Genome-Wide Identification and Analysis of bZIP Transcription Factor Gene Family in Broomcorn Millet (Panicum miliaceum L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome-Wide Identification and Prediction of Physicochemical Properties
2.2. Phylogeny Analyses and bZIP Domain Amino Acid Sequence Alignment
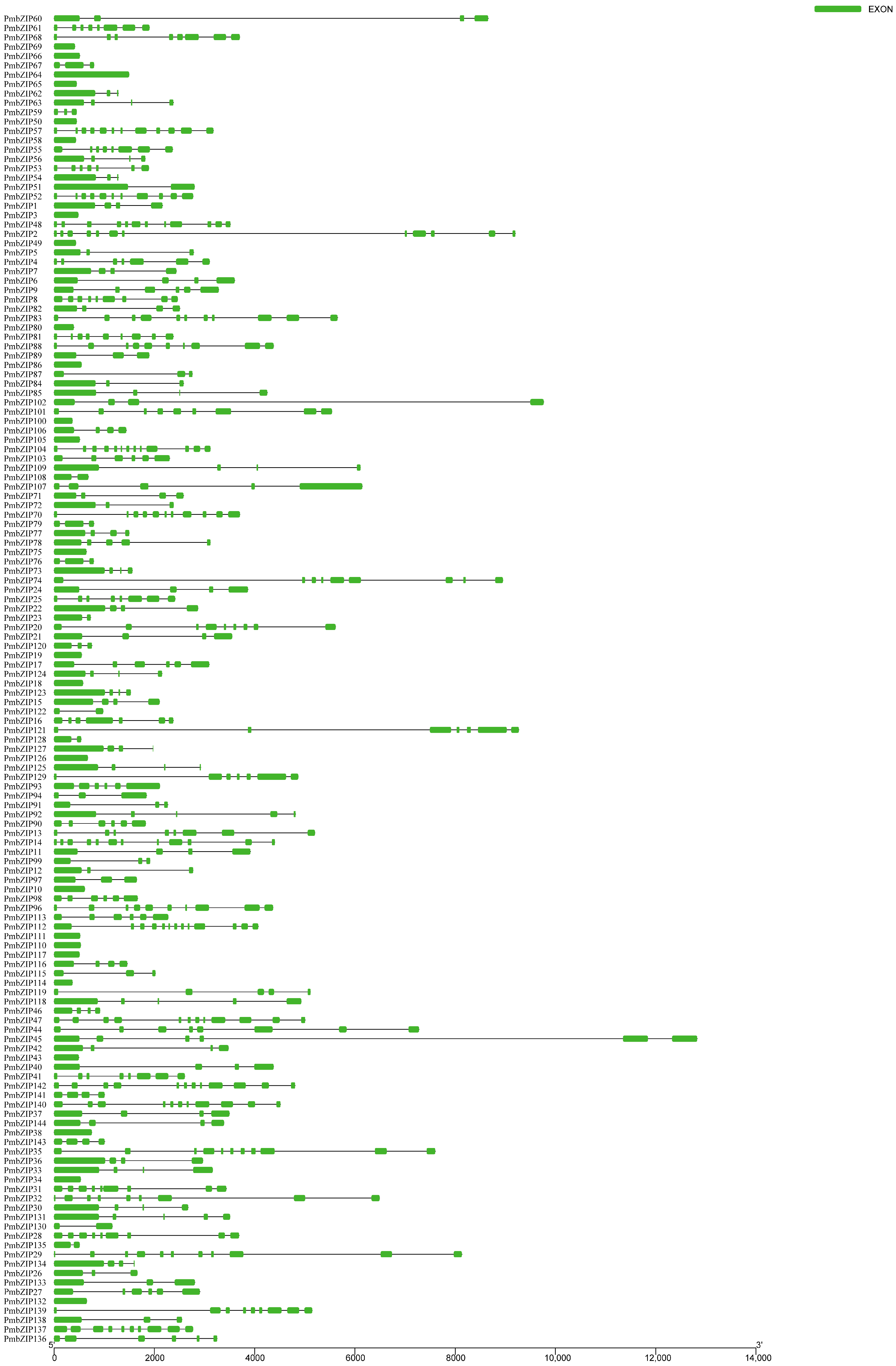
2.3. Motif and Intron/Exon Gene Structure Analysis
2.4. Promoter Analysis
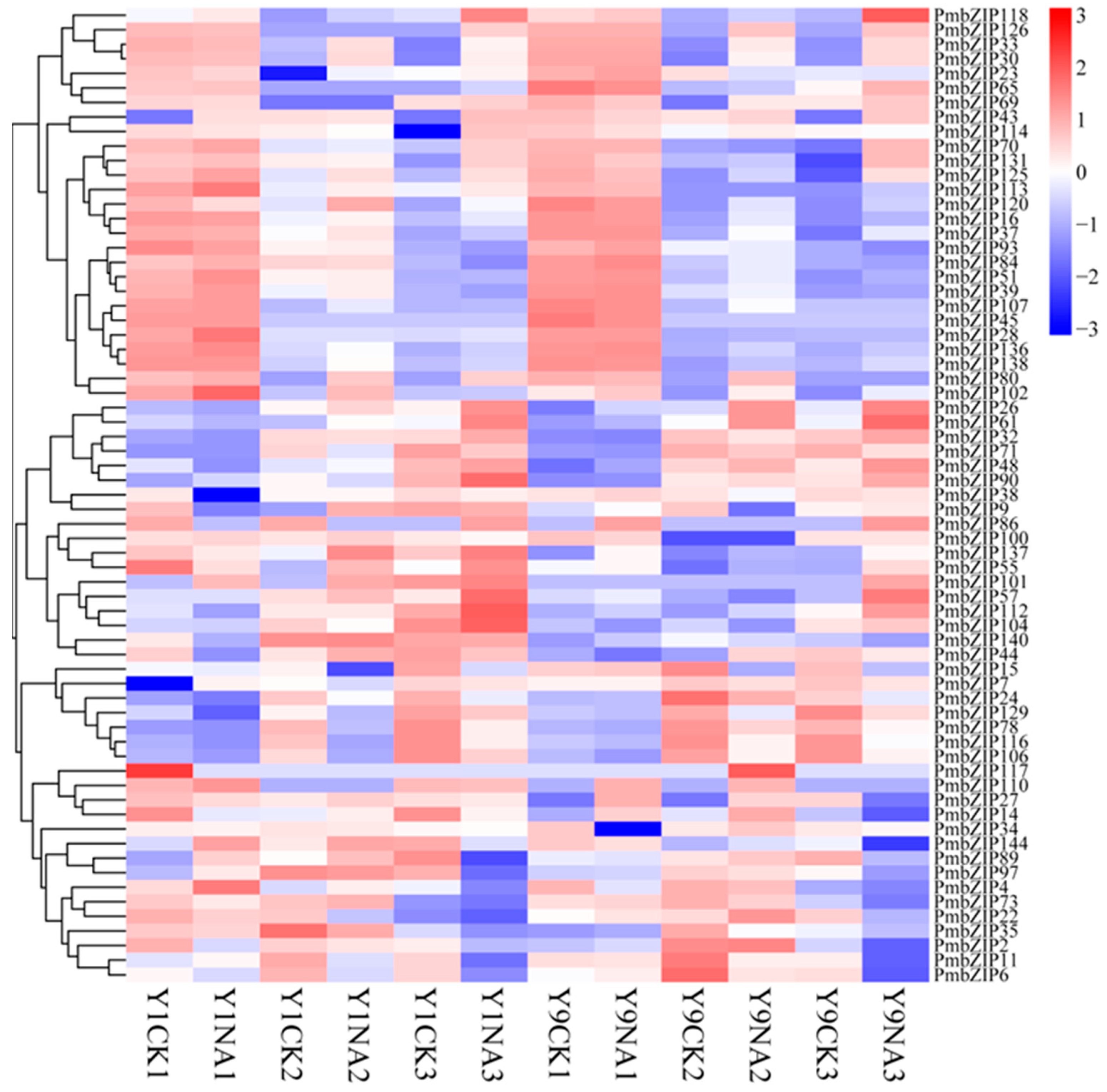
2.5. RNA Isolation and bZIP Gene Expression Analysis
3. Results
3.1. Identification of PmbZIPs in Broomcorn Millet
3.2. Phylogenetic and Sequence Conservation Analysis of PmbZIPs
3.3. Motif and Structural Analysis of PmbZIPs
3.4. Promoter Analysis of PmbZIPs
3.5. Expression Analysis of PmbZIPs in Seed Germination Under Salt Stress
4. Discussion
4.1. Characterization of Broomcorn Millet bZIP Gene Family
4.2. Structural Analysis of PmbZIPs
4.3. Cis-Element Analysis in the Promoters of PmbZIPs
4.4. PmbZIP Involvement in Development and Stress Response
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lu, H.; Zhang, J.; Liu, K.-B.; Wu, N.; Li, Y.; Zhou, K.; Ye, M.; Zhang, T.; Zhang, H.; Yang, X.; et al. Earliest domestication of common millet (Panicum miliaceum) in East Asia extended to 10,000 years ago. Proc. Natl. Acad. Sci. USA 2009, 106, 7367–7372. [Google Scholar] [CrossRef] [PubMed]
- Yue, H.; Wang, M.; Liu, S.; Du, X.; Song, W.; Nie, X. Transcriptome-wide identification and expression profiles of the WRKY transcription factor family in Broomcorn millet (Panicum miliaceum L.). BMC Genom. 2016, 17, 343. [Google Scholar] [CrossRef] [PubMed]
- Rajput, S.G.; Santra, D.K.; Schnable, J.J.M.B. Mapping QTLs for morpho-agronomic traits in proso millet (Panicum miliaceum L.). Mol. Breed. 2016, 36, 37. [Google Scholar] [CrossRef]
- Hunt, H.V.; Badakshi, F.; Romanova, O.; Howe, C.J.; Jones, M.K.; Heslop-Harrison, J.S. Reticulate evolution in Panicum (Poaceae): The origin of tetraploid broomcorn millet, P. miliaceum. J. Exp. Bot. 2014, 65, 3165–3175. [Google Scholar] [CrossRef]
- Wang, H.; Wang, J.; Chen, C.; Chen, L.; Li, M.; Qin, H.; Tian, X.; Hou, S.; Yang, X.; Jian, J.; et al. A complete reference genome of broomcorn millet. Sci. Data 2024, 11, 657. [Google Scholar] [CrossRef]
- Zou, C.; Li, L.; Miki, D.; Li, D.; Tang, Q.; Xiao, L.; Rajput, S.; Deng, P.; Peng, L.; Jia, W.; et al. The genome of broomcorn millet. Nat. Commun. 2019, 10, 436. [Google Scholar] [CrossRef]
- Baltensperger, D.D. Foxtail and Proso Millet. In Progress in New Crops; ASHS Press: Alexandria, VA, USA, 1996. [Google Scholar]
- Washburn, J.D.; Schnable, J.C.; Davidse, G.; Pires, J.C. Phylogeny and photosynthesis of the grass tribe Paniceae. Am. J. Bot. 2015, 102, 1493–1505. [Google Scholar] [CrossRef]
- Cedric, H.; Matanguihan, J.B.; D’Alpoim Guedes, J.; Ganjyal, G.M.; Whiteman, M.R.; Kidwell, K.K.; Murphy, K.M. Proso Millet (Panicum miliaceum L.) and Its Potential for Cultivation in the Pacific Northwest, U.S.: A Review. Front. Plant Sci. 2016, 7, 1961. [Google Scholar]
- Shi, J.; Ma, X.; Zhang, J.; Zhou, Y.; Liu, M.; Huang, L.; Sun, S.; Zhang, X.; Gao, X.; Zhan, W. Chromosome conformation capture resolved near complete genome assembly of broomcorn millet. Nat. Commun. 2019, 10, 464. [Google Scholar] [CrossRef]
- Lai, X.; Chahtane, H.; Martin-Arevalillo, R.; Zubieta, C.; Parcy, F. Contrasted evolutionary trajectories of plant transcription factors. Curr. Opin. Plant Biol. 2020, 54, 101–107. [Google Scholar] [CrossRef]
- de Mendoza, A.; Sebé-Pedrós, A.; Šestak, M.S.; Matejcic, M.; Torruella, G.; Domazet-Loso, T.; Ruiz-Trillo, I. Transcription factor evolution in eukaryotes and the assembly of the regulatory toolkit in multicellular lineages. Proc. Natl. Acad. Sci. USA 2013, 110, E4858–E4866. [Google Scholar] [CrossRef] [PubMed]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Droge-Laser, W.; Snoek, B.L.; Snel, B.; Weiste, C. The Arabidopsis bZIP transcription factor family-an update. Curr. Opin. Plant Biol. 2018, 45, 36–49. [Google Scholar] [CrossRef]
- Deppmann, C.D.; Acharya, A.; Rishi, V.; Wobbes, B.; Smeekens, S.; Taparowsky, E.J.; Vinson, C. Dimerization specificity of all 67 B-ZIP motifs in Arabidopsis thaliana: A comparison to Homo sapiens B-ZIP motifs. Nucleic Acids Res. 2004, 32, 3435–3445. [Google Scholar] [CrossRef]
- Mathura, S.R.; Sutton, F.; Bowrin, V. Characterization and expression analysis of SnRK2, PYL, and ABF/AREB/ABI5 gene families in sweet potato. PLoS ONE 2023, 18, e0288481. [Google Scholar] [CrossRef]
- Nijhawan, A.; Jain, M.; Tyagi, A.K.; Khurana, J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008, 146, 333–350. [Google Scholar] [CrossRef]
- Li, X.; Gao, S.; Tang, Y.; Li, L.; Zhang, F.; Feng, B.; Fang, Z.; Ma, L.; Zhao, C. Genome-wide identification and evolutionary analyses of bZIP transcription factors in wheat and its relatives and expression profiles of anther development related TabZIP genes. BMC Genom. 2015, 16, 976. [Google Scholar] [CrossRef]
- Rong, S.; Wu, Z.; Cheng, Z.; Zhang, S.; Liu, H.; Huang, Q. Genome-Wide Identification, Evolutionary Patterns, and Expression Analysis of bZIP Gene Family in Olive (Olea europaea L.). Genes 2020, 11, 510. [Google Scholar] [CrossRef]
- Pan, F.; Wu, M.; Hu, W.; Liu, R.; Yan, H.; Xiang, Y. Genome-Wide Identification and Expression Analyses of the bZIP Transcription Factor Genes in moso bamboo (Phyllostachys edulis). Int. J. Mol. Sci. 2019, 20, 2203. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, D.; Jia, L.; Huang, X.; Ma, G.; Wang, S.; Zhu, M.; Zhang, A.; Guan, M.; Lu, K.; et al. Genome-Wide Identification and Structural Analysis of bZIP Transcription Factor Genes in Brassica napus. Genes 2017, 8, 288. [Google Scholar] [CrossRef]
- Wang, X.L.; Chen, X.; Yang, T.B.; Cheng, Q.; Cheng, Z.M. Genome-Wide Identification of bZIP Family Genes Involved in Drought and Heat Stresses in Strawberry (Fragaria vesca). Int. J. Genom. 2017, 2017, 3981031. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, X.; Wang, F.; Li, X.; Qin, Z.; Fan, S.; Yan, N.; Xie, Y.; Zhao, R. Metabolomic response of Zizania latifolia to low-temperature stress and identification of the bZIP transcription factor family. GM Crops Food 2025, 16, 413–434. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Sun, J.; Chen, Y.; Zhu, P.; Zhang, L.; Wu, S.; Ma, D.; Cao, Q.; Li, Z.; Xu, T. Genome-wide identification, structural and gene expression analysis of the bZIP transcription factor family in sweet potato wild relative Ipomoea trifida. BMC Genet. 2019, 20, 41. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Roychoudhury, A. Abscisic-acid-dependent basic leucine zipper (bZIP) transcription factors in plant abiotic stress. Protoplasma 2017, 254, 3–16. [Google Scholar] [CrossRef]
- Amir Hossain, M.; Lee, Y.; Cho, J.I.; Ahn, C.H.; Lee, S.K.; Jeon, J.S.; Kang, H.; Lee, C.H.; An, G.; Park, P.B. The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol. Biol. 2010, 72, 557–566. [Google Scholar] [CrossRef]
- Basso, M.F.; Iovieno, P.; Capuana, M.; Contaldi, F.; Ieri, F.; Menicucci, F.; Celso, F.L.; Barone, G.; Martinelli, F. Identification and expression of the AREB/ABF/ABI5 subfamily genes in chickpea and lentil reveal major players involved in ABA-mediated defense response to drought stress. Planta 2025, 262, 22. [Google Scholar] [CrossRef]
- Zou, M.; Guan, Y.; Ren, H.; Zhang, F.; Chen, F. A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol. Biol. 2008, 66, 675–683. [Google Scholar] [CrossRef]
- Hossain, M.A.; Cho, J.I.; Han, M.; Ahn, C.H.; Jeon, J.S.; An, G.; Park, P.B. The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J. Plant Physiol. 2010, 167, 1512–1520. [Google Scholar] [CrossRef]
- Orellana, S.; Yañez, M.; Espinoza, A.; Verdugo, I.; González, E.; Ruiz-Lara, S.; Casaretto, J.A. The transcription factor SlAREB1 confers drought, salt stress tolerance and regulates biotic and abiotic stress-related genes in tomato. Plant Cell Environ. 2010, 33, 2191–2208. [Google Scholar] [CrossRef]
- Kim, S.; Kang, J.Y.; Cho, D.I.; Park, J.H.; Kim, S.Y. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 2004, 40, 75–87. [Google Scholar] [CrossRef]
- Kim, J.H.; Hyun, W.Y.; Nguyen, H.N.; Jeong, C.Y.; Xiong, L.; Hong, S.W.; Lee, H. AtMyb7, a subgroup 4 R2R3 Myb, negatively regulates ABA-induced inhibition of seed germination by blocking the expression of the bZIP transcription factor ABI5. Plant Cell Environ. 2015, 38, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Skubacz, A.; Daszkowska-Golec, A.; Szarejko, I. The Role and Regulation of ABI5 (ABA-Insensitive 5) in Plant Development, Abiotic Stress Responses and Phytohormone Crosstalk. Front. Plant Sci. 2016, 7, 1884. [Google Scholar] [CrossRef] [PubMed]
- Maier, A.T.; Stehling-Sun, S.; Wollmann, H.; Demar, M.; Hong, R.L.; Haubeiss, S.; Weigel, D.; Lohmann, J.U. Dual roles of the bZIP transcription factor PERIANTHIA in the control of floral architecture and homeotic gene expression. Development 2009, 136, 1613–1620. [Google Scholar] [CrossRef]
- Abe, M.; Kobayashi, Y.; Yamamoto, S.; Daimon, Y.; Yamaguchi, A.; Ikeda, Y.; Ichinoki, H.; Notaguchi, M.; Goto, K.; Araki, T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 2005, 309, 1052–1056. [Google Scholar] [CrossRef]
- Shu, K.; Chen, Q.; Wu, Y.; Liu, R.; Zhang, H.; Wang, S.; Tang, S.; Yang, W.; Xie, Q. ABSCISIC ACID-INSENSITIVE 4 negatively regulates flowering through directly promoting Arabidopsis FLOWERING LOCUS C transcription. J. Exp. Bot. 2016, 67, 195–205. [Google Scholar] [CrossRef]
- Howell, S.H. Endoplasmic reticulum stress responses in plants. Annu. Rev. Plant Biol. 2013, 64, 477–499. [Google Scholar] [CrossRef]
- Oñate, L.; Vicente-Carbajosa, J.; Lara, P.; Díaz, I.; Carbonero, P. Barley BLZ2, a seed-specific bZIP protein that interacts with BLZ1 in vivo and activates transcription from the GCN4-like motif of B-hordein promoters in barley endosperm. J. Biol. Chem. 1999, 274, 9175–9182. [Google Scholar] [CrossRef]
- Onodera, Y.; Suzuki, A.; Wu, C.Y.; Washida, H.; Takaiwa, F. A rice functional transcriptional activator, RISBZ1, responsible for endosperm-specific expression of storage protein genes through GCN4 motif. J. Biol. Chem. 2001, 276, 14139–14152. [Google Scholar] [CrossRef]
- Vicente-Carbajosa, J.; Oñate, L.; Lara, P.; Diaz, I.; Carbonero, P. Barley BLZ1: A bZIP transcriptional activator that interacts with endosperm-specific gene promoters. Plant J. 1998, 13, 629–640. [Google Scholar] [CrossRef]
- Mueller, S.; Hilbert, B.; Dueckershoff, K.; Roitsch, T.; Krischke, M.; Mueller, M.J.; Berger, S. General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell 2008, 20, 768–785. [Google Scholar] [CrossRef]
- Zander, M.; La Camera, S.; Lamotte, O.; Métraux, J.P.; Gatz, C. Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. Plant J. 2010, 61, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Choi, S.M.; Kang, M.J.; Yun, S.H.; Kwon, D.J.; Noh, Y.S.; Noh, B. Salicylic acid-induced transcriptional reprogramming by the HAC-NPR1-TGA histone acetyltransferase complex in Arabidopsis. Nucleic Acids Res. 2018, 46, 11712–11725. [Google Scholar] [CrossRef] [PubMed]
- Hussain, R.M.F.; Sheikh, A.H.; Haider, I.; Quareshy, M.; Linthorst, H.J.M. Arabidopsis WRKY50 and TGA Transcription Factors Synergistically Activate Expression of PR1. Front. Plant Sci. 2018, 9, 930. [Google Scholar] [CrossRef]
- Ndamukong, I.; Abdallat, A.A.; Thurow, C.; Fode, B.; Zander, M.; Weigel, R.; Gatz, C. SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 2007, 50, 128–139. [Google Scholar] [CrossRef]
- Gachon, F.; Olela, F.F.; Schaad, O.; Descombes, P.; Schibler, U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006, 4, 25–36. [Google Scholar] [CrossRef]
- Gibalová, A.; Reňák, D.; Matczuk, K.; Dupl’áková, N.; Cháb, D.; Twell, D.; Honys, D. AtbZIP34 is required for Arabidopsis pollen wall patterning and the control of several metabolic pathways in developing pollen. Plant Mol. Biol. 2009, 70, 581–601. [Google Scholar] [CrossRef]
- Assunção, A.G.; Herrero, E.; Lin, Y.F.; Huettel, B.; Talukdar, S.; Smaczniak, C.; Immink, R.G.; van Eldik, M.; Fiers, M.; Schat, H.; et al. Arabidopsis thaliana transcription factors bZIP19 and bZIP23 regulate the adaptation to zinc deficiency. Proc. Natl. Acad. Sci. USA 2010, 107, 10296–10301. [Google Scholar] [CrossRef]
- Inaba, S.; Kurata, R.; Kobayashi, M.; Yamagishi, Y.; Mori, I.; Ogata, Y.; Fukao, Y. Identification of putative target genes of bZIP19, a transcription factor essential for Arabidopsis adaptation to Zn deficiency in roots. Plant J. 2015, 84, 323–334. [Google Scholar] [CrossRef]
- Lopes-da-Silva, M.; McCormack, J.J.; Burden, J.J.; Harrison-Lavoie, K.J.; Ferraro, F.; Cutler, D.F. A GBF1-Dependent Mechanism for Environmentally Responsive Regulation of ER-Golgi Transport. Dev. Cell 2019, 49, 786–801.e786. [Google Scholar] [CrossRef]
- Giri, M.K.; Singh, N.; Banday, Z.Z.; Singh, V.; Ram, H.; Singh, D.; Chattopadhyay, S.; Nandi, A.K. GBF1 differentially regulates CAT2 and PAD4 transcription to promote pathogen defense in Arabidopsis thaliana. Plant J. 2017, 91, 802–815. [Google Scholar] [CrossRef]
- Yang, Y.; Liang, T.; Zhang, L.; Shao, K.; Gu, X.; Shang, R.; Shi, N.; Li, X.; Zhang, P.; Liu, H. UVR8 interacts with WRKY36 to regulate HY5 transcription and hypocotyl elongation in Arabidopsis. Nat. Plants 2018, 4, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Osterlund, M.T.; Hardtke, C.S.; Wei, N.; Deng, X.W. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 2000, 405, 462–466. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Terzaghi, W.; Gong, Y.; Li, C.; Ling, J.J.; Fan, Y.; Qin, N.; Gong, X.; Zhu, D.; Deng, X.W. Modulation of BIN2 kinase activity by HY5 controls hypocotyl elongation in the light. Nat. Commun. 2020, 11, 1592. [Google Scholar] [CrossRef] [PubMed]
- Tsugama, D.; Liu, S.; Takano, T. Analysis of functions of VIP1 and its close homologs in osmosensory responses of Arabidopsis thaliana. PLoS ONE 2014, 9, e103930. [Google Scholar] [CrossRef]
- Tsugama, D.; Liu, S.; Fujino, K.; Takano, T. Calcium signalling regulates the functions of the bZIP protein VIP1 in touch responses in Arabidopsis thaliana. Ann. Bot. 2018, 122, 1219–1229. [Google Scholar] [CrossRef]
- Choi, H.; Hong, J.; Ha, J.; Kang, J.; Kim, S.Y. ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 2000, 275, 1723–1730. [Google Scholar] [CrossRef]
- Fukazawa, J.; Sakai, T.; Ishida, S.; Yamaguchi, I.; Kamiya, Y.; Takahashi, Y. Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 2000, 12, 901–915. [Google Scholar] [CrossRef]
- Izawa, T.; Foster, R.; Nakajima, M.; Shimamoto, K.; Chua, N.H. The rice bZIP transcriptional activator RITA-1 is highly expressed during seed development. Plant Cell 1994, 6, 1277–1287. [Google Scholar] [CrossRef]
- An, P.; Li, X.; Liu, T.; Shui, Z.; Chen, M.; Gao, X.; Wang, Z. The Identification of Broomcorn Millet bZIP Transcription Factors, Which Regulate Growth and Development to Enhance Stress Tolerance and Seed Germination. Int. J. Mol. Sci. 2022, 23, 6448. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, M.; Kong, L.; Lv, Y.; Zou, M.; Lu, G.; Cao, J.; Yu, X. Genome-wide identification, phylogeny, duplication, and expression analyses of two-component system genes in Chinese cabbage (Brassica rapa ssp. pekinensis). DNA Res. Int. J. Rapid Publ. Rep. Genes. Genomes 2014, 21, 379–396. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, P.; Hong, J.K.; Kim, J.A.; Jeong, M.-J.; Lee, Y.-H.; Lee, S.I. Genome-wide analysis of the expansin gene superfamily reveals Brassica rapa-specific evolutionary dynamics upon whole genome triplication. Mol. Genet. Genom. MGG 2015, 290, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Barker, M.S.; Baute, G.J.; Liu, S.L. Duplications and Turnover in Plant Genomes. In Plant Genome Diversity; Springer: Vienna, Austria, 2012; Volume 1, pp. 155–169. [Google Scholar]
- Zhang, J. Evolution by gene duplication: An update. Trends Ecol. Evol. 2003, 18, 292–298. [Google Scholar] [CrossRef]
- Jacob, A.G.; Smith, C.W.J. Intron retention as a component of regulated gene expression programs. Hum. Genet. 2017, 136, 1043–1057. [Google Scholar] [CrossRef]
- Wang, W.; Wang, Y.; Zhang, S.; Xie, K.; Zhang, C.; Xi, Y.; Sun, F. Genome-wide analysis of the abiotic stress-related bZIP family in switchgrass. Mol. Biol. Rep. 2020, 47, 4439–4454. [Google Scholar] [CrossRef]
- Hu, W.; Wang, L.; Tie, W.; Yan, Y.; Ding, Z.; Liu, J.; Li, M.; Peng, M.; Xu, B.; Jin, Z. Genome-wide analyses of the bZIP family reveal their involvement in the development, ripening and abiotic stress response in banana. Sci. Rep. 2016, 6, 30203. [Google Scholar] [CrossRef]
- Yang, S.; Xu, K.; Chen, S.; Li, T.; Xia, H.; Chen, L.; Liu, H.; Luo, L. A stress-responsive bZIP transcription factor OsbZIP62 improves drought and oxidative tolerance in rice. BMC Plant Biol. 2019, 19, 260. [Google Scholar] [CrossRef]
- Ma, H.; Liu, C.; Li, Z.; Ran, Q.; Xie, G.; Wang, B.; Fang, S.; Chu, J.; Zhang, J. ZmbZIP4 Contributes to Stress Resistance in Maize by Regulating ABA Synthesis and Root Development. Plant Physiol. 2018, 178, 753–770. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, Y.; Cai, H.; Guo, M.; Chai, M.; She, Z.; Ye, L.; Cheng, Y.; Wang, B.; Qin, Y. The bZIP Transcription Factor GmbZIP15 Negatively Regulates Salt- and Drought-Stress Responses in Soybean. Int. J. Mol. Sci. 2020, 21, 7778. [Google Scholar] [CrossRef]
- Bi, C.; Yu, Y.; Dong, C.; Yang, Y.; Zhai, Y.; Du, F.; Xia, C.; Ni, Z.; Kong, X.; Zhang, L. The bZIP transcription factor TabZIP15 improves salt stress tolerance in wheat. Plant Biotechnol. J. 2021, 19, 209–211. [Google Scholar] [CrossRef]
- Zhao, P.; Ye, M.; Wang, R.; Wang, D.; Chen, Q. Systematic identification and functional analysis of potato (Solanum tuberosum L.) bZIP transcription factors and overexpression of potato bZIP transcription factor StbZIP-65 enhances salt tolerance. Int. J. Biol. Macromol. 2020, 161, 155–167. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Cai, H.; Bai, M.; Zhang, M.; Chen, F.; Huang, Y.; Priyadarshani, S.V.G.N.; Chai, M.; Liu, L.; Liu, Y.; et al. A Soybean bZIP Transcription Factor Confers Multiple Biotic and Abiotic Stress Responses in Plant. Int. J. Mol. Sci. 2020, 21, 4701. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Yang, Y.; Wang, W.; Guo, G.; Liu, W.; Bi, C. Overexpression of a wheat (Triticum aestivum L.) bZIP transcription factor gene, TabZIP6, decreased the freezing tolerance of transgenic Arabidopsis seedlings by down-regulating the expression of CBFs. Plant Physiol. Biochem. 2018, 124, 100–111. [Google Scholar] [CrossRef] [PubMed]



| Gene ID | Protein Number | Gene Number | Chr | Gene Location | Gene Length/bp | CDS/nt | Amino Acids | PI | Molecular Weight/u |
|---|---|---|---|---|---|---|---|---|---|
| PmbZIP1 | RLN41431.1 | PM01G00370 | 1 | 454801–457374 | 2574 | 1263 | 421 | 5.71 | 44,828.00 |
| PmbZIP2 | RLN41707.1 | PM01G08790 | 1 | 7046270–7059039 | 12,770 | 1149 | 383 | 9.13 | 40,969.23 |
| PmbZIP3 | RLN40362.1 | PM01G12600 | 1 | 10097025–10098402 | 1378 | 486 | 162 | 10.55 | 17,692.55 |
| PmbZIP4 | RLN40088.1 | PM01G13370 | 1 | 10863218–10866321 | 3104 | 912 | 304 | 9.45 | 34,047.74 |
| PmbZIP5 | RLN40734.1 | PM01G13620 | 1 | 11058035–11060820 | 2786 | 684 | 228 | 9.84 | 24,396.40 |
| PmbZIP6 | RLN39254.1 | PM01G14300 | 1 | 11670497–11674442 | 3946 | 1053 | 351 | 6.23 | 37,904.15 |
| PmbZIP7 | RLN41556.1 | PM01G19740 | 1 | 16954898–16957707 | 2810 | 1161 | 387 | 7.23 | 41,543.20 |
| PmbZIP8 | RLN41102.1 | PM01G22630 | 1 | 19957313–19960138 | 2826 | 1056 | 352 | 6.88 | 37,179.95 |
| PmbZIP9 | RLN42614.1 | PM01G23610 | 1 | 20646169–20649807 | 3639 | 1242 | 414 | 5.06 | 42,598.17 |
| PmbZIP10 | RLN40049.1 | PM01G26880 | 1 | 22949190–22950304 | 1115 | 615 | 205 | 10.18 | 22,321.56 |
| PmbZIP11 | RLN19317.1 | PM02G26900 | 2 | 39030059–39034459 | 4401 | 1050 | 350 | 6.28 | 37,679.93 |
| PmbZIP12 | RLN19126.1 | PM02G27550 | 2 | 39595051–39597823 | 2773 | 702 | 234 | 10.00 | 24,691.66 |
| PmbZIP13 | RLN18095.1 | PM02G27800 | 2 | 39766627–39771830 | 5204 | 999 | 333 | 7.04 | 37,066.87 |
| PmbZIP14 | RLN17017.1 | PM02G32070 | 2 | 43256944–43264901 | 7958 | 1149 | 383 | 9.20 | 40,881.04 |
| PmbZIP15 | RLN15790.1 | PM02G38970 | 2 | 48295652–48298282 | 2631 | 1224 | 408 | 5.60 | 43,833.94 |
| PmbZIP16 | RLN15762.1 | PM02G44000 | 2 | 51665787–51668165 | 2379 | 1161 | 387 | 6.10 | 41,310.03 |
| PmbZIP17 | RLN17495.1 | PM02G44930 | 2 | 52341406–52345126 | 3721 | 1260 | 420 | 4.97 | 43,404.97 |
| PmbZIP18 | RLN18647.1 | PM02G46570 | 2 | 53532299–53532877 | 579 | 579 | 193 | 10.48 | 21,390.51 |
| PmbZIP19 | RLN33487.1 | PM03G00660 | 3 | 678937–679488 | 552 | 552 | 184 | 7.21 | 20,564.99 |
| PmbZIP20 | RLN33484.1 | PM03G01530 | 3 | 1468835–1474450 | 5616 | 1014 | 338 | 9.88 | 36,231.59 |
| PmbZIP21 | RLN33244.1 | PM03G03170 | 3 | 2666045–2670259 | 4215 | 1140 | 380 | 6.22 | 40,613.30 |
| PmbZIP22 | RLN32939.1 | PM03G03540 | 3 | 2944837–2947710 | 2874 | 1464 | 488 | 9.20 | 51,669.75 |
| PmbZIP23 | RLN34898.1 | PM03G04810 | 3 | 3889634–3890848 | 1215 | 639 | 213 | 6.84 | 22,223.95 |
| PmbZIP24 | RLN35613.1 | PM03G16340 | 3 | 12865234–12869532 | 4299 | 1107 | 369 | 6.61 | 39,460.24 |
| PmbZIP25 | RLN33050.1 | PM03G16790 | 3 | 13287052–13290598 | 3547 | 1005 | 335 | 8.90 | 37,308.18 |
| PmbZIP26 | RLN35551.1 | PM03G16900 | 3 | 13342155–13343818 | 1664 | 783 | 261 | 10.02 | 28,735.61 |
| PmbZIP27 | RLN35711.1 | PM03G21650 | 3 | 18768221–18771281 | 3061 | 1179 | 393 | 5.01 | 42,558.22 |
| PmbZIP28 | RLN33934.1 | PM03G24730 | 3 | 22107028–22110718 | 3691 | 1161 | 387 | 6.61 | 39,600.43 |
| PmbZIP29 | RLN35629.1 | PM03G29190 | 3 | 35167059–35175866 | 8808 | 1257 | 419 | 8.44 | 46,181.77 |
| PmbZIP30 | RLN35964.1 | PM03G36770 | 3 | 57531413–57534679 | 3267 | 1122 | 374 | 5.10 | 39,436.23 |
| PmbZIP31 | RLM84895.1 | PM04G07270 | 4 | 6860114–6863550 | 3437 | 1233 | 411 | 7.18 | 42,128.33 |
| PmbZIP32 | RLM86983.1 | PM04G16890 | 4 | 29725581–29732074 | 6494 | 1122 | 374 | 9.54 | 41,241.35 |
| PmbZIP33 | RLM85912.1 | PM04G17970 | 4 | 31027628–31030792 | 3165 | 1392 | 464 | 9.15 | 49,993.48 |
| PmbZIP34 | RLM85616.1 | PM04G18960 | 4 | 31857501–31858542 | 1042 | 534 | 178 | 8.95 | 20,269.87 |
| PmbZIP35 | RLM87231.1 | PM04G19820 | 4 | 32472417–32480988 | 8572 | 1491 | 497 | 7.51 | 55,166.25 |
| PmbZIP36 | RLM85816.1 | PM04G21680 | 4 | 33883522–33886493 | 2972 | 1464 | 488 | 9.53 | 51,605.73 |
| PmbZIP37 | RLM86211.1 | PM04G22100 | 4 | 34132892–34136810 | 3919 | 1140 | 380 | 6.38 | 40,629.39 |
| PmbZIP38 | RLM85470.1 | PM04G22810 | 4 | 34716923–34718185 | 1263 | 753 | 251 | 7.83 | 26,101.09 |
| PmbZIP39 | RLM87366.1 | PM04G29540 | 4 | 39691716–39694242 | 2527 | 1491 | 497 | 5.86 | 60,644.54 |
| PmbZIP40 | RLM85147.1 | PM04G34030 | 4 | 42747464–42752113 | 4650 | 1116 | 372 | 6.27 | 39,659.45 |
| PmbZIP41 | RLM86171.1 | PM04G34490 | 4 | 43064867–43067475 | 2609 | 1014 | 338 | 8.90 | 37,565.47 |
| PmbZIP42 | RLM87099.1 | PM04G34620 | 4 | 43157454–43160933 | 3480 | 885 | 295 | 9.59 | 32,209.76 |
| PmbZIP43 | RLN30372.1 | PM05G00470 | 5 | 481032–481791 | 760 | 495 | 165 | 11.57 | 17,411.98 |
| PmbZIP44 | RLN29606.1 | PM05G02460 | 5 | 2429710–2436988 | 7279 | 1305 | 435 | 5.93 | 47,002.20 |
| PmbZIP45 | RLN28633.1 | PM05G04900 | 5 | 4731398–4744225 | 12,828 | 1812 | 604 | 4.88 | 65,083.35 |
| PmbZIP46 | RLN27945.1 | PM05G05230 | 5 | 5015435–5016619 | 1185 | 603 | 201 | 9.19 | 21,729.48 |
| PmbZIP47 | RLN28345.1 | PM05G05380 | 5 | 5101092–5106900 | 5809 | 1449 | 483 | 6.25 | 52,474.70 |
| PmbZIP48 | RLN27488.1 | PM05G08030 | 5 | 7121364–7128050 | 6687 | 1158 | 386 | 7.17 | 40,824.69 |
| PmbZIP49 | RLN29878.1 | PM05G11380 | 5 | 10166559–10169754 | 3196 | 438 | 146 | 9.49 | 16,063.02 |
| PmbZIP50 | RLN30397.1 | PM05G14850 | 5 | 13174076–13175991 | 1916 | 453 | 151 | 6.32 | 16,585.54 |
| PmbZIP51 | RLN29599.1 | PM05G25250 | 5 | 46340685–46343488 | 2804 | 1950 | 650 | 8.15 | 68,282.36 |
| PmbZIP52 | RLN31068.1 | PM05G29120 | 5 | 49501420–49504193 | 2774 | 1152 | 384 | 9.07 | 41,007.77 |
| PmbZIP53 | RLN28247.1 | PM05G33670 | 5 | 53089874–53091765 | 1892 | 543 | 181 | 9.75 | 20,078.72 |
| PmbZIP54 | RLN28944.1 | PM05G33910 | 5 | 53238531–53239814 | 1284 | 933 | 311 | 6.47 | 33,380.34 |
| PmbZIP55 | RLN28901.1 | PM05G36440 | 5 | 55255118–55257485 | 2368 | 1062 | 354 | 6.68 | 39,751.11 |
| PmbZIP56 | RLN27558.1 | PM05G36770 | 5 | 55582519–55584338 | 1820 | 780 | 260 | 9.55 | 26,952.16 |
| PmbZIP57 | RLN01174.1 | PM06G02700 | 6 | 1946640–1949819 | 3180 | 1287 | 429 | 9.30 | 45,780.45 |
| PmbZIP58 | RLN00773.1 | PM06G08160 | 6 | 5838864–5840465 | 1602 | 438 | 146 | 9.93 | 15,983.02 |
| PmbZIP59 | RLN01167.1 | PM06G14080 | 6 | 10158766–10159216 | 451 | 234 | 78 | 10.63 | 9190.60 |
| PmbZIP60 | RLM98944.1 | PM06G14410 | 6 | 10441629–10450559 | 8931 | 999 | 333 | 6.26 | 35,600.39 |
| PmbZIP61 | RLM98085.1 | PM06G17810 | 6 | 12976476–12980172 | 3697 | 999 | 333 | 6.55 | 36,500.10 |
| PmbZIP62 | RLM98033.1 | PM06G17990 | 6 | 13102719–13104001 | 1283 | 921 | 307 | 6.46 | 32,832.58 |
| PmbZIP63 | RLN00565.1 | PM06G20460 | 6 | 14965993–14968992 | 3000 | 777 | 259 | 9.73 | 26,937.16 |
| PmbZIP64 | RLM99485.1 | PM06G21690 | 6 | 16055800–16057296 | 1497 | 1497 | 499 | 7.03 | 52,598.69 |
| PmbZIP65 | RLM99017.1 | PM06G30890 | 6 | 39341652–39342104 | 453 | 453 | 151 | 6.85 | 16,674.63 |
| PmbZIP66 | RLM99272.1 | PM06G35160 | 6 | 43290439–43290954 | 516 | 516 | 172 | 11.51 | 18,029.63 |
| PmbZIP67 | RLN24624.1 | PM07G00440 | 7 | 317276–318073 | 798 | 570 | 190 | 9.98 | 20,307.29 |
| PmbZIP68 | RLN25705.1 | PM07G06850 | 7 | 6745210–6748914 | 3705 | 1080 | 360 | 7.69 | 40,074.24 |
| PmbZIP69 | RLN24094.1 | PM07G10230 | 7 | 8252890–8253306 | 417 | 417 | 139 | 7.65 | 15,279.28 |
| PmbZIP70 | RLN24562.1 | PM07G17750 | 7 | 36088695–36092402 | 3708 | 1110 | 370 | 9.19 | 39,245.04 |
| PmbZIP71 | RLN22304.1 | PM07G23470 | 7 | 41273483–41276065 | 2583 | 804 | 268 | 5.93 | 28,650.69 |
| PmbZIP72 | RLN22621.1 | PM07G25750 | 7 | 43086731–43089115 | 2385 | 981 | 327 | 7.18 | 35,585.99 |
| PmbZIP73 | RLN22673.1 | PM07G29370 | 7 | 45806587–45808150 | 1564 | 1206 | 402 | 6.29 | 42,396.98 |
| PmbZIP74 | RLN23257.1 | PM07G29380 | 7 | 45812241–45821193 | 8953 | 1227 | 409 | 6.90 | 46,071.37 |
| PmbZIP75 | RLN24712.1 | PM07G29810 | 7 | 46066329–46066976 | 648 | 648 | 216 | 9.75 | 21,645.44 |
| PmbZIP76 | RLN25012.1 | PM07G36760 | 7 | 51457907–51459054 | 1148 | 564 | 188 | 10.01 | 20,446.76 |
| PmbZIP77 | RLN24441.1 | PM07G38580 | 7 | 53023815–53026143 | 2329 | 921 | 307 | 6.62 | 32,963.72 |
| PmbZIP78 | RLM93109.1 | PM08G01760 | 8 | 1448785–1451902 | 3118 | 999 | 333 | 6.00 | 36,038.19 |
| PmbZIP79 | RLM93604.1 | PM08G03930 | 8 | 3296164–3296959 | 796 | 585 | 195 | 9.94 | 20,689.71 |
| PmbZIP80 | RLM91692.1 | PM08G15750 | 8 | 30944899–30945294 | 396 | 396 | 132 | 8.40 | 14,577.57 |
| PmbZIP81 | RLM92180.1 | PM08G20990 | 8 | 36451978–36454357 | 2380 | 831 | 277 | 9.31 | 30,542.73 |
| PmbZIP82 | RLM93703.1 | PM08G25890 | 8 | 40248925–40252095 | 3171 | 819 | 273 | 6.01 | 28,971.03 |
| PmbZIP83 | RLM92419.1 | PM08G28810 | 8 | 42140458–42146113 | 5656 | 1401 | 467 | 6.34 | 50,908.00 |
| PmbZIP84 | RLM94116.1 | PM08G29030 | 8 | 42303295–42305878 | 2584 | 981 | 327 | 7.15 | 35,542.86 |
| PmbZIP85 | RLN12842.1 | PM09G06500 | 9 | 5334910–5339163 | 4254 | 1092 | 364 | 8.70 | 39,102.69 |
| PmbZIP86 | RLN12841.1 | PM09G12140 | 9 | 21889497–21890045 | 549 | 549 | 183 | 9.67 | 20,244.55 |
| PmbZIP87 | RLN13025.1 | PM09G17690 | 9 | 43522067–43524827 | 2761 | 423 | 141 | 10.16 | 15,149.82 |
| PmbZIP88 | RLN12905.1 | PM09G18590 | 9 | 44792601–44796981 | 4381 | 1257 | 419 | 9.11 | 44,598.29 |
| PmbZIP89 | RLN12453.1 | PM09G18780 | 9 | 45041072–45044603 | 3532 | 912 | 304 | 4.83 | 32,658.00 |
| PmbZIP90 | RLN13126.1 | PM09G20800 | 9 | 47002029–47003859 | 1831 | 849 | 283 | 6.24 | 30,013.48 |
| PmbZIP91 | RLN12623.1 | PM09G24100 | 9 | 50246233–50248502 | 2270 | 477 | 159 | 9.00 | 17,365.70 |
| PmbZIP92 | RLM55632.1 | PM10G06050 | 10 | 4576359–4581176 | 4818 | 1113 | 371 | 9.00 | 40,165.13 |
| PmbZIP93 | RLM54848.1 | PM10G06130 | 10 | 4623806–4625917 | 2112 | 1524 | 508 | 9.16 | 55,261.12 |
| PmbZIP94 | RLM55196.1 | PM10G10750 | 10 | 10699576–10701422 | 1847 | 735 | 245 | 10.88 | 27,271.78 |
| PmbZIP95 | RLM54794.1 | PM10G15500 | 10 | 26027305–26029674 | 2370 | 525 | 175 | 9.75 | 18,810.58 |
| PmbZIP96 | RLM56218.1 | PM10G16250 | 10 | 26722529–26726897 | 4369 | 1362 | 454 | 9.13 | 48,594.99 |
| PmbZIP97 | RLM55027.1 | PM10G16410 | 10 | 26862899–26864549 | 1651 | 897 | 299 | 5.43 | 32,538.26 |
| PmbZIP98 | RLM55428.1 | PM10G18250 | 10 | 28349450–28351117 | 1668 | 867 | 289 | 6.07 | 30,673.21 |
| PmbZIP99 | RLM56082.1 | PM10G21570 | 10 | 31094548–31096463 | 1916 | 483 | 161 | 8.62 | 17,579.92 |
| PmbZIP100 | RLN09938.1 | PM11G00290 | 11 | 452913–453281 | 369 | 369 | 123 | 8.97 | 13,810.34 |
| PmbZIP101 | RLN08312.1 | PM11G02090 | 11 | 2470988–2476530 | 5543 | 1374 | 458 | 8.92 | 49,484.85 |
| PmbZIP102 | RLN07652.1 | PM11G02220 | 11 | 2606668–2616433 | 9766 | 1035 | 345 | 4.52 | 35,333.37 |
| PmbZIP103 | RLN09078.1 | PM11G02900 | 11 | 3245503–3248289 | 2787 | 939 | 313 | 5.42 | 33,095.76 |
| PmbZIP104 | RLN07125.1 | PM11G13460 | 11 | 25503439–25506558 | 3120 | 1080 | 360 | 5.79 | 38,511.02 |
| PmbZIP105 | RLN09230.1 | PM11G13790 | 11 | 25784422–25786801 | 2380 | 516 | 172 | 6.29 | 17,688.93 |
| PmbZIP106 | RLN07329.1 | PM11G19430 | 11 | 38531228–38532874 | 1647 | 765 | 255 | 5.78 | 27,050.34 |
| PmbZIP107 | RLN08025.1 | PM11G20550 | 11 | 40200637–40206785 | 6149 | 1797 | 599 | 9.76 | 65,299.21 |
| PmbZIP108 | RLN07962.1 | PM11G20760 | 11 | 40430132–40431310 | 1179 | 567 | 189 | 9.02 | 20,553.29 |
| PmbZIP109 | RLN09062.1 | PM11G24760 | 11 | 43626713–43632822 | 6110 | 1062 | 354 | 6.48 | 37,768.04 |
| PmbZIP110 | RLN07622.1 | PM11G27250 | 11 | 46387650–46388183 | 534 | 534 | 178 | 8.65 | 20,643.10 |
| PmbZIP111 | RLM78368.1 | PM12G01810 | 12 | 1318893–1319414 | 522 | 522 | 174 | 6.83 | 17,739.04 |
| PmbZIP112 | RLM78875.1 | PM12G02550 | 12 | 1757193–1761266 | 4074 | 1362 | 454 | 8.20 | 47,603.58 |
| PmbZIP113 | RLM80422.1 | PM12G04340 | 12 | 3257399–3260279 | 2881 | 924 | 308 | 5.20 | 32,615.15 |
| PmbZIP114 | RLM79681.1 | PM12G05380 | 12 | 4052057–4052425 | 369 | 369 | 123 | 9.35 | 13,871.48 |
| PmbZIP115 | RLM80771.1 | PM12G06150 | 12 | 4748641–4750662 | 2022 | 414 | 138 | 10.11 | 15,166.99 |
| PmbZIP116 | RLM79948.1 | PM12G08110 | 12 | 6857609–6859787 | 2179 | 762 | 254 | 5.78 | 26,937.22 |
| PmbZIP117 | RLM79671.1 | PM12G27010 | 12 | 38519464–38520329 | 866 | 510 | 170 | 7.93 | 19,646.96 |
| PmbZIP118 | RLM78209.1 | PM12G28080 | 12 | 39453559–39458489 | 4931 | 1332 | 444 | 9.41 | 47,492.18 |
| PmbZIP119 | RLM79651.1 | PM12G29310 | 12 | 40484976–40490089 | 5114 | 513 | 171 | 4.93 | 18,799.88 |
| PmbZIP120 | RLM80583.1 | PM12G31970 | 12 | 42325452–42326207 | 756 | 519 | 173 | 9.00 | 18,729.10 |
| PmbZIP121 | RLN05398.1 | PM13G04720 | 13 | 5033013–5042282 | 9270 | 1434 | 478 | 7.24 | 51,950.15 |
| PmbZIP122 | RLN05595.1 | PM13G09960 | 13 | 25084063–25085042 | 980 | 261 | 87 | 9.65 | 9705.76 |
| PmbZIP123 | RLN04073.1 | PM13G14010 | 13 | 35015891–35017421 | 1531 | 1209 | 403 | 6.37 | 42,408.97 |
| PmbZIP124 | RLN05004.1 | PM13G14470 | 13 | 35292857–35295671 | 2815 | 810 | 270 | 9.25 | 28,271.05 |
| PmbZIP125 | RLN04831.1 | PM13G19990 | 13 | 41033378–41036302 | 2925 | 999 | 333 | 5.76 | 34,879.32 |
| PmbZIP126 | RLN04579.1 | PM13G20830 | 13 | 41770762–41771759 | 998 | 675 | 225 | 8.54 | 24,211.82 |
| PmbZIP127 | RLN03801.1 | PM13G24510 | 13 | 45184706–45186681 | 1976 | 1218 | 406 | 6.61 | 43,025.01 |
| PmbZIP128 | RLN03549.1 | PM13G25090 | 13 | 45582601–45583142 | 542 | 432 | 144 | 10.02 | 15,452.52 |
| PmbZIP129 | RLM61107.1 | PM14G04080 | 14 | 3139207–3146477 | 7271 | 1263 | 421 | 6.53 | 45,595.62 |
| PmbZIP130 | RLM60470.1 | PM14G09560 | 14 | 20687606–20688770 | 1165 | 441 | 147 | 9.34 | 16,666.93 |
| PmbZIP131 | RLM60601.1 | PM14G16770 | 14 | 29385478–29388989 | 3512 | 1218 | 406 | 5.52 | 42,836.29 |
| PmbZIP132 | RLM61427.1 | PM14G17480 | 14 | 30010114–30011044 | 931 | 654 | 218 | 8.82 | 23,453.05 |
| PmbZIP133 | RLM60401.1 | PM14G19800 | 14 | 31888088–31890896 | 2809 | 1128 | 376 | 6.32 | 39,473.30 |
| PmbZIP134 | RLM61133.1 | PM14G19990 | 14 | 32037397–32038999 | 1603 | 1227 | 409 | 6.43 | 43,304.39 |
| PmbZIP135 | RLM61596.1 | PM14G21450 | 14 | 33079164–33079675 | 512 | 453 | 151 | 9.55 | 16,265.44 |
| PmbZIP136 | RLM74471.1 | PM15G12970 | 15 | 28520231–28523482 | 3252 | 690 | 230 | 9.83 | 24,604.94 |
| PmbZIP137 | RLM74357.1 | PM15G25820 | 15 | 39118732–39121503 | 2772 | 1608 | 536 | 7.97 | 58,416.77 |
| PmbZIP138 | RLM65300.1 | PM16G12130 | 16 | 25119044–25121594 | 2551 | 786 | 262 | 5.10 | 27,207.24 |
| PmbZIP139 | RLM66084.1 | PM16G21310 | 16 | 31781217–31790325 | 9109 | 1203 | 401 | 6.18 | 44,727.90 |
| PmbZIP140 | RLM70069.1 | PM17G01920 | 17 | 1423232–1427747 | 4516 | 1389 | 463 | 6.25 | 50,766.87 |
| PmbZIP141 | RLM69114.1 | PM17G02070 | 17 | 1594724–1595732 | 1009 | 675 | 225 | 8.98 | 24,596.61 |
| PmbZIP142 | RLM59076.1 | PM18G00820 | 18 | 648118–653777 | 5660 | 1431 | 477 | 6.09 | 52,218.33 |
| PmbZIP143 | RLM57820.1 | PM18G00930 | 18 | 761889–762898 | 1010 | 648 | 216 | 8.61 | 23,467.31 |
| PmbZIP144 | RLM58717.1 | PM18G01220 | 18 | 1078921–1083128 | 4208 | 993 | 331 | 6.37 | 35,578.56 |
| Cis-Acting Element | Element Number | Gene Number |
|---|---|---|
| Abscisic acid responsiveness element | 618 | 135 |
| Anoxic specific inducibility element | 220 | 90 |
| Auxin-responsive element | 95 | 66 |
| Defense and stress responsiveness element | 39 | 33 |
| Endosperm expression element | 29 | 25 |
| Gibberellin-responsive element | 108 | 75 |
| Light responsive element | 1655 | 144 |
| Low-temperature responsiveness element | 95 | 64 |
| MeJA-responsiveness element | 752 | 128 |
| Meristem expression element | 154 | 99 |
| MYB binding site | 175 | 97 |
| Protein binding site | 4 | 4 |
| Rcircadian control element | 40 | 30 |
| Root specific element | 7 | 5 |
| Salicylic acid responsiveness element | 72 | 50 |
| Seed-specific regulation element | 30 | 26 |
| Wound-responsive element | 8 | 8 |
| Zein metabolism regulation element | 61 | 46 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
An, P.; Liu, T.; Shui, Z.; Ren, P.; Duan, S. Genome-Wide Identification and Analysis of bZIP Transcription Factor Gene Family in Broomcorn Millet (Panicum miliaceum L.). Genes 2025, 16, 734. https://doi.org/10.3390/genes16070734
An P, Liu T, Shui Z, Ren P, Duan S. Genome-Wide Identification and Analysis of bZIP Transcription Factor Gene Family in Broomcorn Millet (Panicum miliaceum L.). Genes. 2025; 16(7):734. https://doi.org/10.3390/genes16070734
Chicago/Turabian StyleAn, Peipei, Tianxiang Liu, Zhijie Shui, Panrong Ren, and Shan Duan. 2025. "Genome-Wide Identification and Analysis of bZIP Transcription Factor Gene Family in Broomcorn Millet (Panicum miliaceum L.)" Genes 16, no. 7: 734. https://doi.org/10.3390/genes16070734
APA StyleAn, P., Liu, T., Shui, Z., Ren, P., & Duan, S. (2025). Genome-Wide Identification and Analysis of bZIP Transcription Factor Gene Family in Broomcorn Millet (Panicum miliaceum L.). Genes, 16(7), 734. https://doi.org/10.3390/genes16070734






