Integration of Newer Genomic Technologies into Clinical Cytogenetics Laboratories
Abstract
1. Introduction
2. Classical Cytogenetics
3. FISH
4. Microarrays
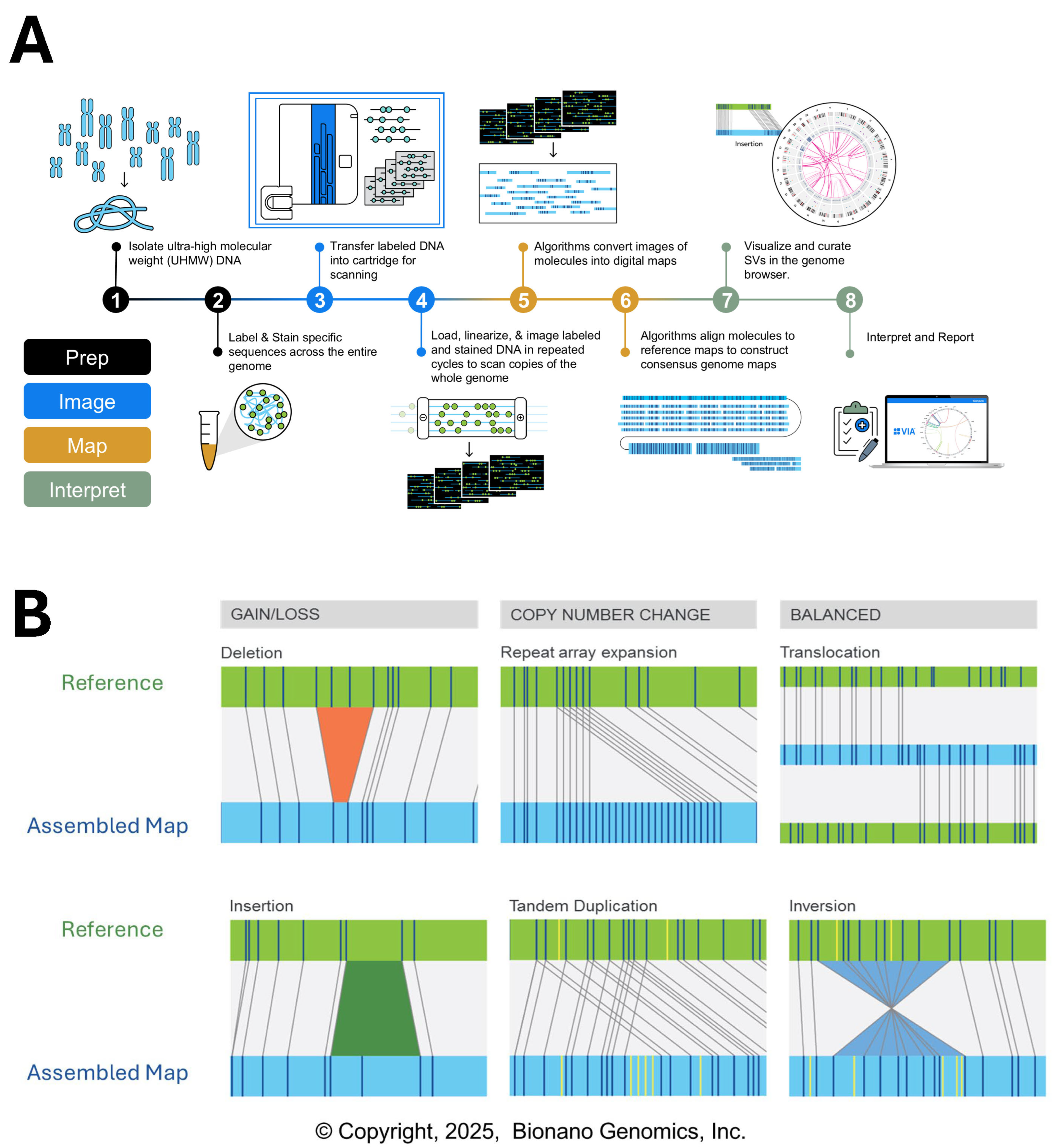
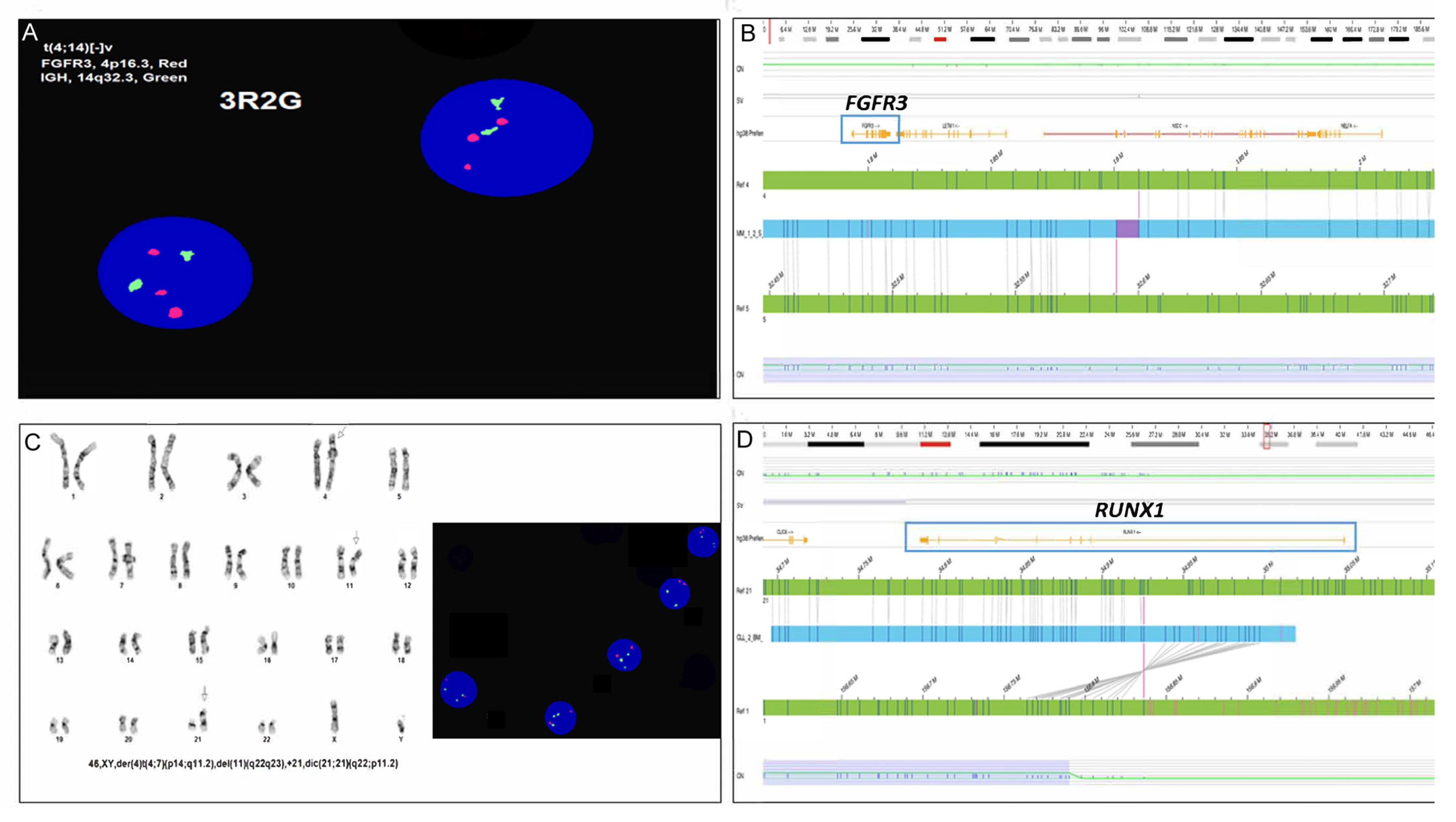
5. Optical Genome Mapping
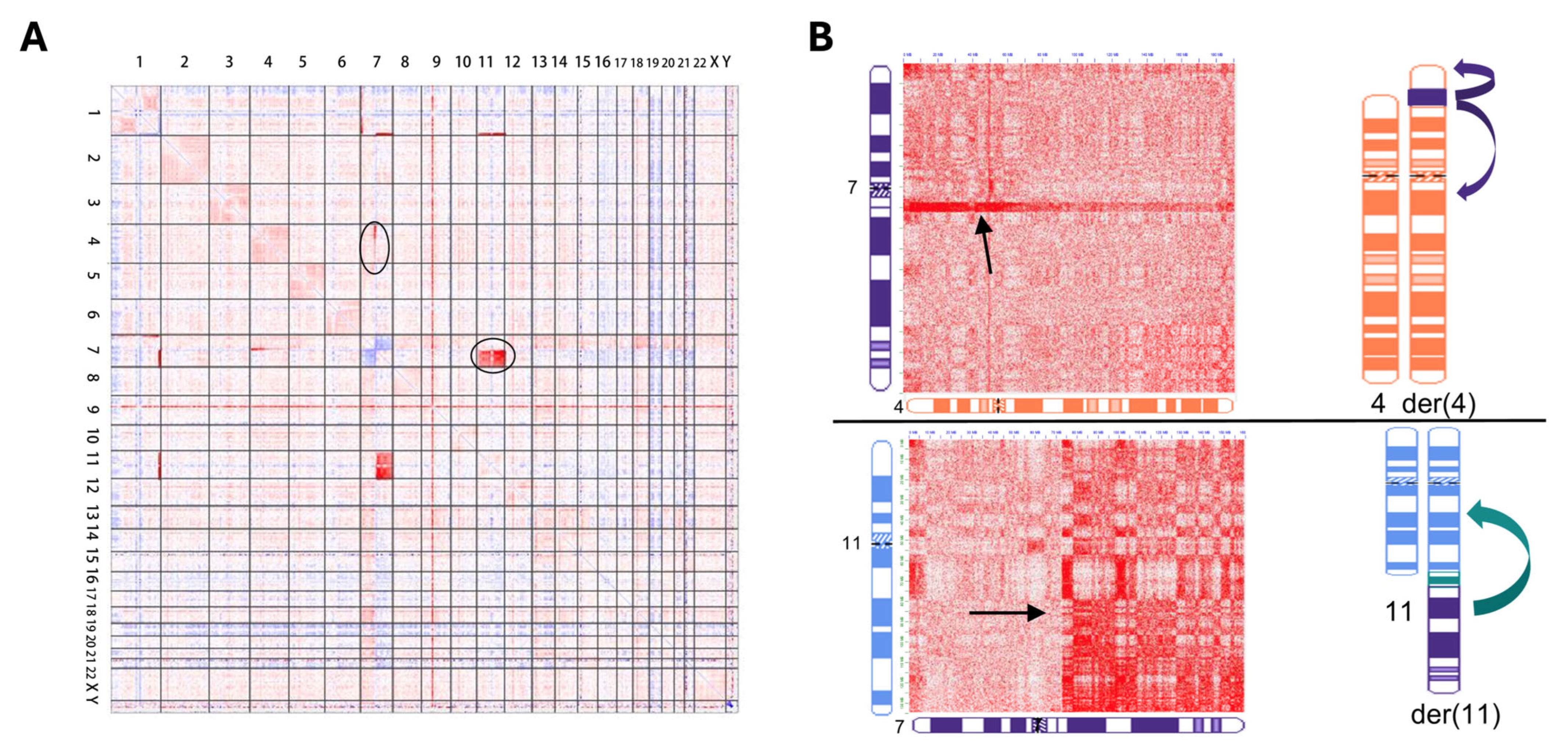
6. Genomic Proximity Mapping
7. Discussion/Summary
Funding
Acknowledgments
Conflicts of Interest
References
- The Principles of Clinical Cytogenetics, 3rd ed.; Gersen, S.L., Keagle, M.B., Eds.; Springer: New York, NY, USA, 2013. [Google Scholar] [CrossRef][Green Version]
- Crotwell, P.L.; Hoyme, H.E. Advances in Whole-Genome Genetic Testing: From Chromosomes to Microarrays. Curr. Probl. Pediatr. Adolesc. Health Care 2012, 42, 47–73. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Eacker, S.M.; Wu, Y.; Neufeld–Kaiser, W.; Laurino, M.; Keel, S.; Horwitz, M.S.; Liu, Y.J. Genetic and Functional Characterization of Inherited Complex Chromosomal Rearrangements in a Family with Multisystem Anomalies. Genet. Med. Open 2025, 1–39. [Google Scholar] [CrossRef]
- Lejeune, J.; Gautier, M.; Turpin, R. [Study of somatic chromosomes from 9 mongoloid children]. C. R. Hebd. Seances Acad. Sci. 1959, 248, 1721–1722. [Google Scholar] [PubMed]
- Nowell, P.C.; Hungerford, D.A. Chromosome Studies on Normal and Leukemic Human Leukocytes. J. Natl. Cancer Inst. 1960, 25, 85–109. [Google Scholar]
- Trask, B.J. Human Cytogenetics: 46 Chromosomes, 46 Years and Counting. Nat. Rev. Genet. 2002, 3, 769–778. [Google Scholar] [CrossRef]
- Boles, B.; Gardner, J.-A.; Rehder, C.W.; Levy, B.; Velagaleti, G.V.; Toydemir, R.M.; Sukov, W.R.; Larson, D.P.; Cao, Y.; Mixon, C.; et al. Conventional Cytogenetic Analysis of Constitutional Abnormalities. Arch. Pathol. Lab. Med. 2025, 149, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Eacker, S.M.; Wu, Y.; Paschal, C.; Wood, M.; Nelson, B.; Muratov, A.; Liu, Y. Evaluation of Genomic Proximity Mapping (GPM) for Detecting Genomic and Chromosomal Structural Variants in Constitutional Disorders. medRxiv 2025, 1–29. Available online: https://www.medrxiv.org/content/10.1101/2025.04.23.25326303v1 (accessed on 29 May 2025).
- Fluorescence In Situ Hybridization (FISH): Protocols and Applications; Bridger, J.M., Volpi, E.V., Eds.; Humana Press: New York, NY, USA, 2010; Volume 1. [Google Scholar] [CrossRef]
- ACMG; ASHG. Technical and Clinical Assessment of Fluorescence in Situ Hybridization: An ACMG/ASHG Position Statement. I. Technical Considerations. Genet. Med. 2000, 2, 356–361. [Google Scholar] [CrossRef]
- Zneimer, S.M. Cytogenetic Abnormalities: Chromosomal, FISH and Microarray-Based Clinical Reporting; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2014. [Google Scholar] [CrossRef]
- Varella-Garcia, M. Chromosomal and Genomic Changes in Lung Cancer. Cell Adhes. Migr. 2010, 4, 100–106. [Google Scholar] [CrossRef]
- Cytogenomics; Liehr, T., Ed.; Elsevier: Cambridge, MA, USA, 2021. [Google Scholar] [CrossRef]
- Guo, B.; Han, X.; Wu, Z.; Da, W.; Zhu, H. Spectral Karyotyping: An Unique Technique for the Detection of Complex Genomic Rearrangements in Leukemia. Transl. Pediatr. 2014, 3, 135–139. [Google Scholar] [CrossRef]
- Smeets, D.F.C.M. Historical Prospective of Human Cytogenetics: From Microscope to Microarray. Clin. Biochem. 2004, 37, 439–446. [Google Scholar] [CrossRef]
- Thorland, E.C.; Gonzales, P.R.; Gliem, T.J.; Wiktor, A.E.; Ketterling, R.P. Comprehensive Validation of Array Comparative Genomic Hybridization Platforms: How Much Is Enough? Genet. Med. 2007, 9, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.T.; Shen, Y.; Wu, B.-L. Oligonucleotide Microarrays for Clinical Diagnosis of Copy Number Variation and Zygosity Status. Curr. Protoc. Hum. Genet. 2012, 74, 8.12.1–8.12.17. [Google Scholar] [CrossRef] [PubMed]
- Peterson, J.F.; Aggarwal, N.; Smith, C.A.; Gollin, S.M.; Surti, U.; Rajkovic, A.; Swerdlow, S.H.; Yatsenko, S.A. Integration of Microarray Analysis into the Clinical Diagnosis of Hematological Malignancies: How Much Can We Improve Cytogenetic Testing? Oncotarget 2015, 6, 18845–18862. [Google Scholar] [CrossRef]
- Shaffer, L.G.; Beaudet, A.L.; Brothman, A.R.; Hirsch, B.; Levy, B.; Martin, C.L.; Mascarello, J.T.; Rao, K.W. Microarray Analysis for Constitutional Cytogenetic Abnormalities. Genet. Med. 2007, 9, 654–662. [Google Scholar] [CrossRef]
- Miller, D.T.; Adam, M.P.; Aradhya, S.; Biesecker, L.G.; Brothman, A.R.; Carter, N.P.; Church, D.M.; Crolla, J.A.; Eichler, E.E.; Epstein, C.J.; et al. Consensus Statement: Chromosomal Microarray Is a First-Tier Clinical Diagnostic Test for Individuals with Developmental Disabilities or Congenital Anomalies. Am. J. Hum. Genet. 2010, 86, 749–764. [Google Scholar] [CrossRef]
- LaFramboise, T. Single Nucleotide Polymorphism Arrays: A Decade of Biological, Computational and Technological Advances. Nucleic Acids Res. 2009, 37, 4181–4193. [Google Scholar] [CrossRef] [PubMed]
- Kearney, H.M.; Kulkarni, S.; Bijwaard, K.E.; Hegde, M.R.; Kalman, L.; Shi, L.; Shippy, R.; South, S.T.; Thorland, E.C.; Vitazka, P.; et al. Genomic Copy Number Microarrays for Constitutional Genetic and Oncology Applications, 1st ed.; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2015. [Google Scholar]
- Gordeeva, V.; Sharova, E.; Arapidi, G. Progress in Methods for Copy Number Variation Profiling. Int. J. Mol. Sci. 2022, 23, 2143. [Google Scholar] [CrossRef]
- Hoppman, N.; Rumilla, K.; Lauer, E.; Kearney, H.; Thorland, E. Patterns of Homozygosity in Patients with Uniparental Disomy: Detection Rate and Suggested Reporting Thresholds for SNP Microarrays. Genet. Med. 2018, 20, 1522–1527. [Google Scholar] [CrossRef]
- Smith, A.C.; Hoischen, A.; Raca, G. Cytogenetics Is a Science, Not a Technique! Why Optical Genome Mapping Is So Important to Clinical Genetic Laboratories. Cancers 2023, 15, 5470. [Google Scholar] [CrossRef]
- Dremsek, P.; Schwarz, T.; Weil, B.; Malashka, A.; Laccone, F.; Neesen, J. Optical Genome Mapping in Routine Human Genetic Diagnostics—Its Advantages and Limitations. Genes 2021, 12, 1958. [Google Scholar] [CrossRef]
- Mantere, T.; Neveling, K.; Pebrel-Richard, C.; Benoist, M.; van der Zande, G.; Kater-Baats, E.; Baatout, I.; van Beek, R.; Yammine, T.; Oorsprong, M.; et al. Optical Genome Mapping Enables Constitutional Chromosomal Aberration Detection. Am. J. Hum. Genet. 2021, 108, 1409–1422. [Google Scholar] [CrossRef] [PubMed]
- Sahajpal, N.S.; Mondal, A.K.; Tvrdik, T.; Hauenstein, J.; Shi, H.; Deeb, K.K.; Saxe, D.; Hastie, A.; Chaubey, A.; Savage, N.M.; et al. Optical Genome Mapping: Clinical Validation and Diagnostic Utility for Enhanced Cytogenomic Analysis of Hematological Neoplasms. J. Mol. Diagnostics 2022, 24, 1279–1291. [Google Scholar] [CrossRef] [PubMed]
- Sahajpal, N.S.; Mondal, A.K.; Singh, H.; Vashisht, A.; Ananth, S.; Saul, D.; Hastie, A.R.; Hilton, B.; DuPont, B.R.; Savage, N.M.; et al. Clinical Utility of Optical Genome Mapping and 523-Gene Next Generation Sequencing Panel for Comprehensive Evaluation of Myeloid Cancers. Cancers 2023, 15, 3214. [Google Scholar] [CrossRef] [PubMed]
- Sahajpal, N.S.; Barseghyan, H.; Kolhe, R.; Hastie, A.; Chaubey, A. Optical Genome Mapping as a Next-Generation Cytogenomic Tool for Detection of Structural and Copy Number Variations for Prenatal Genomic Analyses. Genes 2021, 12, 398. [Google Scholar] [CrossRef]
- Ma, Y.; Gui, C.; Shi, M.; He, J.; Xie, B.; Zheng, H.; Lei, X.; Cheng, Z.; Zhou, X.; Chen, S. The Cryptic Complex Rearrangements Involving the DMD Gene Causing Completely Opposite Manifestations: Etiologic Clues Revealed by Optical Genome Mapping. Hum. Genomics 2024, 18, 103. [Google Scholar] [CrossRef]
- Cope, H.; Barseghyan, H.; Bhattacharya, S.; Fu, Y.; Hoppman, N.; Marcou, C.; Walley, N.; Rehder, C.; Deak, K.; Alkelai, A.; et al. Detection of a Mosaic CDKL5 Deletion and Inversion by Optical Genome Mapping Ends an Exhaustive Diagnostic Odyssey. Mol. Genet. Genomic. Med. 2021, 9, e1665. [Google Scholar] [CrossRef]
- Sund, K.L.; Liu, J.; Lee, J.; Garbe, J.; Abdelhamed, Z.; Maag, C.; Hallinan, B.; Wu, S.W.; Sperry, E.; Deshpande, A.; et al. Long-Read Sequencing and Optical Genome Mapping Identify Causative Gene Disruptions in Noncoding Sequence in Two Patients with Neurologic Disease and Known Chromosome Abnormalities. Am. J. Hum. Genet. Part A 2024, 194, e63818. [Google Scholar] [CrossRef]
- Stence, A.A.; Thomason, J.G.; Pruessner, J.A.; Sompallae, R.R.; Snow, A.N.; Ma, D.; Moore, S.A.; Bossler, A.D. Validation of Optical Genome Mapping for the Molecular Diagnosis of Facioscapulohumeral Muscular Dystrophy. J. Mol. Diagnostics 2021, 23, 1506–1514. [Google Scholar] [CrossRef]
- Puiggros, A.; Ramos-Campoy, S.; Kamaso, J.; de la Rosa, M.; Salido, M.; Melero, C.; Rodríguez-Rivera, M.; Bougeon, S.; Collado, R.; Gimeno, E.; et al. Optical Genome Mapping: A Promising New Tool to Assess Genomic Complexity in Chronic Lymphocytic Leukemia (CLL). Cancers 2022, 14, 3376. [Google Scholar] [CrossRef]
- Soler, G.; Ouedraogo, Z.G.; Goumy, C.; Lebecque, B.; Aspas Requena, G.; Ravinet, A.; Kanold, J.; Véronèse, L.; Tchirkov, A. Optical Genome Mapping in Routine Cytogenetic Diagnosis of Acute Leukemia. Cancers 2023, 15, 2131. [Google Scholar] [CrossRef]
- Giguère, A.; Raymond-Bouchard, I.; Collin, V.; Claveau, J.S.; Hébert, J.; LeBlanc, R. Optical Genome Mapping Reveals the Complex Genetic Landscape of Myeloma. Cancers 2023, 15, 4687. [Google Scholar] [CrossRef] [PubMed]
- Klos, C.; Roynard, P.; Berthon, C.; Soenen, V.; Marceau, A.; Fournier, E.; Fenwarth, L.; Daudignon, A.; Roche, C.; Guermouche, H. Unravelling a KMT2A::ARHGEF12 Fusion within Chromoanagenesis in Acute Myeloid Leukemia Using Optical Genome Mapping. Ann. Hematol. 2024, 4793–4795. [Google Scholar] [CrossRef]
- Yeung, C.C.S.; Eacker, S.M.; Sala-torra, O.; Beppu, L.; Woolston, D.W.; Liachko, I.; Malig, M.; Stirewalt, D.; Fang, M.; Radich, J. Evaluation of Acute Myeloid Leukemia Genomes Using Genomic Proximity Mapping. medRxiv 2024. [Google Scholar] [CrossRef]
- Mathew, M.T.; Babcock, M.; Hou, Y.C.C.; Hunter, J.M.; Leung, M.L.; Mei, H.; Schieffer, K.; Akkari, Y. Clinical Cytogenetics: Current Practices and Beyond. J. Appl. Lab. Med. 2024, 9, 61–75. [Google Scholar] [CrossRef] [PubMed]
- Lacoste, S.A.; Gagnon, V.; Béliveau, F.; Lavallée, S.; Collin, V.; Hébert, J. Unveiling the Complexity of KMT2A Rearrangements in Acute Myeloid Leukemias with Optical Genome Mapping. Cancers 2024, 16, 4171. [Google Scholar] [CrossRef]
- Tembrink, M.; Gerding, W.M.; Wieczorek, S.; Mika, T.; Schroers, R.; Nguyen, H.P.; Vangala, D.B.; Nilius-Eliliwi, V. Novel NUP98::ASH1L Gene Fusion in Acute Myeloid Leukemia Detected by Optical Genome Mapping. Cancers 2023, 15, 2942. [Google Scholar] [CrossRef]
- Valkama, A.; Vorimo, S.; Kumpula, T.A.; Räsänen, H.; Savolainen, E.R.; Pylkäs, K.; Mantere, T. Optical Genome Mapping as an Alternative to FISH-Based Cytogenetic Assessment in Chronic Lymphocytic Leukemia. Cancers 2023, 15, 1294. [Google Scholar] [CrossRef] [PubMed]
- Suttorp, J.; Lühmann, J.L.; Behrens, Y.L.; Göhring, G.; Steinemann, D.; Reinhardt, D.; von Neuhoff, N.; Schneider, M. Optical Genome Mapping as a Diagnostic Tool in Pediatric Acute Myeloid Leukemia. Cancers 2022, 14, 2058. [Google Scholar] [CrossRef]
- Seto, A.; Downs, G.; King, O.; Salehi-Rad, S.; Baptista, A.; Chin, K.; Grenier, S.; Nwachukwu, B.; Tierens, A.; Minden, M.D.; et al. Genomic Characterization of Partial Tandem Duplication Involving the KMT2A Gene in Adult Acute Myeloid Leukemia. Cancers 2024, 16, 1693. [Google Scholar] [CrossRef]
- Lühmann, J.L.; Stelter, M.; Wolter, M.; Kater, J.; Lentes, J.; Bergmann, A.K.; Schieck, M.; Göhring, G.; Möricke, A.; Cario, G.; et al. The Clinical Utility of Optical Genome Mapping for the Assessment of Genomic Aberrations in Acute Lymphoblastic Leukemia. Cancers 2021, 13, 4388. [Google Scholar] [CrossRef]
- Fang, H.; Liu, Y.; Eacker, S.; Neufeld-Kaiser, W. 11. Evaluation of Hi-C versus Optical Genome Mapping for Diagnosing Constitutional Genomic Structural Variants. Cancer Genet. 2023, 278–279, 4. [Google Scholar] [CrossRef]
- Malig, M.; Sala-Torra, O.; Wood, M.; Langford, K.; Beppu, L.W.; Madarang, E.C.; Woolston, D.; Nong, T.; Wu, Y.; Fang, H.; et al. Genomic Proximity Mapping (GPM): Evaluation of a Next Generation Cytogenomic Assay for Improved Risk Stratification in Acute Myeloid Leukemia. Blood 2024, 144, 6157–6158. [Google Scholar] [CrossRef]
- Boyd, B.M.; Fang, H.; Allingham-Hawkins, D.; Fischer, G.J.; Peng, S.; Puryear, L.; Liu, Y.J.; Hisama, F.M. Chromosomal Translocation Resolves a Diagnostic Odyssey for Familial Ruvalcaba Syndrome. Am. J. Med. Genet. Part A 2024, 197, e63847. [Google Scholar] [CrossRef] [PubMed]
- Dennis, M.J.; Pavlick, D.C.; Kacew, A.; Wotman, M.; MacConaill, L.E.; Jones, S.M.; Pfaff, K.L.; Rodig, S.J.; Eacker, S.; Malig, M.; et al. Low PD-L1 Expression, MAP2K2 Alterations, and Enriched HPV Gene Signatures Characterize Brain Metastases in Head and Neck Squamous Cell Carcinoma. J. Transl. Med. 2024, 22, 960. [Google Scholar] [CrossRef]
- Yuan, Y.; Chung, C.Y.L.; Chan, T.F. Advances in Optical Mapping for Genomic Research. Comput. Struct. Biotechnol. J. 2020, 18, 2051–2062. [Google Scholar] [CrossRef]
- Wang, Y.; Liehr, T. The Need for a Concert of Cytogenomic Methods in Chromosomic Research and Diagnostics. Genes 2025, 16, 533. [Google Scholar] [CrossRef]


| Karyotype | FISH | CMA | OGM | GPM | |
|---|---|---|---|---|---|
| Region | Genome-Wide | Targeted * | Genome-Wide | Genome-Wide | Genome-Wide |
| Resolution | 5–10 Mb | 100 kb * | 40 kb | 5–500 kb | 5–50 kb |
| Detection of CNV | Yes | Yes | Yes | Yes | Yes |
| Detection of Ploidy | Yes | Yes | Yes * | No | No |
| Detection of Mosaicism | Yes | Yes | Yes * | Yes * | Yes * |
| Detection of LOH/ROH | No | No | Yes | Yes * | Yes |
| Detection of UPD | No | No | Yes * | Yes * | Yes * |
| Unbalanced SV | Yes | Yes | Yes | Yes | Yes |
| Balanced SV | Yes | Yes | No | Yes | Yes |
| Reconstruction of Complex CR | Yes | No | No | Yes | Yes |
| Robertsonian Translocations | Yes | No * | No | No | Yes |
| Culturing Required? | Yes | No * | No | No | No |
| Cost | Low | Low | Low | Medium-High | Medium-High |
| TAT | Days-Weeks | Days | Days | Days-Weeks | Days-Weeks |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonzales, P.R. Integration of Newer Genomic Technologies into Clinical Cytogenetics Laboratories. Genes 2025, 16, 688. https://doi.org/10.3390/genes16060688
Gonzales PR. Integration of Newer Genomic Technologies into Clinical Cytogenetics Laboratories. Genes. 2025; 16(6):688. https://doi.org/10.3390/genes16060688
Chicago/Turabian StyleGonzales, Patrick R. 2025. "Integration of Newer Genomic Technologies into Clinical Cytogenetics Laboratories" Genes 16, no. 6: 688. https://doi.org/10.3390/genes16060688
APA StyleGonzales, P. R. (2025). Integration of Newer Genomic Technologies into Clinical Cytogenetics Laboratories. Genes, 16(6), 688. https://doi.org/10.3390/genes16060688






