis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Z.; Huang, B.; Ye, J.; He, Y.; Tang, S.; Wang, H.; Wen, Q. Comparative transcriptomic analysis reveals genes related to the rapid accumulation of oleic acid in Camellia chekiangoleosa; an oil tea plant with early maturity and large fruit. Plant Physiol. Biochem. 2022, 171, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Quan, W.; Wang, A.; Gao, C.; Li, C. Applications of Chinese Camellia oleifera and its By-Products: A Review. Front. Chem. 2022, 10, 921246. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Song, Q.; Ji, K.; Gong, S.; Wang, L.; Chen, L.; Zhang, J.; Yuan, D. Full-Length Transcriptome from Camellia oleifera Seed Provides Insight into the Transcript Variants Involved in Oil Biosynthesis. J. Agric. Food Chem. 2020, 68, 14670–14683. [Google Scholar] [CrossRef] [PubMed]
- Qin, P.; Shen, J.; Wei, J.; Chen, Y. A critical review of the bioactive ingredients and biological functions of camellia oleifera oil. Curr. Res. Food Sci. 2024, 8, 100753. [Google Scholar] [CrossRef]
- Geigenberger, P.; Thormählen, I.; Daloso, D.M.; Fernie, A.R. The Unprecedented Versatility of the Plant Thioredoxin System. Trends Plant Sci. 2017, 22, 249–262. [Google Scholar] [CrossRef]
- Yoshida, K.; Hisabori, T. Current Insights into the Redox Regulation Network in Plant Chloroplasts. Plant Cell Physiol. 2023, 64, 704–715. [Google Scholar] [CrossRef]
- Chibani, K.; Pucker, B.; Dietz, K.J.; Cavanagh, A. Genome-wide analysis and transcriptional regulation of the typical and atypical thioredoxins in Arabidopsis thaliana. FEBS Lett. 2021, 595, 2715–2730. [Google Scholar] [CrossRef]
- Meyer, Y.; Reichheld, J.P.; Vignols, F. Thioredoxins in Arabidopsis and other plants. Photosynth. Res. 2005, 86, 419–433. [Google Scholar] [CrossRef]
- Delorme-Hinoux, V.; Bangash, S.A.; Meyer, A.J.; Reichheld, J.P. Nuclear thiol redox systems in plants. Plant Sci. 2016, 243, 84–95. [Google Scholar] [CrossRef]
- de Bont, L.; Donnay, N.; Favier, F.; Couturier, J.; Didierjean, C.; Rouhier, N. Potential unrecognized redox-active atypical thioredoxins in eukaryote photosynthetic organisms. J. Exp. Bot. 2025, 21, eraf066. [Google Scholar] [CrossRef]
- Thormählen, I.; Meitzel, T.; Groysman, J.; Öchsner, A.B.; von Roepenack-Lahaye, E.; Naranjo, B.; Cejudo, F.J.; Geigenberger, P. Thioredoxin f1 and NADPH-Dependent Thioredoxin Reductase C Have Overlapping Functions in Regulating Photosynthetic Metabolism and Plant Growth in Response to Varying Light Conditions. Plant Physiol. 2015, 169, 1766–1786. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Wong, J.H.; Feldman, L.J.; Lemaux, P.G.; Buchanan, B.B. A membrane-associated thioredoxin required for plant growth moves from cell to cell, suggestive of a role in intercellular communication. Proc. Natl. Acad. Sci. USA 2010, 107, 3900–3905. [Google Scholar] [CrossRef]
- Fan, Y.; Li, M.; Wu, Y.; Wang, X.; Wang, P.; Zhang, L.; Meng, X.; Meng, F.; Li, Y. Characterization of thioredoxin gene TaTrxh9 associated with heading-time regulation in wheat. Plant Physiol. Biochem. 2023, 201, 107903. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Sharoni, A.M.; Satoh, K.; Al-Shammari, T.; Shimizu, T.; Sasaya, T.; Omura, T.; Kikuchi, S. The thioredoxin gene family in rice: Genome-wide identification and expression profiling under different biotic and abiotic treatments. Biochem. Biophys. Res. Commun. 2012, 423, 417–423. [Google Scholar] [CrossRef]
- Tong, L.; Lin, M.; Zhu, L.; Liao, B.; Lu, L.; Lu, Y.; Chen, J.; Shi, J.; Hao, Z. Unraveling the Role of the Liriodendron Thioredoxin (TRX) Gene Family in an Abiotic Stress Response. Plants 2024, 13, 1674. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, Y.; Huang, X. Genome-Wide Analysis of the Thioredoxin Gene Family in Gossypium hirsutum L. and the Role of the Atypical Thioredoxin Gene GhTRXL3-2 in Flowering. J. Plant Biol. 2021, 64, 461–473. [Google Scholar] [CrossRef]
- Zhou, J.; Song, T.; Zhou, H.; Zhang, M.; Li, N.; Xiang, J.; Zhang, X. Genome-wide identification; characterization; evolution; and expression pattern analyses of the typical thioredoxin gene family in wheat (Triticum aestivum L.). Front. Plant Sci. 2022, 13, 1020584. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Wang, F.; Xu, Z.; Wang, G.; Hu, L.; Cheng, J.; Ge, X.; Liu, J.; Chen, W.; Li, Q.; et al. The complex hexaploid oil-Camellia genome traces back its phylogenomic history and multi-omics analysis of Camellia oil biosynthesis. Plant Biotechnol. J. 2024, 22, 2890–2906. [Google Scholar] [CrossRef]
- Kim, J.; Shiu, S.H.; Thoma, S.; Li, W.H.; Patterson, S.E. Patterns of expansion and expression divergence in the plant polygalacturonase gene family. Genome Biol. 2006, 7, R87. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE; a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Du, B.; Zhou, J.; Cao, Y.; Zhang, L. Camellia oleifera CoSWEET10 Is Crucial for Seed Development and Drought Resistance by Mediating Sugar Transport in Transgenic Arabidopsis. Plants 2023, 12, 2818. [Google Scholar] [CrossRef]
- Su, W.; Zhou, Z.; Zeng, J.; Cao, R.; Zhang, Y.; Hu, D.; Liu, J. Genome-wide identification of the WRKY gene family in Camellia oleifera and expression analysis under phosphorus deficiency. Front. Plant Sci. 2023, 14, 1082496. [Google Scholar] [CrossRef]
- Zhang, X.; He, W.; Wang, X.; Duan, Y.; Li, Y.; Wang, Y.; Jiang, Q.; Liao, B.; Zhou, S.; Li, Y. Genome-Wide Analyses of MADS-Box Genes Reveal Their Involvement in Seed Development and Oil Accumulation of Tea-Oil Tree (Camellia oleifera). Int J. Genom. 2024, 2024, 3375173. [Google Scholar] [CrossRef]
- Fu, C.; Xiao, Y.; Jiang, N.; Yang, Y. Genome-wide identification and molecular evolution of Dof gene family in Camellia oleifera. BMC Genom. 2024, 25, 702. [Google Scholar] [CrossRef]
- Bretani, G.; Rossini, L.; Ferrandi, C.; Russell, J.; Waugh, R.; Kilian, B.; Bagnaresi, P.; Cattivelli, L.; Fricano, A. Segmental duplications are hot spots of copy number variants affecting barley gene content. Plant J. 2020, 103, 1073–1088. [Google Scholar] [CrossRef]
- Mascher, M.; Gundlach, H.; Himmelbach, A.; Beier, S.; Twardziok, S.O.; Wicker, T.; Radchuk, V.; Dockter, C.; Hedley, P.E.; Russell, J.; et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 2017, 544, 427–433. [Google Scholar] [CrossRef]
- Zuo, X.; Yang, C.; Yan, Y.; Huang, G.; Li, R. Systematic analysis of the thioredoxin gene family in Citrus sinensis: Identification, phylogenetic analysis, and gene expression patterns. Plant Signal. Behav. 2023, 18, 2294426. [Google Scholar] [CrossRef]
- Du, W.; Ding, J.; Li, J.; Li, H.; Ruan, C. Co-regulatory effects of hormone and mRNA-miRNA module on flower bud formation of Camellia oleifera. Front. Plant Sci. 2023, 14, 1109603. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Locus ID | Chromosome Number | Predicted Size (aa) | Molecular Weight (kDa) | Isoelectric Points (pI) | Exon | Subcellular Localization Prediction | Tertiary Structure Prediction Template Protein Data Bank Molecule | |
|---|---|---|---|---|---|---|---|---|---|
| CoTRX1 | A1.1G231500 | chr1a1 | 113 | 12.51 | 5.51 | 3 | Nucleus | c1xflA | Thioredoxin h1 |
| CoTRX2 | A1.2G146400 | chr2a1 | 87 | 9.50 | 9.83 | 3 | Chloroplast | c6x0bA | Thioredoxin H |
| CoTRX3 | A1.5G005400 | chr5a1 | 153 | 17.25 | 6.07 | 5 | Chloroplast | c3d22A | Thioredoxin H-type |
| CoTRX4 | A1.5G295300 | chr5a1 | 124 | 13.67 | 5.29 | 3 | Cytoplasm | c1xflA | Thioredoxin h1 |
| CoTRX5 | A1.6G124800 | chr6a1 | 191 | 20.64 | 9.36 | 2 | Chloroplast outer membrane | c8qpdA | Thioredoxin M-type |
| CoTRX6 | A1.8G025600 | chr8a1 | 121 | 13.24 | 4.95 | 3 | Nucleus | c1xflA | Thioredoxin h1 |
| CoTRX7 | A1.9G067200 | chr9a1 | 189 | 21.15 | 9.05 | 6 | Chloroplast | c6g62A | Thioredoxin O2 |
| CoTRX8 | A1.15G023400 | chr15a1 | 79 | 9.09 | 4.6 | 4 | Extracellular space | c3d22A | Thioredoxin H-type |
| CoTRX9 | A2.1G086600 | chr1a2 | 149 | 17.31 | 9.27 | 4 | Endomembrane system | c6x0bA | Thioredoxin H |
| CoTRX10 | A2.4G068900 | chr4a2 | 115 | 12.84 | 5.65 | 3 | Nucleus | c1xflA | Thioredoxin h1 |
| CoTRX11 | A2.6G043700 | chr6a2 | 93 | 10.28 | 9.82 | 3 | Chloroplast | c6x0bA | Thioredoxin H |
| CoTRX12 | A2.6G201400 | chr6a2 | 85 | 9.30 | 6.24 | 2 | Chloroplast | c6x0bA | Thioredoxin H |
| CoTRX13 | A2.8G139200 | chr8a2 | 191 | 20.84 | 8.77 | 2 | Chloroplast | c8qpdA | Thioredoxin M-type |
| CoTRX14 | B1.2G091900 | chr2b1 | 189 | 20.95 | 8.57 | 6 | Chloroplast | c6g61A | Thioredoxin O1 |
| CoTRX15 | B1.2G117300 | chr2b1 | 180 | 20.29 | 5.15 | 4 | Chloroplast | c8w9zP | Thioredoxin-like CITRX1 |
| CoTRX16 | B1.4G234200 | chr4b1 | 140 | 15.34 | 6.42 | 3 | Chloroplast | c6x0bA | Thioredoxin H |
| CoTRX17 | B1.5G132500 | chr5b1 | 172 | 19.15 | 9.14 | 2 | Chloroplast outer membrane | c8qpdA | Thioredoxin M-type |
| CoTRX18 | B1.6G199900 | chr6b1 | 185 | 19.86 | 9.36 | 2 | Chloroplast outer membrane | c8qpdA | Thioredoxin M-type |
| CoTRX19 | B1.7G135100 | chr7b1 | 181 | 19.70 | 9.35 | 3 | Chloroplast outer membrane | c7c2bC | Thioredoxin F2 |
| CoTRX20 | B2.11G006300 | chr11b2 | 179 | 20.25 | 8.96 | 2 | Chloroplast | c5vo7A | Thioredoxin |
| CoTRX21 | C2.4G306700 | chr4c2 | 139 | 15.53 | 4.98 | 3 | Chloroplast thylakoid membrane | c6x0bA | Thioredoxin H |
| CoTRX22 | C2.5G224200 | chr5c2 | 98 | 10.97 | 5.17 | 4 | Mitochondrion | c7bzkB | Thioredoxin Y1 |
| CoTRX23 | C2.7G132800 | chr7c2 | 85 | 9.30 | 6.24 | 2 | Chloroplast | c6x0bA | Thioredoxin H |
| CoTRX24 | C2.7G235200 | chr7c2 | 122 | 14.04 | 9.96 | 2 | Chloroplast outer membrane | c8qpdA | Thioredoxin M-type |
| CoTRX25 | C2.10G026400 | chr10c2 | 172 | 19.30 | 9.07 | 4 | Cytoplasm | c3d22A | Thioredoxin H-type |
| CoTRX26 | C2.15G002900 | chr15c2 | 85 | 92.97 | 6.24 | 2 | Extracellular space | c3d22A | Thioredoxin H-type |
| CoTRX27 | C2.15G092100 | chr15c2 | 191 | 20.65 | 9.36 | 2 | Chloroplast | c6x0bA | Thioredoxin H |
| Paralog Pairs | Duplication | Ka | Ks | ω (Ka/Ks) | |
|---|---|---|---|---|---|
| Type | |||||
| CoTRX25.1 | CoTRX3.1 | Segmental | 0.0961 | 0.2755 | 0.3487 |
| CoTRX1.1 | CoTRX4.1 | Segmental | 0.2312 | 1.4515 | 0.1593 |
| CoTRX1.1 | CoTRX6.1 | Segmental | 0.2412 | 0.9251 | 0.2608 |
| CoTRX1.1 | CoTRX10.1 | Segmental | 0.1554 | 0.6061 | 0.2564 |
| CoTRX4.1 | CoTRX10.1 | Segmental | 0.1614 | 1.0115 | 0.1596 |
| CoTRX4.1 | CoTRX6.1 | Segmental | 0.1004 | 0.6293 | 0.1595 |
| CoTRX6.1 | CoTRX10.1 | Segmental | 0.1648 | 1.1239 | 0.1467 |
| CoTRX18.1 | CoTRX24.1 | Segmental | 0.1027 | 0.5517 | 0.1861 |
| CoTRX3.2 | CoTRX25.2 | Segmental | 0.1135 | 0.3081 | 0.3684 |
| CoTRX18.2 | CoTRX24.2 | Segmental | 0.0987 | 0.4328 | 0.2281 |
| CoTRX1.3 | CoTRX4.3 | Segmental | 0.2820 | 1.4789 | 0.1907 |
| CoTRX1.3 | CoTRX6.3 | Segmental | 0.2704 | 0.9981 | 0.2709 |
| CoTRX3.3 | CoTRX25.3 | Segmental | 0.0961 | 0.2902 | 0.3313 |
| CoTRX4.3 | CoTRX6.3 | Segmental | 0.1004 | 0.5743 | 0.1748 |
| CoTRX4.3 | CoTRX10.3 | Segmental | 0.3152 | 1.2577 | 0.2507 |
| CoTRX18.3 | CoTRX24.3 | Segmental | 0.1644 | 0.6501 | 0.2528 |
| CoTRX1.4 | CoTRX4.4 | Segmental | 0.2898 | 1.3189 | 0.2197 |
| CoTRX1.4 | CoTRX6.4 | Segmental | 0.2726 | 0.9843 | 0.2770 |
| CoTRX1.4 | CoTRX10.4 | Segmental | 0.1980 | 0.5525 | 0.3585 |
| CoTRX3.4 | CoTRX25.4 | Segmental | 0.1175 | 0.3071 | 0.3827 |
| CoTRX4.4 | CoTRX6.4 | Segmental | 0.1308 | 0.5757 | 0.2271 |
| CoTRX1.5 | CoTRX4.5 | Segmental | 0.2309 | 1.2991 | 0.1778 |
| CoTRX1.5 | CoTRX6.5 | Segmental | 0.2433 | 0.9142 | 0.2661 |
| CoTRX1.5 | CoTRX10.5 | Segmental | 0.1598 | 0.6415 | 0.2491 |
| CoTRX3.5 | CoTRX25.5 | Segmental | 0.3167 | 0.7476 | 0.4237 |
| CoTRX4.5 | CoTRX10.5 | Segmental | 0.1640 | 1.0816 | 0.1516 |
| CoTRX6.5 | CoTRX10.5 | Segmental | 0.1670 | 1.1601 | 0.1440 |
| CoTRX18.5 | CoTRX24.5 | Segmental | 0.1105 | 0.4981 | 0.2219 |
| CoTRX1.6 | CoTRX6.6 | Segmental | 0.2418 | 0.9597 | 0.2519 |
| CoTRX1.6 | C2.4G094900 | Segmental | 0.1554 | 0.6370 | 0.2439 |
| CoTRX3.6 | CoTRX25.6 | Segmental | 0.1121 | 0.2685 | 0.4174 |
| CoTRX6.6 | C2.4G094900 | Segmental | 0.1652 | 1.1667 | 0.1416 |
| CoTRX13.6 | CoTRX22.6 | Segmental | 0.7456 | NaN | NaN |
| C2.6G214900 | CoTRX24.6 | Segmental | 0.1071 | 0.5110 | 0.2096 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, L.; Song, P.; Xia, Y.; Min, M.; Xu, T.; Cheng, J.; Cheng, J.; Zhu, H. Genome-Wide Analysis of the Typical Thioredoxin Gene Family in Hexaploid Oil-Camellia: Identification, Phylogenetic Analysis, and Gene Expression Patterns. Genes 2025, 16, 790. https://doi.org/10.3390/genes16070790
Wu L, Song P, Xia Y, Min M, Xu T, Cheng J, Cheng J, Zhu H. Genome-Wide Analysis of the Typical Thioredoxin Gene Family in Hexaploid Oil-Camellia: Identification, Phylogenetic Analysis, and Gene Expression Patterns. Genes. 2025; 16(7):790. https://doi.org/10.3390/genes16070790
Chicago/Turabian StyleWu, Lan, Peipei Song, Yifan Xia, Min Min, Tingting Xu, Junyong Cheng, Jihua Cheng, and Huaguo Zhu. 2025. "Genome-Wide Analysis of the Typical Thioredoxin Gene Family in Hexaploid Oil-Camellia: Identification, Phylogenetic Analysis, and Gene Expression Patterns" Genes 16, no. 7: 790. https://doi.org/10.3390/genes16070790
APA StyleWu, L., Song, P., Xia, Y., Min, M., Xu, T., Cheng, J., Cheng, J., & Zhu, H. (2025). Genome-Wide Analysis of the Typical Thioredoxin Gene Family in Hexaploid Oil-Camellia: Identification, Phylogenetic Analysis, and Gene Expression Patterns. Genes, 16(7), 790. https://doi.org/10.3390/genes16070790






