The Single Nucleotide Substitution T → A rs2072580 Damages the CREB1 Binding Site in the Bidirectional SART3/ISCU Promoter
Abstract
1. Introduction
2. Materials and Methods
2.1. Oligonucleotides
2.2. Cell Cultures
2.3. Genomic DNA Isolation and Genotyping
2.4. Preparation of Nuclear Extracts and Electrophoretic Mobility Shift Assay (EMSA)
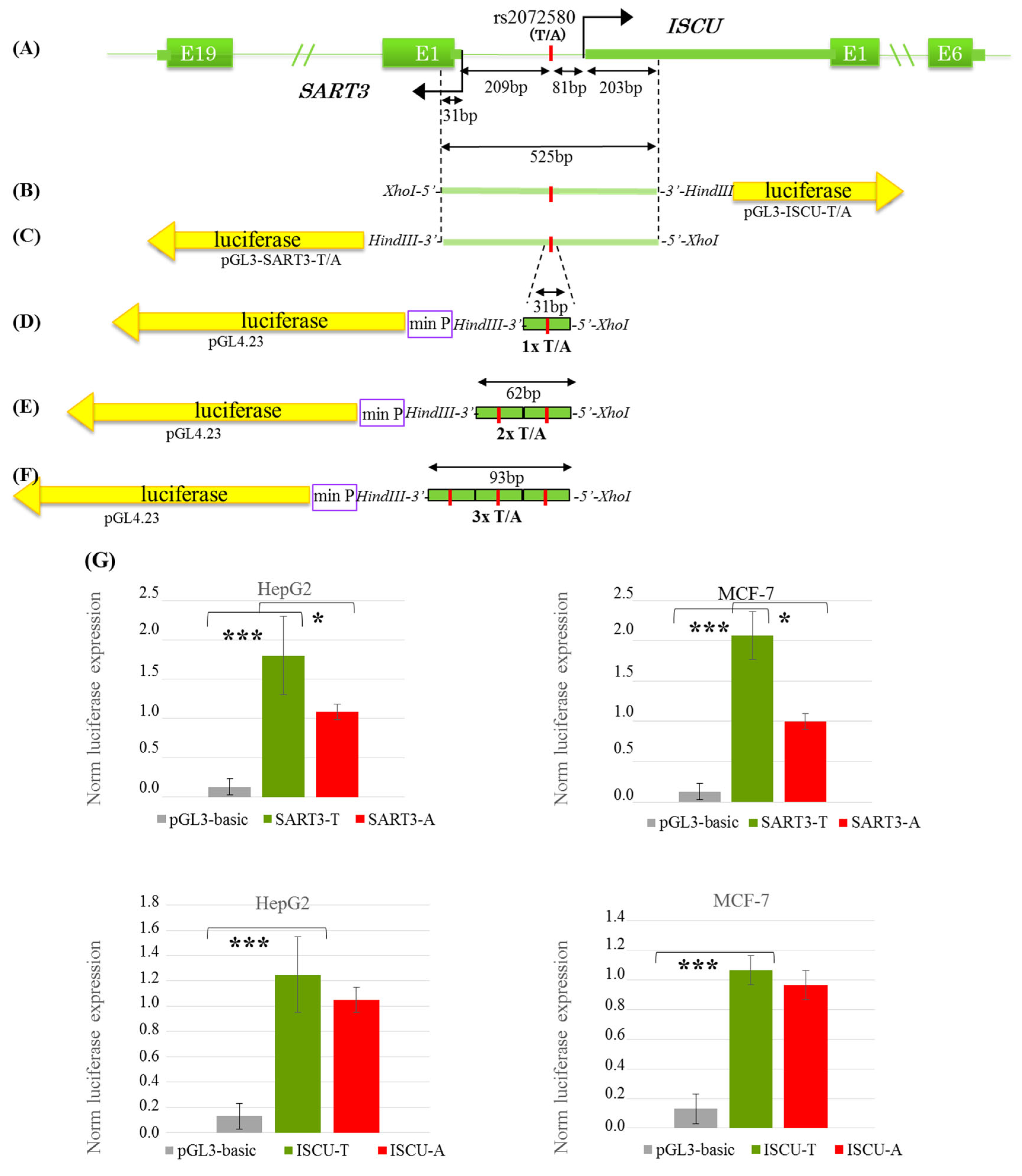
2.5. Plasmid Construction, Transfection, and Luciferase Reporter Assay
2.6. Statistical Analysis
3. Results
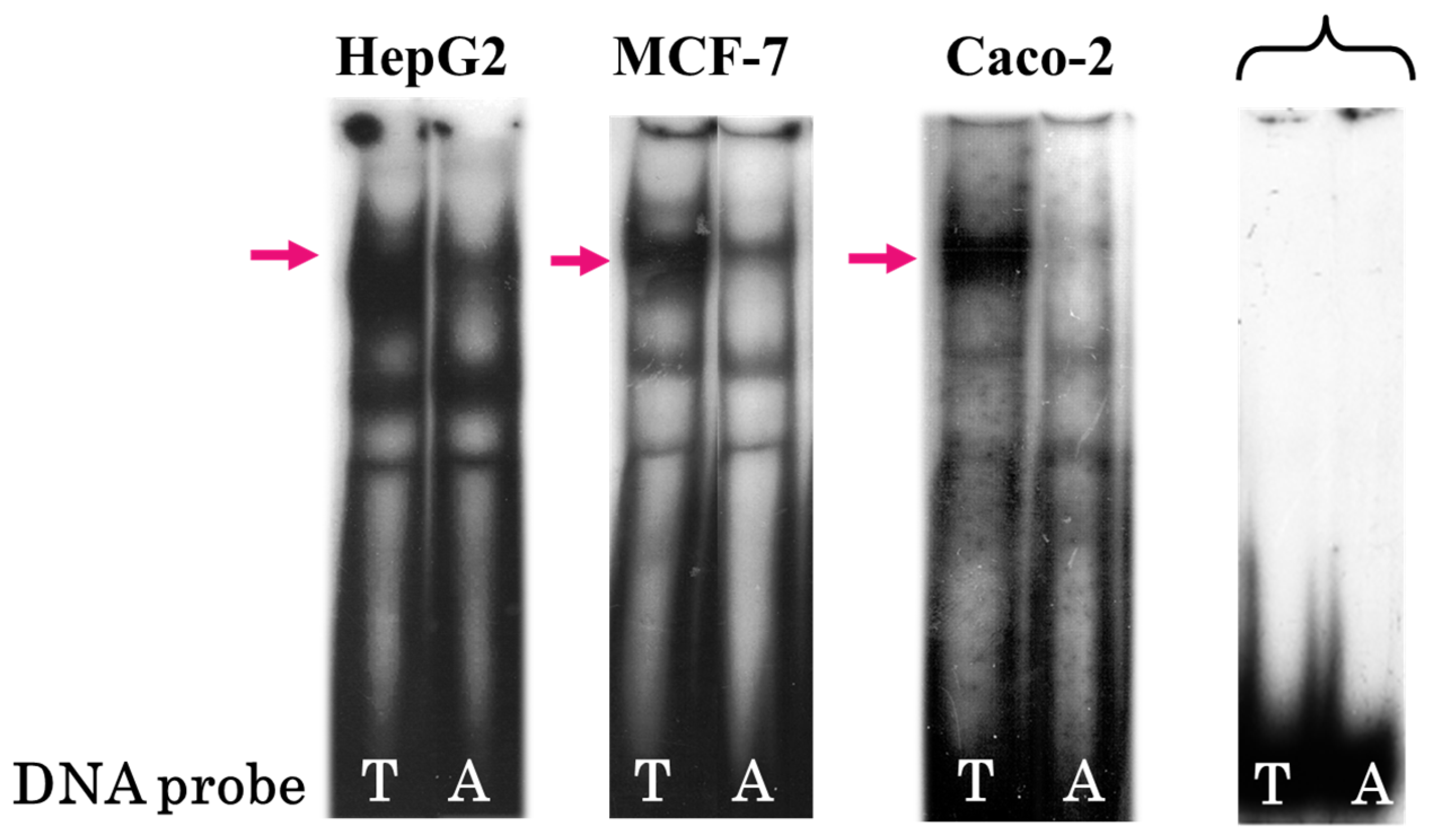
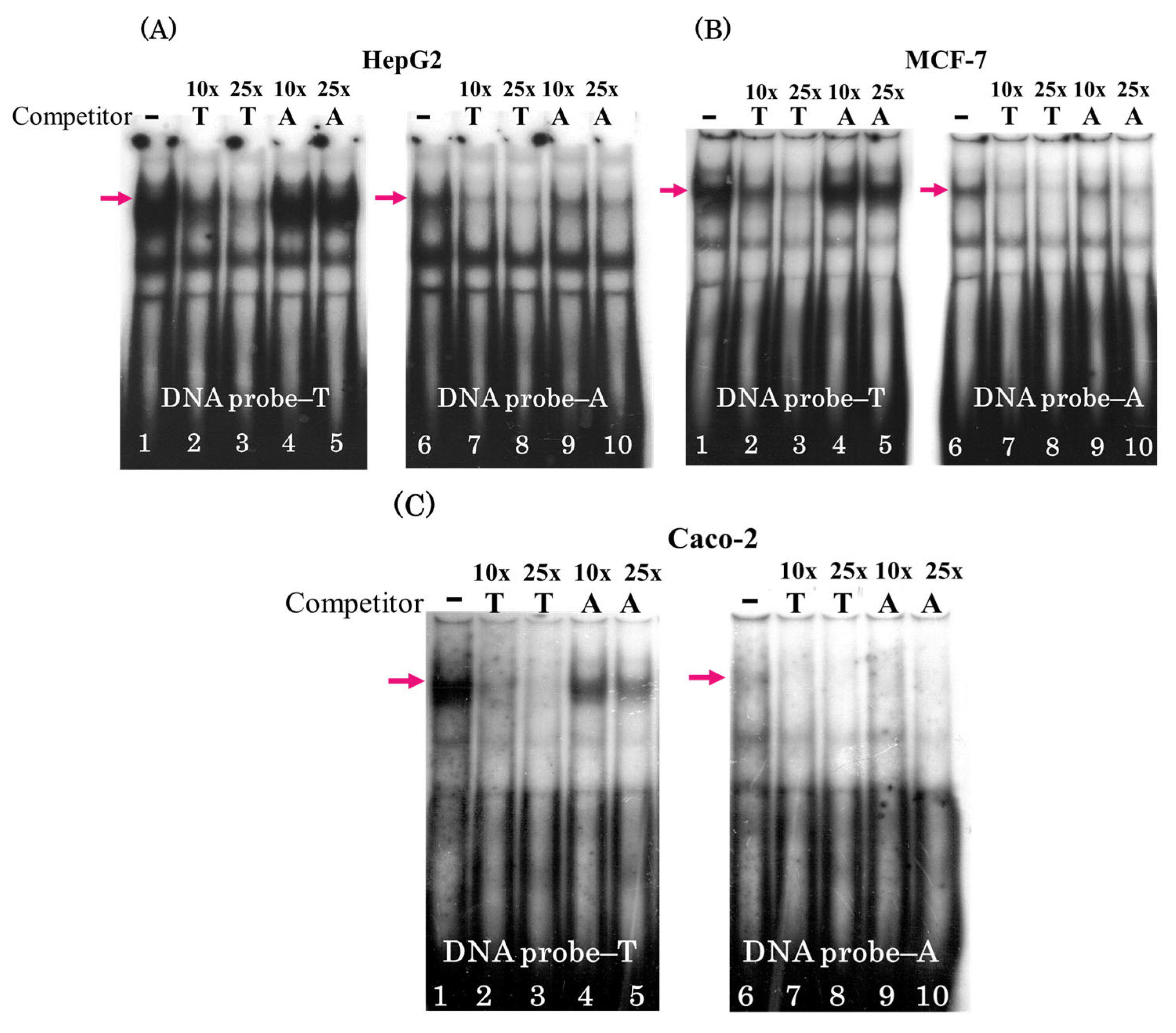
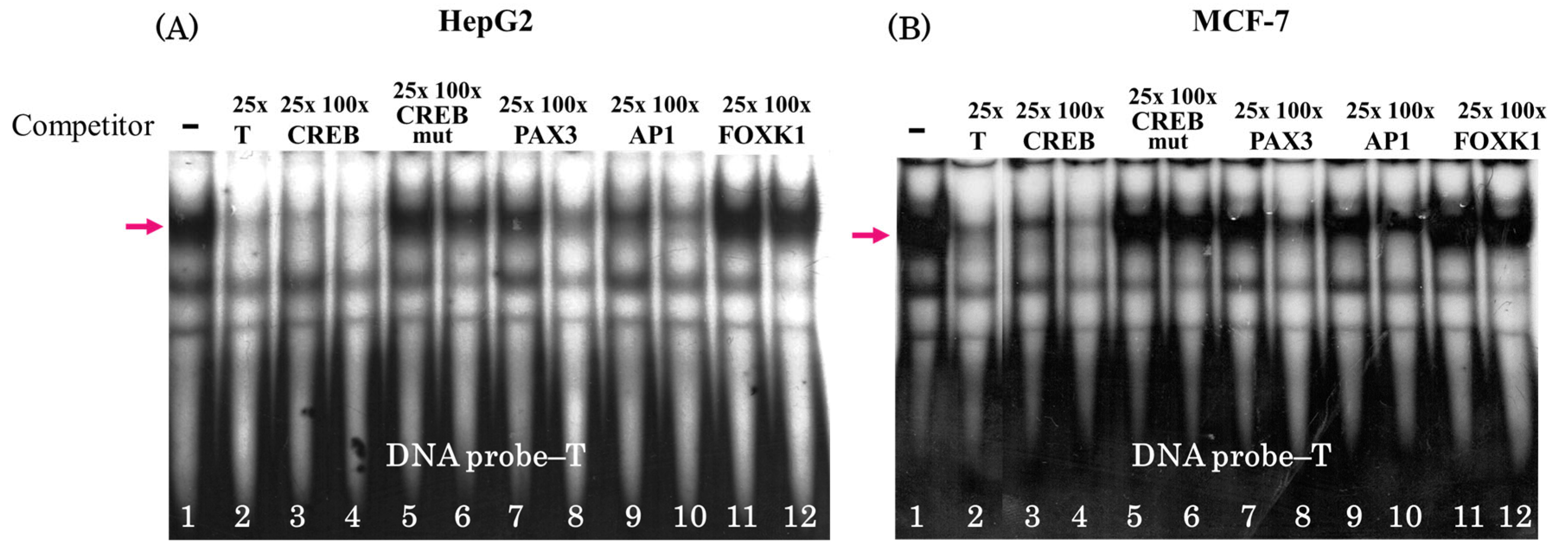
3.1. Single Nucleotide Substitution T → A rs2072580 Disrupts CREB1 Transcription Factor Binding Site
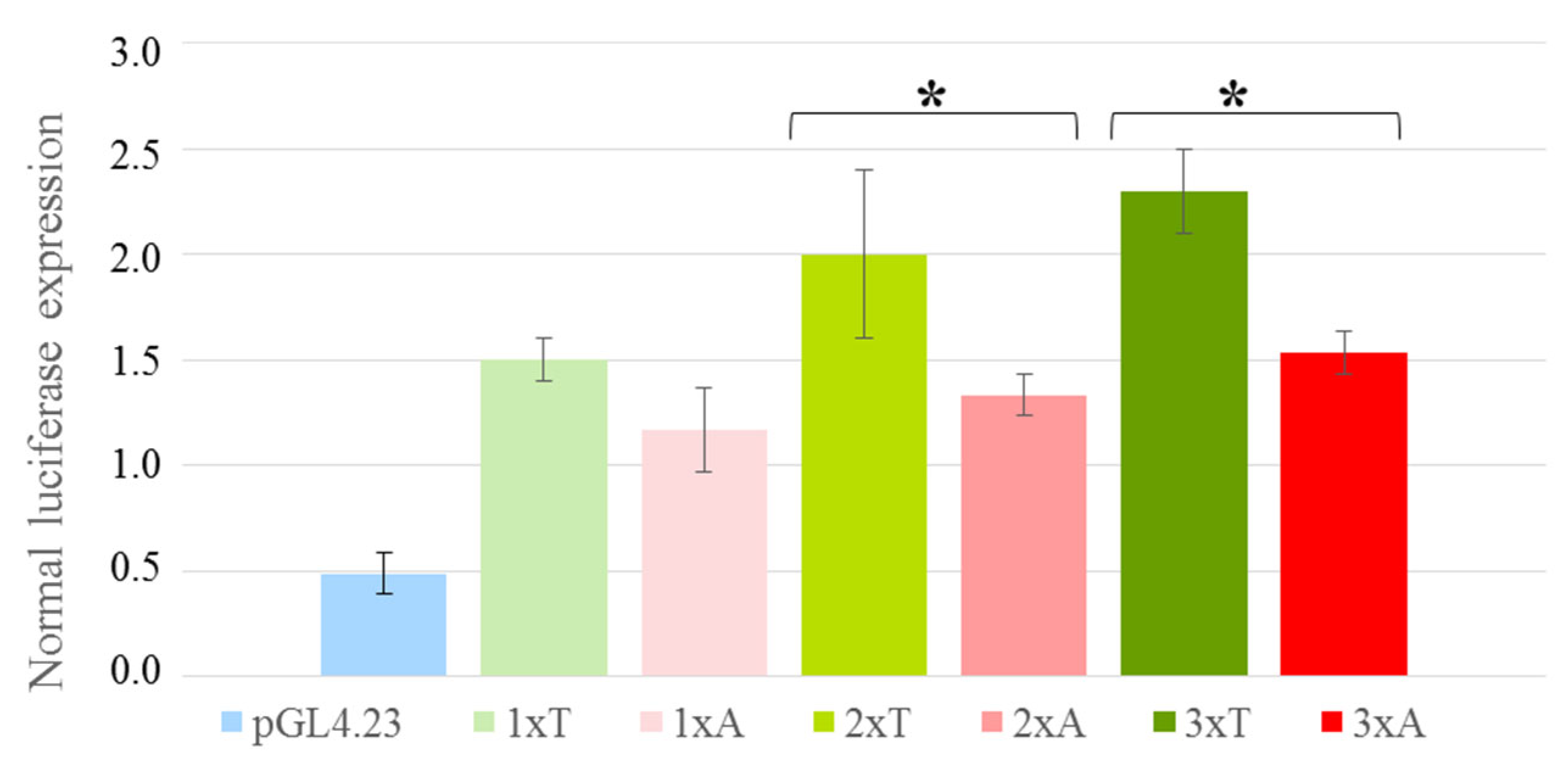
3.2. The Oligonucleotide T → A rs2072580 Substitution Destroys the CREB1 Binding Site and Decreases the Activity of the Corresponding Regulatory Element
3.3. The T → A rs2072580 Substitution Destroys the CREB1 Binding Site abd Decreases the Activity of SART3 Promoter but Has No Effect on the Activity of the ISCU Promoter Within the Bidirectional SART3/ISCU Promoter
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meddens, C.A.; van der List, A.C.J.; Nieuwenhuis, E.E.S.; Mokry, M. Non-coding DNA in IBD: From sequence variation in DNA regulatory elements to novel therapeutic potential. Gut 2019, 68, 928–941. [Google Scholar] [CrossRef] [PubMed]
- Zeggini, E.; Gloyn, A.L.; Barton, A.C.; Wain, L.V. Translational genomics and precision medicine: Moving from the lab to the clinic. Science 2019, 365, 1409–1413. [Google Scholar] [CrossRef] [PubMed]
- Maurano, M.T.; Humbert, R.; Rynes, E.; Thurman, R.E.; Haugen, E.; Wang, H.; Reynolds, A.P.; Sandstrom, R.; Qu, H.; Brody, J.; et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 2012, 337, 1190–1195. [Google Scholar] [CrossRef]
- Farh, K.K.; Marson, A.; Zhu, J.; Kleinewietfeld, M.; Housley, W.J.; Beik, S.; Shoresh, N.; Whitton, H.; Ryan, R.J.; Shishkin, A.A.; et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 2015, 518, 337–343. [Google Scholar] [CrossRef] [PubMed]
- French, J.D.; Edwards, S.L. The role of noncoding variants in heritable disease. Trends Genet. 2020, 36, 880–889. [Google Scholar] [CrossRef]
- Degtyareva, A.O.; Antontseva, E.V.; Merkulova, T.I. Regulatory SNPs: Altered Transcription Factor Binding Sites Implicated in Complex Traits and Diseases. Int. J. Mol. Sci. 2021, 22, 6454. [Google Scholar] [CrossRef]
- Fabo, T.; Khavari, P. Functional characterization of human genomic variation linked to polygenic diseases. Trends Genet. 2023, 39, 462–490. [Google Scholar] [CrossRef]
- Mudappathi, R.; Patton, T.; Chen, H.; Yang, P.; Sun, Z.; Wang, P.; Shi, C.X.; Wang, J.; Liu, L. reg-eQTL: Integrating transcription factor effects to unveil regulatory variants. Am. J. Hum. Genet. 2025, 112, 659–674. [Google Scholar] [CrossRef]
- Tam, V.; Patel, N.; Turcotte, M.; Bossé, Y.; Paré, G.; Meyre, D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef]
- Claussnitzer, M.; Cho, J.H.; Collins, R.; Cox, N.J.; Dermitzakis, E.T.; Hurles, M.E.; Kathiresan, S.; Kenny, E.E.; Lindgren, C.M.; MacArthur, D.G.; et al. A brief history of human disease genetics. Nature 2020, 577, 179–189. [Google Scholar] [CrossRef]
- Alsheikh, A.J.; Wollenhaupt, S.; King, E.A.; Reeb, J.; Ghosh, S.; Stolzenburg, L.R.; Tamim, S.; Lazar, J.; Davis, J.W.; Jacob, H.J. The landscape of GWAS validation; systematic review identifying 309 validated non-coding variants across 130 human diseases. BMC Med. Genom. 2022, 15, 74. [Google Scholar] [CrossRef] [PubMed]
- GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020, 369, 1318–1330. [Google Scholar] [CrossRef]
- Fan, J.; Hu, J.; Xue, C.; Zhang, H.; Susztak, K.; Reilly, M.P.; Xiao, R.; Li, M. ASEP: Gene-based detection of allele-specific expression across individuals in a population by RNA sequencing. PLoS Genet. 2020, 16, e1008786. [Google Scholar] [CrossRef]
- Maurano, M.T.; Haugen, E.; Sandstrom, R.; Vierstra, J.; Shafer, A.; Kaul, R.; Stamatoyannopoulos, J.A. Large-scale identification of sequence variants influencing human transcription factor occupancy in vivo. Nat. Genet. 2015, 47, 1393–1401. [Google Scholar] [CrossRef]
- Xu, S.; Feng, W.; Lu, Z.; Yu, C.Y.; Shao, W.; Nakshatri, H.; Reiter, J.L.; Gao, H.; Chu, X.; Wang, Y.; et al. regSNPs-ASB: A Computational Framework for Identifying Allele-Specific Transcription Factor Binding From ATAC-seq Data. Front. Bioeng. Biotechnol. 2020, 8, 886. [Google Scholar] [CrossRef] [PubMed]
- Boytsov, A.; Abramov, S.; Aiusheeva, A.Z.; Kasianova, A.M.; Baulin, E.; Kuznetsov, I.A.; Aulchenko, Y.S.; Kolmykov, S.; Yevshin, I.; Kolpakov, F.; et al. ANANASTRA: Annotation and enrichment analysis of allele-specific transcription factor binding at SNPs. Nucleic Acids Res. 2022, 50, W51–W56. [Google Scholar] [CrossRef]
- Bryzgalov, L.O.; Korbolina, E.E.; Merkulova, T.I. Exploring the Genetic Predisposition to Epigenetic Changes in Alzheimer’s Disease. Int. J. Mol. Sci. 2023, 24, 7955. [Google Scholar] [CrossRef]
- Damarov, I.S.; Korbolina, E.E.; Rykova, E.Y.; Merkulova, T.I. Multi-Omics Analysis Revealed the rSNPs Potentially Involved in T2DM Pathogenic Mechanism and Metformin Response. Int. J. Mol. Sci. 2024, 25, 9297. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Huang, Y.; Zhang, P.; Han, C.; Lu, Y.; Mo, Z.; Zhang, Z.; Li, X.; Zhao, S.; Cai, F.; et al. Characterization of a pathogenic variant in GBA for Parkinson’s disease with mild cognitive impairment patients. Mol. Brain 2020, 13, 102. [Google Scholar] [CrossRef]
- Li, S.; Li, Y.; Li, X.; Liu, J.; Huo, Y.; Wang, J.; Liu, Z.; Li, M.; Luo, X.J. Regulatory mechanisms of major depressive disorder risk variants. Mol. Psychiatry 2020, 25, 1926–1945. [Google Scholar] [CrossRef]
- Pan, G.; Cavalli, M.; Carlsson, B.; Skrtic, S.; Kumar, C.; Wadelius, C. rs953413 regulates polyunsaturated fatty acid metabolism by modulating ELOVL2 expression. iScience 2020, 23, 100808. [Google Scholar] [CrossRef] [PubMed]
- Korbolina, E.E.; Brusentsov, I.I.; Bryzgalov, L.O.; Leberfarb, E.Y.; Degtyareva, A.O.; Merkulova, T.I. Novel approach to functional SNPs discovery from genome-wide data reveals promising variants for colon cancer risk. Hum. Mutat. 2018, 39, 851–859. [Google Scholar] [CrossRef] [PubMed]
- Leberfarb, E.Y.; Degtyareva, A.O.; Brusentsov, I.I.; Maximov, V.N.; Voevoda, M.I.; Autenshlus, A.I.; Morozov, D.V.; Sokolov, A.V.; Merkulova, T.I. Potential regulatory SNPs in the ATXN7L3B and KRT15 genes are associated with gender-specific colorectal cancer risk. Per. Med. 2020, 17, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Degtyareva, A.O.; Leberfarb, E.Y.; Efimova, E.G.; Brusentsov, I.I.; Usova, A.V.; Lushnikova, E.L.; Merkulova, T.I. rs2072580T>A Polymorphism in the Overlapping Promoter Regions of the SART3 and ISCU Genes Associated with the Risk of Breast Cancer. Bull. Exp. Biol. Med. 2020, 169, 81–84. [Google Scholar] [CrossRef]
- Kawagoe, N.; Shintaku, I.; Yutani, S.; Etoh, H.; Matuoka, K.; Noda, S.; Itoh, K. Expression of the SART3 Tumor Rejection Antigen in Renal Cell Carcinoma. J. Urol. 2000, 164, 2090–2095. [Google Scholar] [CrossRef]
- Murayama, K.; Kobayashi, T.; Imaizumi, T.; Matsunaga, K.; Kuramoto, T.; Shigemori, M.; Shichijo, S.; Itoh, K. Expression of the SART3 Tumor-Rejection Antigen in Brain Tumors and Induction of Cytotoxic T Lymphocytes by its Peptides. J. Immunother. 2000, 23, 511–518. [Google Scholar] [CrossRef]
- Kaji, K.; Mizukoshi, E.; Yamashita, T.; Arai, K.; Sunagozaka, H.; Fushimi, K.; Nakagawa, H.; Yamada, K.; Terashima, T.; Kitahara, M.; et al. Cellular Immune Responses for Squamous Cell Carcinoma Antigen Recognized by T Cells 3 in Patients with Hepatocellular Carcinoma. PLoS ONE 2017, 12, e0170291. [Google Scholar] [CrossRef]
- Whitmill, A.; Timani, K.A.; Liu, Y.; He, J.J. Tip110: Physical properties, primary structure, and biological functions. Life Sci. 2016, 149, 79–95. [Google Scholar] [CrossRef]
- Nong, J.; Yang, K.; Li, T.; Lan, C.; Zhou, X.; Liu, J.; Xie, H.; Luo, J.; Liao, X.; Zhu, G.; et al. SART3, regulated by p53, is a biomarker for diagnosis, prognosis and immune infiltration in hepatocellular carcinoma. Aging 2023, 15, 8408–8432. [Google Scholar] [CrossRef]
- Wang, P.S.; Liu, Z.; Sweef, O.; Xie, J.; Chen, J.; Zhu, H.; Zeidler-Erdely, P.C.; Yang, C.; Wang, Z. Long noncoding RNA ABHD11-AS1 interacts with SART3 and regulates CD44 RNA alternative splicing to promote lung carcinogenesis. Environ. Int. 2024, 185, 108494. [Google Scholar] [CrossRef]
- Favaro, E.; Ramachandran, A.; McCormick, R.; Gee, H.; Blancher, C.; Crosby, M.; Devlin, C.; Blick, C.; Buffa, F.; Li, J.L.; et al. MicroRNA-210 regulates mitochondrial free radical response to hypoxia and krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PLoS ONE 2010, 5, e10345. [Google Scholar] [CrossRef] [PubMed]
- Funauchi, Y.; Tanikawa, C.; Yi Lo, P.H.; Mori, J.; Daigo, Y.; Takano, A.; Miyagi, Y.; Okawa, A.; Nakamura, Y.; Matsuda, K. Regulation of iron homeostasis by the p53-ISCU pathway. Sci. Rep. 2015, 5, 16497. [Google Scholar] [CrossRef] [PubMed]
- Rouault, T.A.; Maio, N. Biogenesis and functions of mammalian iron-sulfur proteins in the regulation of iron homeostasis and pivotal metabolic pathways. J. Biol. Chem. 2017, 292, 12744–12753. [Google Scholar] [CrossRef] [PubMed]
- Suwei, D.; Zhen, L.; Zhimin, L.; Mei, L.; Jianping, K.; Zhuohui, P.; Yanbin, X.; Xiang, M. Hypoxia Modulates Melanoma Cells Proliferation and Apoptosis via miRNA-210/ISCU/ROS Signaling. Bull. Exp. Biol. 2022, 173, 645–650. [Google Scholar] [CrossRef]
- Bernardo-Castiñeira, C.; Valdés, N.; Sierra, M.I.; Sáenz-de-Santa-María, I.; Bayón, G.F.; Perez, R.F.; Fernández, A.F.; Fraga, M.F.; Astudillo, A.; Menéndez, R.; et al. SDHC Promoter Methylation, a Novel Pathogenic Mechanism in Parasympathetic Paragangliomas. J. Clin. Endocrinol. Metab. 2018, 103, 295–305. [Google Scholar] [CrossRef]
- Veronez, L.C.; Xavier, A.E.T.; Nagano, L.F.; Correa, C.A.P.; Borges, K.S.; Santos, P.; Baroni, M.; Silva Queiroz, R.P.; Antonini, S.R.R.; Yunes, J.A.; et al. Identifying prognostic hub genes and key pathways in pediatric adrenocortical tumors through RNA sequencing and Co-expression analysis. Mol. Cell. Endocrinol. 2024, 594, 112383. [Google Scholar] [CrossRef]
- Sáenz-de-Santa-María, I.; Bernardo-Castiñeira, C.; Secades, P.; Bernaldo-de-Quirós, S.; Rodrigo, J.P.; Astudillo, A.; Chiara, M.D. Clinically relevant HIF-1α-dependent metabolic reprogramming in oropharyngeal squamous cell carcinomas includes coordinated activation of CAIX and the miR-210/ISCU signaling axis, but not MCT1 and MCT4 upregulation. Oncotarget 2017, 8, 13730–13746. [Google Scholar] [CrossRef]
- Ullmann, P.; Qureshi-Baig, K.; Rodriguez, F.; Ginolhac, A.; Nonnenmacher, Y.; Ternes, D.; Weiler, J.; Gäbler, K.; Bahlawane, C.; Hiller, K.; et al. Hypoxia-responsive miR-210 promotes self-renewal capacity of colon tumor-initiating cells by repressing ISCU and by inducing lactate production. Oncotarget 2016, 7, 65454–65470. [Google Scholar] [CrossRef]
- Abudusalam, K.; Xu, Y.; Keyumu, P.; Cheng, T.; Xu, M.; Lu, B.; Sun, P.; Musha, K.; Huang, J. WSCD2 Expression: Its Relevance to Tumor-Infiltrating Immune Cells and Glioma Prognosis. Curr. Med. Chem. 2024. [Google Scholar] [CrossRef]
- Bushel, P.R.; Ward, J.; Burkholder, A.; Li, J.; Anchang, B. Mitochondrial-nuclear epistasis underlying phenotypic variation in breast cancer pathology. Sci. Rep. 2022, 12, 1393. [Google Scholar] [CrossRef]
- Wang, X.C.; Yue, X.; Zhang, R.X.; Liu, T.Y.; Pan, Z.Z.; Yang, M.J.; Lu, Z.H.; Wang, Z.Y.; Peng, J.H.; Le, L.Y.; et al. Genome-wide RNAi Screening Identifies RFC4 as a Factor That Mediates Radioresistance in Colorectal Cancer by Facilitating Nonhomologous End Joining Repair. Clin. Cancer Res. 2019, 25, 4567–4579. [Google Scholar] [CrossRef] [PubMed]
- Bryzgalov, L.O.; Antontseva, E.V.; Matveeva, M.Y.; Shilov, A.G.; Kashina, E.V.; Mordvinov, V.A.; Merkulova, T.I. Detection of regulatory SNPs in human genome using ChIP-seq ENCODE data. PLoS ONE 2013, 8, e78833. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Coetzee, S.G.; Hazelett, D.J. motifbreakR v2: Expanded variant analysis including indels and integrated evidence from transcription factor binding databases. Bioinform. Adv. 2024, 4, vbae162. [Google Scholar] [CrossRef]
- Uvarova, A.N.; Tkachenko, E.A.; Stasevich, E.M.; Zheremyan, E.A.; Korneev, K.V.; Kuprash, D.V. Methods for Functional Characterization of Genetic Polymorphisms of Non-Coding Regulatory Regions of the Human Genome. Biochemistry 2024, 89, 1002–1013. [Google Scholar] [CrossRef]
- Hai, T.; Hartman, M.G. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: Activating transcription factor proteins and homeostasis. Gene 2001, 273, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, M.A.R.; Haq, M.M.; Lee, J.H.; Jeong, S. Multi-faceted regulation of CREB family transcription factors. Front. Mol. Neurosci. 2024, 17, 1408949. [Google Scholar] [CrossRef]
- Cui, A.; Ding, D.; Li, Y. Regulation of hepatic metabolism and cell growth by the ATF/CREB family of transcription factors. Diabetes 2021, 70, 653–664. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, M.A.R.; An, J.; Jeong, S. The Pleiotropic Face of CREB Family Transcription Factors. Moll. Cells 2023, 46, 399–413. [Google Scholar] [CrossRef]
- Wang, L.; Nie, Q.; Gao, M.; Yang, L.; Xiang, J.W.; Xiao, Y.; Liu, F.Y.; Gong, X.D.; Fu, J.L.; Wang, Y.; et al. The transcription factor CREB acts as an important regulator mediating oxidative stress-induced apoptosis by suppressing αB-crystallin expression. Aging 2020, 12, 13594–13617. [Google Scholar] [CrossRef]
- Zheng, T.; Huang, J.; Xiang, X.; Li, S.; Yu, J.; Qu, K.; Xu, Z.; Han, P.; Dong, Z.; Liu, Y.; et al. Systematical analysis reveals a strong cancer relevance of CREB1-regulated genes. Cancer Cell Int. 2021, 21, 530. [Google Scholar] [CrossRef]
- Wang, W.D.; Gralla, J.D. Differential ability of proximal and remote element pairs to cooperate in activating RNA polymerase II transcription. Mol. Cell. Biol. 1991, 11, 4561–4571. [Google Scholar] [CrossRef] [PubMed]
- Plisov, S.Y.; Nichiporenko, M.G.; Shkapenko, A.L.; Kumarev, V.P.; Baranova, L.V.; Merkulova, T.I. The immediate vicinity of mouse metallothionein-I gene contains two sites conferring glucocorticoid inducibility to the heterologous promoter. FEBS Lett. 1995, 358, 104. [Google Scholar] [CrossRef] [PubMed]
- Kietzmann, T.; Samoylenko, A.; Roth, U.; Jungermann, K. Hypoxia-inducible factor-1 and hypoxia response elements mediate the induction of plasminogen activator inhibitor-1 gene expression by insulin in primary rat hepatocytes. Blood 2003, 101, 907–914. [Google Scholar] [CrossRef]
- Porter, R.S.; Murata-Nakamura, Y.; Nagasu, H.; Kim, H.G.; Iwase, S. Transcriptome Analysis Revealed Impaired cAMP Responsiveness in PHF21A-Deficient Human Cells. Neuroscience 2018, 370, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Nemsick, S.; Hansen, A.S. Molecular models of bidirectional promoter regulation. Curr. Opin. Struct. Biol. 2024, 87, 102865. [Google Scholar] [CrossRef]
- Chen, Y.Q.; Yu, H.; Li, Y.X.; Li, Y.Y. Sorting out inherent features of head-to-head gene pairs by evolutionary conservation. BMC Bioinform. 2010, 11, S16. [Google Scholar] [CrossRef]
- Ahmad, S.S.; Samia, N.S.N.; Khan, A.S.; Turjya, R.R.; Khan, M.A. Bidirectional promoters: An enigmatic genome architecture and their roles in cancers. Mol. Biol. Rep. 2021, 48, 6637–6644. [Google Scholar] [CrossRef]
- Klimešová, K.; Petržílková, H.; Bařinka, C.; Staněk, D. SART3 associates with a post-splicing complex. J. Cell. Sci. 2023, 136, jcs260380. [Google Scholar] [CrossRef]
- Kim, J.; Taketomi, T.; Yamada, A.; Uematsu, Y.; Ueda, K.; Chiba, T.; Tsuruta, F. USP4 regulates TUT1 ubiquitination status in concert with SART3. Biochem. Biophys. Res. Commun. 2024, 701, 149557. [Google Scholar] [CrossRef]
- Freibert, S.A.; Boniecki, M.T.; Stümpfig, C.; Schulz, V.; Krapoth, N.; Winge, D.R.; Mühlenhoff, U.; Stehling, O.; Cygler, M.; Lill, R. N-terminal tyrosine of ISCU2 triggers [2Fe-2S] cluster synthesis by ISCU2 dimerization. Nat. Commun. 2021, 12, 6902. [Google Scholar] [CrossRef]
- Srour, B.; Gervason, S.; Monfort, B.; D’Autréaux, B. Mechanism of Iron-Sulfur Cluster Assembly: In the Intimacy of Iron and Sulfur Encounter. Inorganics 2020, 8, 55. [Google Scholar] [CrossRef]
- Bartha, Á.; Győrffy, B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef] [PubMed]
- Sherman, E.J.; Mitchell, D.C.; Garner, A.L. The RNA-binding protein SART3 promotes miR-34a biogenesis and G1 cell cycle arrest in lung cancer cells. J. Biol. Chem. 2019, 294, 17188–17196. [Google Scholar] [CrossRef]
- Fishilevich, S.; Nudel, R.; Rappaport, N.; Hadar, R.; Plaschkes, I.; Iny Stein, T.; Rosen, N.; Kohn, A.; Twik, M.; Safran, M.; et al. GeneHancer: Genome-wide integration of enhancers and target genes in GeneCards. Database 2017, 2017, bax028. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, X.; Wang, M.; Wang, Y.; Wang, S.; Jin, L.; Liu, M.; Zhou, J.; Chen, Y. UBE3B promotes breast cancer progression by antagonizing HIF-2α degradation. Oncogene 2023, 42, 3394–3406. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Li, P.; Luo, Y.; Zhang, M.; Yan, T.; Yang, Y.; Han, Y.; Liu, S.; Wang, E. PWP1 Promotes the Malignant Phenotypes of Lung Cancer Cells by Interacting with DVL2 and Merlin. Onco Targets Ther. 2020, 13, 10025–10037. [Google Scholar] [CrossRef]
- Jiang, M.; Wang, S.; Ji, J.; Baral, S.; Sun, Q.; Wang, Y.; Liu, B.; Ren, J.; Wang, W.; Wang, D. PWP1 transcriptionally regulates p53, modulating apoptosis and cell cycle to promote gastric cancer progression. Apoptosis 2024, 30, 693–709. [Google Scholar] [CrossRef]
- Huang, R.; Xu, F.; Su, L.; Lu, Y.; Liu, W.; Liu, S.; Yang, L.; Su, L.; Song, W. PWP1 is overexpressed in hepatocellular carcinoma and facilitates liver cancer cell proliferation. Heliyon 2024, 10, e32409. [Google Scholar] [CrossRef]
- Yan, Z.; Xiong, Y.; Xu, W.; Li, M.; Cheng, Y.; Chen, F.; Ding, S.; Xu, H.; Zheng, G. Identification of recurrence-related genes by integrating microRNA and gene expression profiling of gastric cancer. Int. J. Oncol. 2012, 41, 2166–2174. [Google Scholar] [CrossRef][Green Version]
- Liu, H.; Dai, X.; Cao, X.; Yan, H.; Ji, X.; Zhang, H.; Shen, S.; Si, Y.; Zhang, H.; Chen, J.; et al. PRDM4 mediates YAP-induced cell invasion by activating leukocyte-specific integrin β2 expression. EMBO Rep. 2018, 19, e45180. [Google Scholar] [CrossRef]
- Yang, W.T.; Chen, M.; Xu, R.; Zheng, P.S. PRDM4 inhibits cell proliferation and tumorigenesis by inactivating the PI3K/AKT signaling pathway through targeting of PTEN in cervical carcinoma. Oncogene 2021, 40, 3318–3330. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.H.; Peng, J.; Yang, H.L.; Fu, X.L.; Wang, J.Z.; Liu, L.; Jiang, J.N.; Tan, Y.F.; Ge, Z.J. Upregulation and biological function of transmembrane protein 119 in osteosarcoma. Exp. Mol. Med. 2017, 49, e329. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Bi, F.; Liu, Z.; Yang, Q. TMEM119 facilitates ovarian cancer cell proliferation, invasion, and migration via the PDGFRB/PI3K/AKT signaling pathway. J. Transl. Med. 2021, 19, 111. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Wang, F.; Zheng, G. Transmembrane protein TMEM119 facilitates the stemness of breast cancer cells by activating Wnt/β-catenin pathway. Bioengineered 2021, 12, 4856–4867. [Google Scholar] [CrossRef]
- Lin, X.; Dong, Y.; Gu, Y.; Wei, F.; Peng, J.; Su, Y.; Wang, Y.; Yang, C.; Neira, S.V.; Kapoor, A.; et al. Taxifolin Inhibits the Growth of Non-Small-Cell Lung Cancer via Downregulating Genes Displaying Novel and Robust Associations with Immune Evasion Factors. Cancers 2023, 15, 4818. [Google Scholar] [CrossRef]
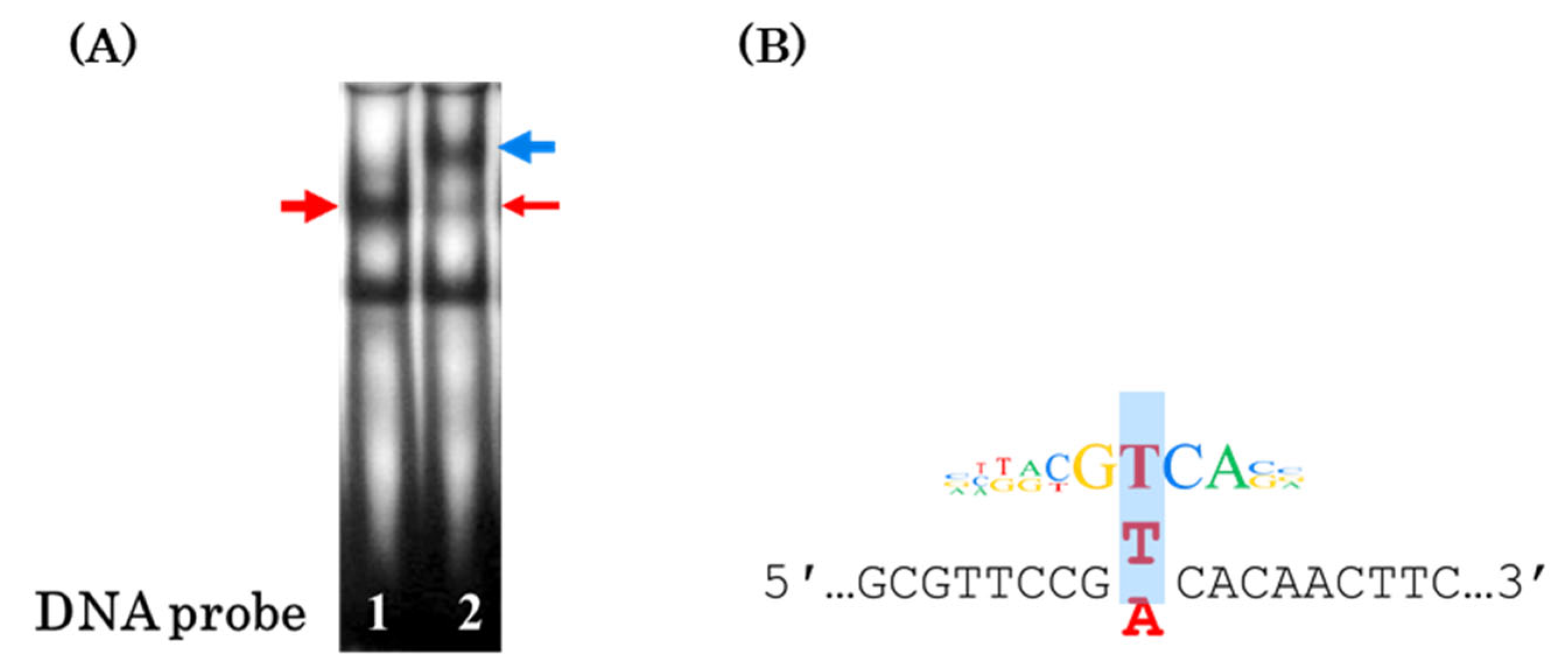
| REF | ALT | scoreRef | scoreAlt | alleleDiff | Effect | TF |
|---|---|---|---|---|---|---|
| T | A | 12.639493 | 14.623237 | 1.9837437 | Strong | FOXK1 |
| T | A | 9.203210 | 7.564058 | −1.6391518 | Strong | CREB1 |
| T | A | 9.069532 | 7.481721 | −1.5878104 | Strong | PAX3 |
| T | A | 5.061990 | 4.043738 | −1.0182518 | Strong | AP1 (FOSL1:JUND) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Degtyareva, A.; Antontseva, E.; Evseenko, A.; Orishchenko, K.; Merkulova, T. The Single Nucleotide Substitution T → A rs2072580 Damages the CREB1 Binding Site in the Bidirectional SART3/ISCU Promoter. Genes 2025, 16, 713. https://doi.org/10.3390/genes16060713
Degtyareva A, Antontseva E, Evseenko A, Orishchenko K, Merkulova T. The Single Nucleotide Substitution T → A rs2072580 Damages the CREB1 Binding Site in the Bidirectional SART3/ISCU Promoter. Genes. 2025; 16(6):713. https://doi.org/10.3390/genes16060713
Chicago/Turabian StyleDegtyareva, Arina, Elena Antontseva, Anastasia Evseenko, Konstantin Orishchenko, and Tatiana Merkulova. 2025. "The Single Nucleotide Substitution T → A rs2072580 Damages the CREB1 Binding Site in the Bidirectional SART3/ISCU Promoter" Genes 16, no. 6: 713. https://doi.org/10.3390/genes16060713
APA StyleDegtyareva, A., Antontseva, E., Evseenko, A., Orishchenko, K., & Merkulova, T. (2025). The Single Nucleotide Substitution T → A rs2072580 Damages the CREB1 Binding Site in the Bidirectional SART3/ISCU Promoter. Genes, 16(6), 713. https://doi.org/10.3390/genes16060713






