Selection and Validation of Stable Reference Genes for RT-qPCR Analyses of Rumex patientia (Polygonaceae) Under Four Abiotic Stresses
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Experimental Treatments
2.2. RNA Extraction, Quality Assessment, and cDNA Synthesis
2.3. Reference Gene Selection and Primer Design
2.4. Real-Time Quantitative PCR Analysis
2.5. Gene Expression Stability Analysis
2.6. Reference Gene Validation
3. Results
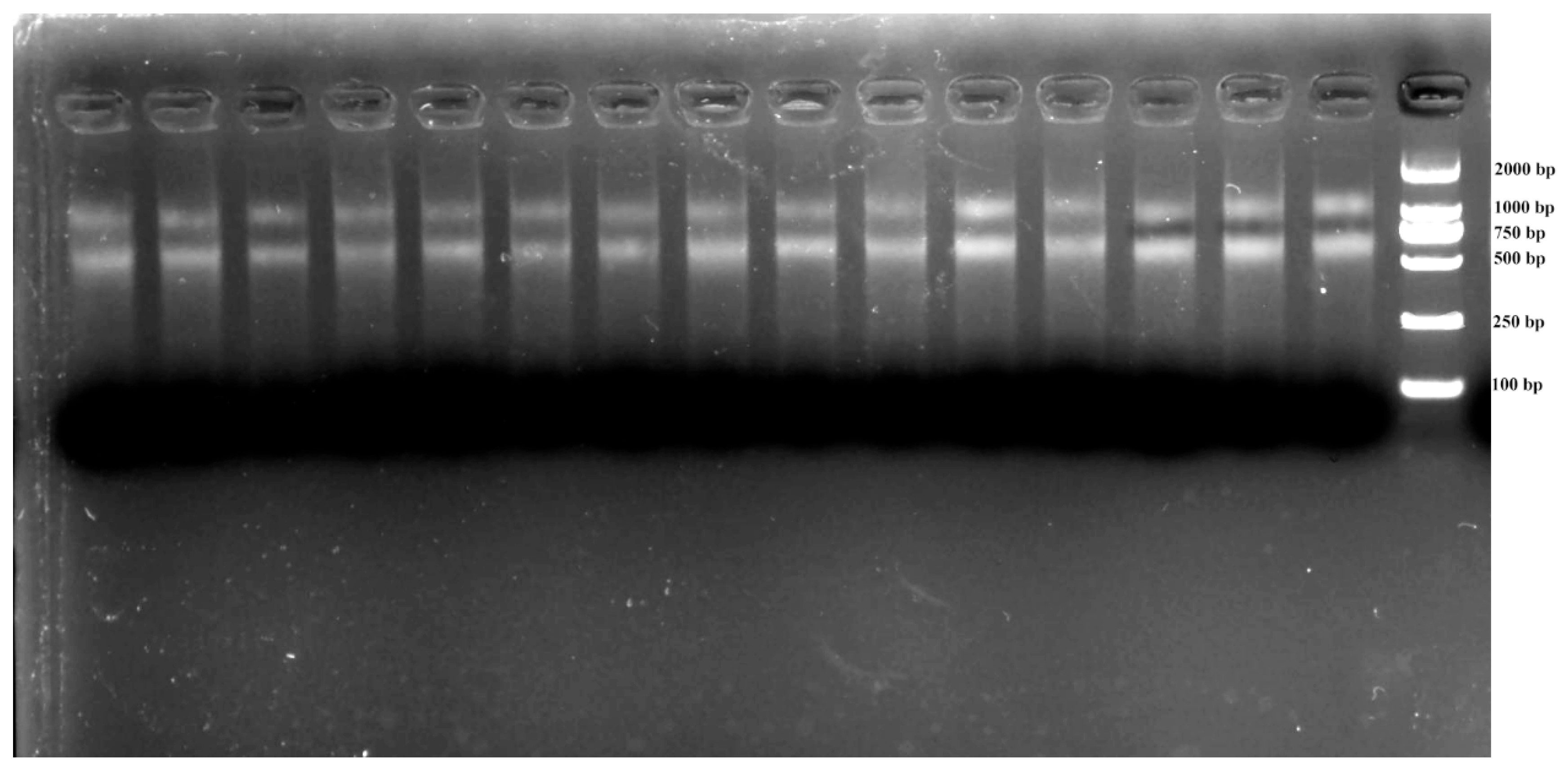
3.1. RNA Integrity and Primer Specificity
3.2. Expression Profile of Reference Genes
3.3. Expression Stability of Candidate Reference Genes
3.3.1. The Comparative Delta-Ct Method Analysis
3.3.2. geNorm Analysis
3.3.3. NormFinder Analysis
3.3.4. BestKeeper Analysis
3.3.5. RefFinder Analysis
3.4. Validation of the Stability for Candidate Reference Genes Based on MYB
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nawaz, M.; Sun, J.; Shabbir, S. A review of plants strategies to resist biotic and abiotic environmental stressors. Sci. Total Environ. 2023, 900, 165832. [Google Scholar] [CrossRef] [PubMed]
- Baweja, P.; Kumar, G. Abiotic stress in plants: An overview. Plant Stress Biol. Strateg. Trends 2021, 1–15. [Google Scholar] [CrossRef]
- Muluneh, M.G. Impact of climate change on biodiversity and food security: A global perspective—A review article. Agric. Food Secur. 2021, 10, 36. [Google Scholar] [CrossRef]
- Ghori, N.H.; Ghori, T.; Hayat, M.Q.; Imadi, S.R.; Gul, A.; Altay, V.; Ozturk, M. Heavy metal stress and responses in plants. Int. J. Environ. Sci. Technol. 2019, 16, 1807–1828. [Google Scholar] [CrossRef]
- Lamalakshmi, D.E.; Kumar, S.; Basanta, S.T.; Sharma, S.K.; Beemrote, A.; Devi, C.P.; Chongtham, S.K.; Singh, C.H.; Yumlembam, R.A.; Haribhushan, A.; et al. Adaptation strategies and defense mechanisms of plants during environmental stress. In Medicinal Plants and Environmental Challenges; Springer: Cham, Switzerland, 2017; pp. 359–413. [Google Scholar] [CrossRef]
- Son, S.; Im, J.H.; Ko, J.H. Role of plant cell wall in biotic and abiotic stress resistance. Front. Plant Sci. 2025, 16, 1617371. [Google Scholar] [CrossRef]
- Qi, J.N.; Yu, S.C.; Zhang, F.L.; Shen, X.Q.; Zhao, X.Y.; Yu, Y.J.; Zhang, D.S. Reference gene selection for real-time quantitative polymerase chain reaction of mRNA transcript levels in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Plant Mol. Biol. Rep. 2010, 28, 597–604. [Google Scholar] [CrossRef]
- Dean, J.D.; Goodwin, P.H.; Hsiang, T. Comparison of relative RT-PCR and northern blot analyses to measure expression of β-1,3-glucanase in Nicotiana benthamiana infected with Colltotrichum destructivum. Plant Mol. Biol. Rep. 2002, 20, 347–356. [Google Scholar] [CrossRef]
- Zhand, S.; Goss, D.M.; Cheng, Y.Y. Recent advances in microfluidics for nucleic acid analysis of small extracellular vesicles in cancer. Adv. Healthc. Mater. 2025, 14, 2401295. [Google Scholar] [CrossRef] [PubMed]
- Grätz, C.; Bui, M.L.U.; Thaqi, G. Obtaining reliable RT-qPCR results in molecular diagnostics—MIQE goals and pitfalls for transcriptional biomarker discovery. Life 2022, 12, 386. [Google Scholar] [CrossRef]
- Zheng, H.; Zhao, H.; Zhang, X. Systematic identification and validation of suitable reference genes for the normalization of gene expression in Prunella vulgaris under different organs and spike development stages. Genes 2022, 13, 1947. [Google Scholar] [CrossRef]
- Gasmi, A.; Triki, T.; Bennani, L. Identification of reference genes and analysis of heat shock protein gene expression (Hsp90) in arta (Calligonum comosum L.) leaf under heat stress. S. Afr. J. Bot. 2024, 167, 570–577. [Google Scholar] [CrossRef]
- Luan, Y.; Xu, C.; Wang, X. Selection of stable reference genes for quantitative real-time PCR on Paeonia ostii T. Hong et JX Zhang leaves exposed to different drought stress conditions. Phyton 2022, 91, 1045. [Google Scholar] [CrossRef]
- Zhao, Y.Q. Anti-tumor Activity Composition Screening of Rumex patientia L. Root. Master’s thesis, Shanxi University of Chinese Medicine, Taiyuan, China, 2018. [Google Scholar]
- Demirezer, L.Ö.; Kuruüzüm-Uz, A.; Bergere, I.; Schiewe, H.J.; Zeeck, A. The structures of antioxidant and cytotoxic agents from natural source: Anthraquinones and tannins from roots of Rumex patientia. Phytochemistry 2001, 58, 1213–1217. [Google Scholar] [CrossRef] [PubMed]
- Demirezer, Ö.; Kuruüzüm, A.; Bergere, I.; Schiewe, H.J.; Zeeck, A. Five naphthalene glycosides from the roots of Rumex patientia. Phytochemistry 2001, 56, 399–402. [Google Scholar] [CrossRef]
- Yuan, Y.; Chen, W.S.; Zheng, S.Q.; Yang, G.J.; Zhang, W.D.; Zhang, H.M. Studies on chemical constituents in root of Rumex patientia L. Chung-kuo Chung Yao Tsa Chih. 2001, 26, 256–258. Available online: https://pubmed.ncbi.nlm.nih.gov/12525048/ (accessed on 3 June 2025).
- Liu, J.; Xia, Z.T.; Zhou, G.R.; Zhang, L.L.; Kong, L.Y. Study on the chemical constituents of Rumex patientia. Zhong Yao Cai Zhongyaocai J. Chin. Med. Mater. 2011, 34, 893–895. Available online: https://pubmed.ncbi.nlm.nih.gov/22017004/ (accessed on 3 June 2025).
- Süleyman, H.; Demirezer, L.Ö.; Kuruüzüm, A.; Banoğlu, Z.N.; Göçer, F.; Özbakir, O.; Gepdiremen, A. Antiinflammatory effect of the aqueous extract from Rumex patientia L. roots. J. Ethnopharmacol. 1999, 65, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.J.; Wang, E.; Xing, M.; Zhao, W.; Chen, X. Rhizosphere and non-rhizosphere bacterial community composition of the wild medicinal plant Rumex patientia. World J. Microbiol. Biotechnol. 2012, 28, 2257–2265. [Google Scholar] [CrossRef]
- Sayre, J.D. Physiology of stomata of Rumex patientia. Science 1923, 57, 205–206. [Google Scholar] [CrossRef]
- Dickson, J.G.; Caldwell, M.M. Leaf development of Rumex patientia L. (Polygonaceae) exposed to UV irradiation (280–320 nm). Am. J. Bot. 1978, 65, 857–863. [Google Scholar] [CrossRef]
- Liu, J.; Xu, Y.; Zhang, L.; Li, W.; Cai, Z.X.; Li, F.; Peng, M.; Li, F.L.; Hu, B.Z. De novo assembly and analysis of the transcriptome of Rumex patientia L. during cold stress. PLoS ONE 2017, 12, e0186470. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Ju, Y.L.; Yang, Q.C.; Li, B.L.; Yang, Z.M. Genetic diversity of Rumex k-1(Rumex patientia × R.tianschanicus cv.Rumex k-1) and Wild Rumex spp. Acta Agrestia Sinica 2004, 12, 298–302. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Roy, N.V.; Paepe, A.D.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. Quantification strategies in real-time PCR. AZ Quant. PCR 2004, 1, 89–113. [Google Scholar]
- Xie, F.L.; Xiao, P.; Chen, D.L.; Xu, L.; Zhang, B.H. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, S.; Yang, L. Reference genes for qRT-PCR normalisation in different tissues, developmental stages, and stress conditions of Hypericum perforatum. Peer J. 2019, 7, e7133. [Google Scholar] [CrossRef]
- Niu, K.; Shi, Y.; Ma, H. Selection of candidate reference genes for gene expression analysis in Kentucky Bluegrass (Poa pratensis L.) under abiotic stress. Front. Plant Sci. 2017, 8, 193. [Google Scholar] [CrossRef]
- Baldoni, E.; Genga, A.; Cominelli, E. Plant MYB transcription factors: Their role in drought response mechanisms. Int. J. Mol. Sci. 2015, 16, 15811–15851. [Google Scholar] [CrossRef]
- Derveaux, S.; Vandesompele, J.; Hellemans, J. How to do successful gene expression analysis using real-time PCR. Methods 2010, 50, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.; Wakem, M.; Dijkman, G.; Alsarraj, M.; Nguyen, M. A practical approach to RT-qPCR-publishing data that conform to the MIQE guidelines. Methods 2010, 50, S1–S5. [Google Scholar] [CrossRef] [PubMed]
- Sreedharan, S.P.; Kumar, A.; Giridhar, P. Primer design and amplification efficiencies are crucial for reliability of quantitative PCR studies of caffeine biosynthetic N-methyltransferases in coffee. 3 Biotech 2018, 8, 467. [Google Scholar] [CrossRef] [PubMed]
- Suslov, O.; Steindler, D.A. PCR inhibition by reverse transcriptase leads to an overestimation of amplification efficiency. Nucleic Acids Res. 2005, 33, e181. [Google Scholar] [CrossRef]
- Ramezani, A. CtNorm: Real time PCR cycle of threshold (Ct) normalization algorithm. J. Microbiol. Methods 2021, 187, 106267. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Wang, J.; Zhang, B. RefFinder: A web-based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genom. 2023, 23, 125. [Google Scholar] [CrossRef]
- Li, Y.; Ye, X.; Wang, B.; Chen, M.; Liu, J.; Zhu, H.; Wen, Q. Cloning and selection evaluation of reference gene for quantitative real-time PCR in Hibiscus esculentus L. J. Nucl. Agric. Sci. 2021, 35, 60–71. [Google Scholar]
- Wang, X.W.; Wu, Z.J.; Bao, W.Q.; Hu, H.Y.; Chen, M.; Chai, T.Y.; Wang, H. Identification and evaluation of reference genes for quantitative real-time PCR analysis in Polygonum cuspidatum based on transcriptome data. BMC Plant Biol. 2019, 19, 498. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Cheng, K.X.; Qin, Z.Q.; Wu, T.; Li, X.M.; Tu, J.F.; Yang, F.C.; Zhu, H.Y.; Yang, L. Selection and validation of suitable reference genes for qRT-PCR analysis in pear leaf tissues under distinct training systems. PLoS ONE 2018, 13, e0202472. [Google Scholar] [CrossRef]
- Buono, R.A.; Paez-Valencia, J.; Miller, N.D.; Goodman, K.; Spitzer, C.; Spalding, E.P.; Otegui, M.S. Role of SKD1 regulators LIP5 and IST1-LIKE1 in endosomal sorting and plant development. Plant Physiol. 2016, 171, 251–264. [Google Scholar] [CrossRef]
- Xia, Z.; Wei, Y.; Sun, K.; Wu, J.; Wang, Y.; Wu, K. The maize AAA-type protein SKD1 confers enhanced salt and drought stress tolerance in transgenic tobacco by interacting with Lyst-interacting protein 5. PLoS ONE 2013, 8, e69787. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Fan, C.; Li, H. Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol. Biol. 2009, 10, 93. [Google Scholar] [CrossRef] [PubMed]
- Chi, X.Y.; Hu, R.B.; Yang, Q.L.; Zhang, X.W.; Pan, L.J.; Chen, M.N.; Yang, Z.; Wang, T.; He, Y.N.; Yu, S.L. Validation of reference genes for gene expression studies in peanut by quantitative real-time RT-PCR. Mol. Genet. Genom. 2012, 287, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Long, X.Y.; He, B.; Gao, X.S.; Qin, Y.X.; Yang, J.H.; Fang, Y.J.; Qi, J.Y.; Tang, C.R. Validation of reference genes for quantitative real-time PCR during latex regeneration in rubber tree. Gene 2015, 563, 190–195. [Google Scholar] [CrossRef]
- Mascia, T.; Santovito, E.; Gallitelli, D.; Cillo, F. Evaluation of reference genes for quantitative reverse-transcription polymerase chain reaction normalization in infected tomato plants. Mol. Plant Pathol. 2010, 11, 805–816. [Google Scholar] [CrossRef]
- Singh, V.V.; Naseer, A.; Sellamuthu, G. Robust reference gene selection in Norway spruce: Essential for real-time quantitative PCR across different tissue, stress and developmental conditions. Front. For. Glob. Change 2024, 7, 1458554. [Google Scholar] [CrossRef]
- Vijayakumar, S.; Sakuntala, M. Validation of reference gene stability for normalization of RT-qPCR in Phytophthora capsici Leonian during its interaction with Piper nigrum L. Sci. Rep. 2024, 14, 7331. [Google Scholar] [CrossRef]
- Liu, Y.P.; Zhang, Y.; Liu, F.; Liu, T.; Chen, J.Y.; Fu, G.; Zheng, C.Y.; Su, D.D.; Wang, Y.N.; Zhou, H.K.; et al. Establishment of reference (housekeeping) genes via quantitative real-time PCR for investigation of the genomic basis of abiotic stress resistance in Psammochloa villosa (Poaceae). J. Plant Physiol. 2022, 268, 153575. [Google Scholar] [CrossRef]
- Jain, M.; Nijhawan, A.; Tyagi, A.K.; Khurana, J.P. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2006, 345, 646–651. [Google Scholar] [CrossRef]
- Tong, Z.G.; Gao, Z.H.; Wang, F.; Zhou, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 2009, 10, 71. [Google Scholar] [CrossRef]
- Reid, K.E.; Olsson, N.; Schlosser, J.; Peng, F.; Lund, S.T. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 2006, 6, 27. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Zhang, N.; Si, H. Selection and validation of reference genes for RT-qPCR analysis in potato under abiotic stress. Plant Methods 2017, 13, 85. [Google Scholar] [CrossRef] [PubMed]
- Stephan, L.; Tilmes, V.; Hülskamp, M. Selection and validation of reference genes for quantitative Real-Time PCR in Arabisalpina. PLoS ONE 2019, 14, e0211172. [Google Scholar] [CrossRef] [PubMed]










| Gene Name | Gene ID | Primer Sequence (5′-3′) (Forward/Reverse) | Amplicon Length | Tm (°C) | RT-qPCR Efficiency(%) | R2 |
|---|---|---|---|---|---|---|
| ACT | TRINITY_DN197 | GGGCTGGCTGATAGGATGAG/ GCACTTGCGATGAACGATT | 201 | 56 | 95.251 | 0.999 |
| GAPDH | TRINITY_DN2913 | AACTTCGGCATTGTTGAGGGAT/ TGGGAATGATGTTGAATGAGGC | 127 | 56 | 127.364 | 0.975 |
| YLS | TRINITY_DN2081 | GTCTTACCTTCTCCCGCATCT/ CAAAGTTCTTTATAGTCTCAGCCAC | 164 | 56 | 113.547 | 0.988 |
| SKD1 | TRINITY_DN115 | AGGAAGCCATCACGCAGAAAT/ GCGGAGTTCAACCCAGACCTAA | 217 | 58 | 101.845 | 0.998 |
| UBQ | TRINITY_DN16 | GCTCCCAATACCAACCCTC/ GAATCAGACATAGCACGCAGTT | 275 | 58 | 97.462 | 0.992 |
| UBC | TRINITY_DN11678 | GGACTGACTGGGAAGGTGG/ GACAAACAGTTCCTGATGGGT | 132 | 58 | 92.583 | 0.987 |
| EF-1α | TRINITY_DN2194 | GAAGGAGGCTGCTGAGATG/ GGTGTAGGCAAGAAGAGCG | 298 | 59 | 108.566 | 0.986 |
| TUA | TRINITY_DN2714 | GCTGTGGCTACAATCAAGACC/ AGAACTCACCTTCCTCCATACC | 265 | 59 | 105.632 | 0.984 |
| Group | Gene | SD | CV | r | Group | Gene | SD | CV | r | Group | Gene | SD | CV | r |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S-R | ACT | 3.79 | 12.76 | 0.983 | S-ST | ACT | 0.98 | 3.81 | 0.984 | S-L | ACT | 1.18 | 4.58 | 0.126 |
| GAPDH | 4.35 | 16.60 | 0.993 | GAPDH | 0.74 | 3.60 | 0.539 | GAPDH | 0.72 | 3.28 | −0.782 | |||
| YLS | 2.20 | 7.47 | 0.978 | YLS | 1.46 | 5.04 | 0.87 | YLS | 1.06 | 3.56 | 0.729 | |||
| SKD1 | 4.01 | 14.48 | 0.978 | SKD1 | 0.34 | 1.42 | 0.461 | SKD1 | 0.71 | 2.89 | −0.104 | |||
| UBQ | 3.50 | 12.46 | 0.981 | UBQ | 3.37 | 12.54 | 0.898 | UBQ | 2.46 | 8.97 | 0.876 | |||
| UBC | 1.57 | 5.81 | 0.542 | UBC | 2.44 | 9.43 | 0.189 | UBC | 1.82 | 6.92 | −0.367 | |||
| EF-1α | 4.67 | 19.61 | 0.99 | EF-1α | 4.03 | 18.66 | 0.881 | EF-1α | 3.66 | 16.25 | 0.675 | |||
| TUA | 5.29 | 19.23 | 0.994 | TUA | 0.91 | 4.35 | 0.686 | TUA | 4.09 | 15.84 | 0.312 | |||
| D-R | ACT | 2.97 | 10.18 | 0.978 | D-ST | ACT | 1.05 | 4.01 | 0.763 | D-L | ACT | 1.13 | 4.59 | 0.975 |
| GAPDH | 3.13 | 12.33 | 0.986 | GAPDH | 0.62 | 2.94 | 0.069 | GAPDH | 0.45 | 2.21 | 0.278 | |||
| YLS | 1.10 | 3.65 | 0.946 | YLS | 1.22 | 4.25 | 0.959 | YLS | 1.50 | 5.39 | 0.952 | |||
| SKD1 | 3.00 | 10.72 | 0.993 | SKD1 | 0.36 | 1.48 | 0.353 | SKD1 | 0.96 | 4.12 | 0.922 | |||
| UBQ | 1.82 | 6.32 | 0.987 | UBQ | 2.33 | 8.75 | 0.89 | UBQ | 2.63 | 10.19 | 0.945 | |||
| UBC | 0.20 | 0.73 | 0.295 | UBC | 2.20 | 8.49 | 0.053 | UBC | 2.06 | 7.89 | −0.751 | |||
| EF-1α | 3.91 | 16.22 | 0.991 | EF-1α | 3.83 | 17.33 | 0.866 | EF-1α | 4.76 | 21.22 | 0.972 | |||
| TUA | 3.23 | 11.99 | 0.981 | TUA | 0.81 | 3.84 | 0.608 | TUA | 0.97 | 4.52 | 0.32 | |||
| C-R | ACT | 2.77 | 9.32 | 0.993 | C-ST | ACT | 0.94 | 3.54 | 0.706 | C-L | ACT | 1.25 | 4.85 | 0.931 |
| GAPDH | 3.72 | 14.86 | 0.998 | GAPDH | 0.88 | 4.17 | 0.114 | GAPDH | 0.97 | 4.67 | 0.408 | |||
| YLS | 1.62 | 5.38 | 0.952 | YLS | 1.32 | 4.55 | 0.986 | YLS | 1.57 | 5.48 | 0.98 | |||
| SKD1 | 3.25 | 11.62 | 0.986 | SKD1 | 0.38 | 1.52 | −0.113 | SKD1 | 1.12 | 4.57 | 0.558 | |||
| UBQ | 2.6 | 9.03 | 0.922 | UBQ | 2.32 | 8.7 | 0.861 | UBQ | 2.52 | 9.77 | 0.92 | |||
| UBC | 0.25 | 0.92 | 0.305 | UBC | 1.94 | 7.37 | −0.134 | UBC | 1.24 | 4.61 | −0.133 | |||
| EF-1α | 3.48 | 14.9 | 0.991 | EF-1α | 3.60 | 15.86 | 0.797 | EF-1α | 3.89 | 17.05 | 0.96 | |||
| TUA | 3.78 | 14.78 | 0.936 | TUA | 0.74 | 3.38 | 0.13 | TUA | 2.32 | 10.03 | 0.982 | |||
| CD-R | ACT | 3.63 | 12.45 | 0.985 | CD-ST | ACT | 1.6 | 5.95 | 0.8 | CD-L | ACT | 1.13 | 4.410 | 0.727 |
| GAPDH | 3.33 | 12.93 | 0.985 | GAPDH | 1.29 | 5.69 | −0.561 | GAPDH | 1.29 | 5.63 | −0.445 | |||
| YLS | 1.36 | 4.45 | 0.805 | YLS | 1.38 | 4.6 | 0.966 | YLS | 1.77 | 6.18 | 0.629 | |||
| SKD1 | 3.49 | 12.48 | 0.995 | SKD1 | 0.46 | 1.81 | −0.371 | SKD1 | 0.4 | 1.63 | 0.315 | |||
| UBQ | 2.1 | 7.61 | 0.95 | UBQ | 2.11 | 7.78 | 0.515 | UBQ | 2.94 | 11.29 | 0.993 | |||
| UBC | 0.19 | 0.68 | 0.494 | UBC | 1.45 | 5.45 | −0.365 | UBC | 1.77 | 6.71 | −0.747 | |||
| EF-1α | 4.24 | 17.38 | 0.987 | EF-1α | 3.29 | 13.57 | 0.419 | EF-1α | 3.76 | 16.05 | 0.914 | |||
| TUA | 3.43 | 12.37 | 0.995 | TUA | 1.48 | 6.08 | −0.138 | TUA | 1 | 4.19 | −0.883 |
| Software | Reference Gene | C-R(R) | C-S (R) | C-L (R) | CD-R (R) | CD-S (R) | CD-L (R) | ST-R (R) | ST-S (R) | ST-L (R) | D-R(R) | D-S(R) | D-L(R) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| geNorm | UBC | 8 | 6 | 7 | 8 | 1 | 2 | 8 | 6 | 5 | 8 | 6 | 7 |
| TUA | 7 | 1 | 5 | 1 | 4 | 1 | 6 | 1 | 8 | 1 | 3 | 3 | |
| YLS | 6 | 4 | 4 | 7 | 5 | 6 | 7 | 5 | 4 | 7 | 5 | 5 | |
| UBQ | 5 | 7 | 6 | 6 | 7 | 7 | 1 | 7 | 6 | 6 | 7 | 6 | |
| ACT | 4 | 4 | 3 | 4 | 6 | 5 | 2 | 3 | 2 | 5 | 4 | 4 | |
| SKD1 | 3 | 3 | 1 | 3 | 3 | 4 | 3 | 4 | 1 | 3 | 1 | 1 | |
| GAPDH | 2 | 2 | 2 | 2 | 2 | 3 | 4 | 2 | 3 | 2 | 2 | 2 | |
| EF-1α | 1 | 8 | 8 | 5 | 8 | 8 | 8 | 8 | 7 | 4 | 8 | 8 | |
| NormFinder | UBC | 8 | 7 | 7 | 8 | 5 | 6 | 8 | 7 | 6 | 8 | 7 | 6 |
| TUA | 7 | 4 | 3 | 4 | 6 | 4 | 7 | 3 | 8 | 5 | 3 | 8 | |
| YLS | 5 | 2 | 2 | 7 | 2 | 3 | 6 | 1 | 1 | 6 | 2 | 1 | |
| UBQ | 3 | 6 | 5 | 2 | 7 | 7 | 2 | 6 | 5 | 2 | 6 | 5 | |
| ACT | 1 | 1 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 3 | 1 | 3 | |
| SKD1 | 2 | 3 | 4 | 1 | 1 | 2 | 3 | 5 | 2 | 1 | 4 | 2 | |
| GAPDH | 4 | 5 | 6 | 5 | 4 | 5 | 4 | 4 | 4 | 4 | 5 | 4 | |
| EF-1α | 6 | 8 | 8 | 6 | 8 | 8 | 5 | 8 | 7 | 7 | 8 | 7 | |
| BestKeeper | UBC | 1 | 6 | 3 | 1 | 3 | 6 | 1 | 6 | 5 | 1 | 6 | 6 |
| TUA | 7 | 2 | 7 | 4 | 6 | 2 | 8 | 4 | 8 | 6 | 3 | 2 | |
| YLS | 2 | 5 | 4 | 2 | 2 | 5 | 2 | 5 | 3 | 2 | 5 | 5 | |
| UBQ | 3 | 7 | 6 | 3 | 7 | 7 | 3 | 7 | 6 | 3 | 7 | 7 | |
| ACT | 4 | 3 | 5 | 6 | 5 | 3 | 4 | 3 | 4 | 4 | 4 | 4 | |
| SKD1 | 5 | 1 | 2 | 5 | 1 | 1 | 5 | 1 | 1 | 5 | 1 | 3 | |
| GAPDH | 8 | 4 | 1 | 7 | 4 | 4 | 6 | 2 | 2 | 7 | 2 | 1 | |
| EF-1α | 6 | 8 | 8 | 8 | 8 | 8 | 7 | 8 | 7 | 8 | 8 | 8 | |
| RefFinder | UBC | 7 | 6 | 6 | 6 | 4 | 4 | 6 | 7 | 6 | 6 | 7 | 7 |
| TUA | 8 | 2 | 5 | 2 | 5 | 3 | 8 | 1 | 8 | 5 | 4 | 5 | |
| YLS | 6 | 5 | 4 | 7 | 3 | 5 | 5 | 4 | 2 | 7 | 5 | 4 | |
| UBQ | 4 | 7 | 7 | 4 | 7 | 7 | 2 | 6 | 5 | 3 | 6 | 6 | |
| ACT | 1 | 1 | 1 | 5 | 6 | 2 | 1 | 2 | 3 | 4 | 1 | 2 | |
| SKD1 | 2 | 3 | 3 | 1 | 1 | 1 | 3 | 5 | 1 | 1 | 2 | 1 | |
| GAPDH | 3 | 4 | 2 | 3 | 2 | 6 | 4 | 3 | 4 | 2 | 3 | 3 | |
| EF-1α | 5 | 8 | 8 | 8 | 8 | 8 | 7 | 8 | 7 | 8 | 8 | 8 | |
| ΔCt | UBC | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 7 | 6 |
| TUA | 4 | 3 | 3 | 5 | 2 | 3 | 4 | 3 | 3 | 2 | 1 | 2 | |
| YLS | 6 | 7 | 7 | 7 | 7 | 7 | 6 | 6 | 7 | 6 | 8 | 8 | |
| UBQ | 2 | 5 | 5 | 3 | 5 | 5 | 2 | 5 | 5 | 3 | 6 | 5 | |
| ACT | 7 | 6 | 6 | 6 | 6 | 6 | 7 | 6 | 6 | 7 | 4 | 3 | |
| SKD1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | 5 | 1 | |
| GAPDH | 5 | 2 | 2 | 2 | 3 | 2 | 5 | 2 | 2 | 5 | 3 | 4 | |
| EF-1α | 3 | 4 | 4 | 4 | 4 | 4 | 3 | 4 | 4 | 1 | 2 | 7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Q.; Li, X.; Qu, R.; Liu, Y.; Su, X.; Jin, J.; Yu, M.; Cairang, Z.; Zhang, P.; Zheng, Y.; et al. Selection and Validation of Stable Reference Genes for RT-qPCR Analyses of Rumex patientia (Polygonaceae) Under Four Abiotic Stresses. Genes 2025, 16, 787. https://doi.org/10.3390/genes16070787
Yang Q, Li X, Qu R, Liu Y, Su X, Jin J, Yu M, Cairang Z, Zhang P, Zheng Y, et al. Selection and Validation of Stable Reference Genes for RT-qPCR Analyses of Rumex patientia (Polygonaceae) Under Four Abiotic Stresses. Genes. 2025; 16(7):787. https://doi.org/10.3390/genes16070787
Chicago/Turabian StyleYang, Qian, Xiaoli Li, Rongju Qu, Yuping Liu, Xu Su, Jiarui Jin, Mingjun Yu, Zhaxi Cairang, Penghui Zhang, Yinghui Zheng, and et al. 2025. "Selection and Validation of Stable Reference Genes for RT-qPCR Analyses of Rumex patientia (Polygonaceae) Under Four Abiotic Stresses" Genes 16, no. 7: 787. https://doi.org/10.3390/genes16070787
APA StyleYang, Q., Li, X., Qu, R., Liu, Y., Su, X., Jin, J., Yu, M., Cairang, Z., Zhang, P., Zheng, Y., Gao, X., & Caraballo-Ortiz, M. A. (2025). Selection and Validation of Stable Reference Genes for RT-qPCR Analyses of Rumex patientia (Polygonaceae) Under Four Abiotic Stresses. Genes, 16(7), 787. https://doi.org/10.3390/genes16070787








