Progress in the Genetics of Myelodysplastic Syndromes with a Latin American Perspective
Abstract
1. Introduction
2. Overview of Molecular Characterization Studies
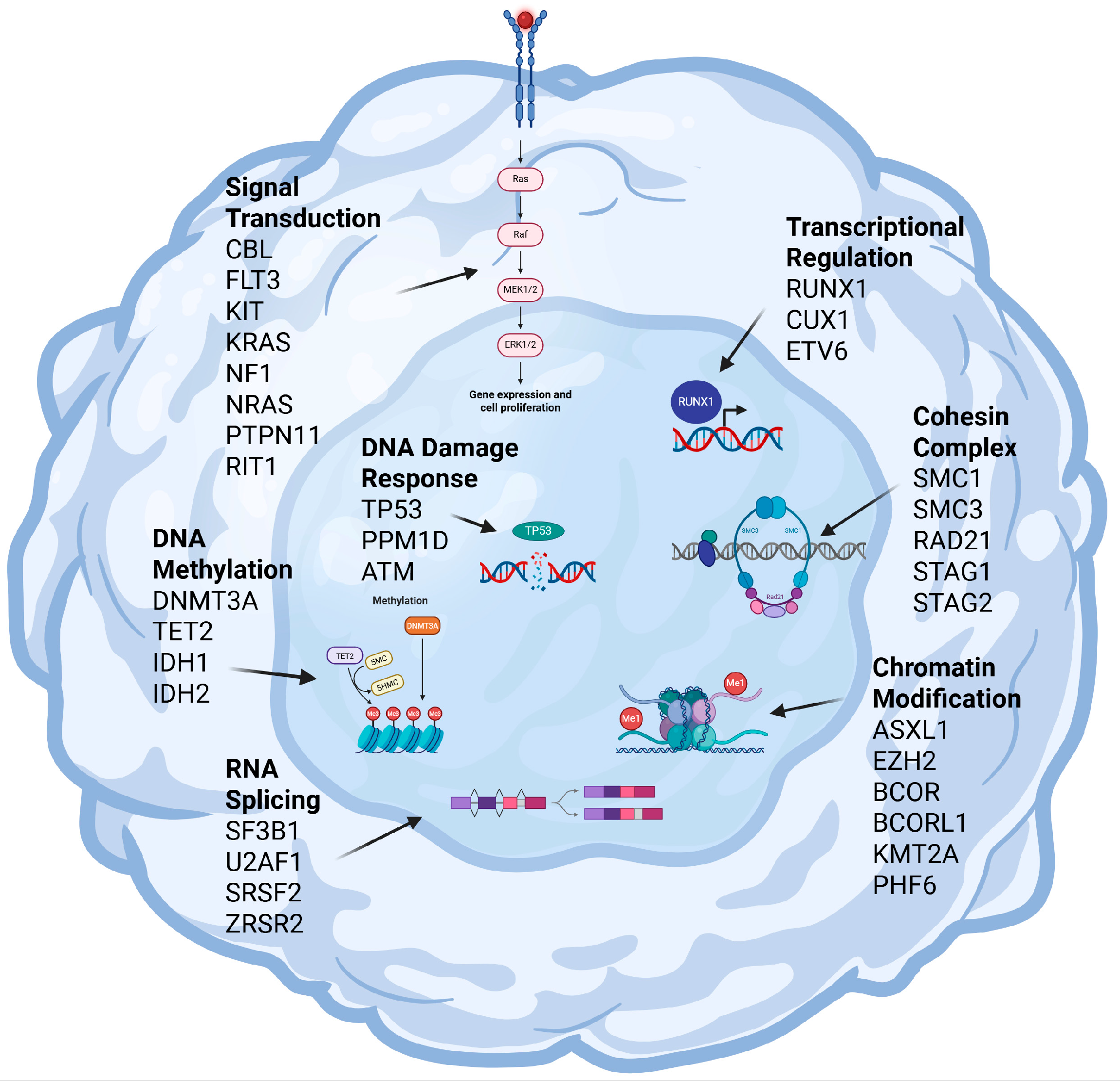
3. Key Molecular Mechanisms Driving MDS Pathogenesis
3.1. RNA Splicing
3.2. Chromatin Modification
3.3. DNA Methylation
3.4. Transcriptional Regulation
3.5. DNA Damage Response
3.6. Signal Transduction
3.7. Other Mechanisms
4. Molecular Genetics Data Integrated for Prognostic Risk Stratification in MDS, Including Artificial Intelligence
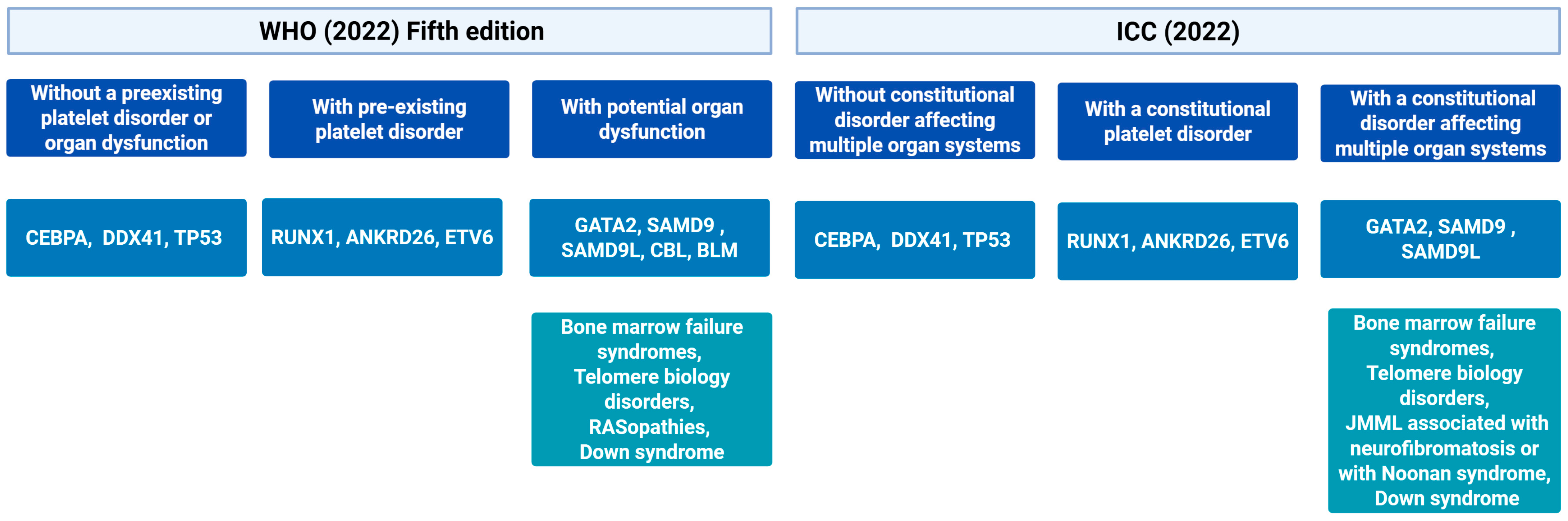
5. Molecular Data Incorporated in Current Diagnostic Classifications and New Proposals
6. Molecular Data to Tailor Treatment Strategies
7. Germline Predisposition: Implications and Challenges in Latin America
7.1. Main Syndromes
7.2. When to Suspect and How to Diagnose a Germline Predisposition
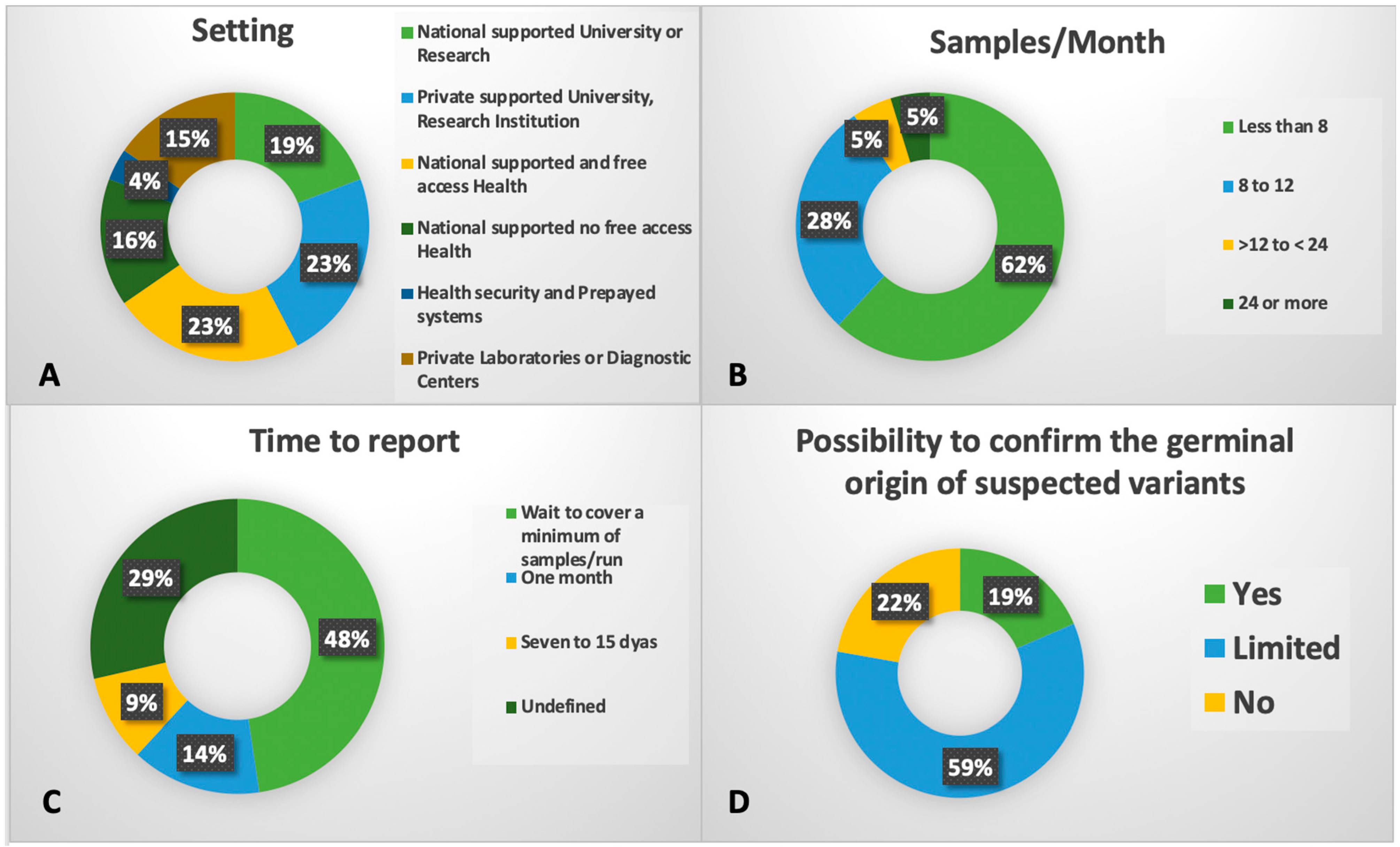
8. Molecular Tools in Latin America
9. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MDS | Myelodysplastic syndromes |
| AML | Acute myeloid leukemia |
| AL | Acute leukemia |
| WHO | World Health Organization |
| ICC | International Consensus Classification |
| SEER | Surveillance, Epidemiology, and End Results |
| CMML | Chronic myelomonocytic leukemia |
| NGS | Next-generation sequencing |
| CNV | Copy number variations |
| IPSS-R | International Prognostic Scoring System Revised |
| IPSS-M | International Prognostic Scoring System Molecular |
| OS | Overall Survival |
| LFS | Leukemia Free Survival |
| VAF | Variant allele frequency |
| HMA | Hypomethylating agents |
| CHIP | Clonal hematopoiesis of indeterminate potential |
| PFS | Progression free survival |
| AIPSS | Artificial Intelligence Prognostic Scoring System |
References
- Hasserjian, R.P.; Germing, U.; Malcovati, L. Diagnosis and classification of myelodysplastic syndromes. Blood 2023, 142, 2247–2257. [Google Scholar] [CrossRef] [PubMed]
- Belli, C.B.; Bestach, Y.; Giunta, M.; Iastrebner, M.; Santos, I.; Pintos, N.; Arbelbide, J.; Basquiera, A.L.; Bengio, R.; Larripa, I. Application of the revised International Prognostic Scoring System for myelodysplastic syndromes in Argentinean patients. Ann. Hematol. 2014, 93, 705–707. [Google Scholar] [CrossRef]
- Naqvi, K.; Garcia-Manero, G.; Sardesai, S.; Oh, J.; Vigil, C.E.; Pierce, S.; Lei, X.; Shan, J.; Kantarjian, H.M.; Suarez-Almazor, M.E. Association of comorbidities with overall survival in myelodysplastic syndrome: Development of a prognostic model. J. Clin. Oncol. 2011, 29, 2240–2246. [Google Scholar] [CrossRef]
- Basquiera, A.; Enrico, A.; Arbelbide, J.; Nucifora, E.; Iastrebner, M.; Flores, G.; Gonzalez, J.; Crisp, R.; Novoa, V.; Alfonso, G.; et al. Myelodysplasia-Related Mortality Remains the Main Cause of Death Along Different Groups of Risks: An Analysis from MDS Argentinean Study Group. Haematologica 2017, 102 (Suppl. S2), 485. [Google Scholar]
- National Comprehensive Cancer Network. Myelodysplastic Syndromes (Version 2.2025). Available online: https://www.nccn.org/professionals/physician_gls/pdf/mds.pdf (accessed on 13 April 2025).
- Luo, B.; Dong, F.; Qin, T.; Zhang, Q.; Bai, H.; Wang, J.; Jia, Y.; Ma, S.; Jiang, E.; Cheng, T.; et al. Myelodysplastic syndromes are multiclonal diseases derived from hematopoietic stem and progenitor cells. Exp. Hematol. Oncol. 2022, 11, 28. [Google Scholar] [CrossRef] [PubMed]
- Khoury, J.D.; Solary, E.; Abla, O.; Akkari, Y.; Alaggio, R.; Apperley, J.F.; Bejar, R.; Berti, E.; Busque, L.; Chan, J.K.C.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia 2022, 36, 1703–1719. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.P.; Borowitz, M.J.; Calvo, K.R.; Kvasnicka, H.M.; Wang, S.A.; Bagg, A.; Barbui, T.; Branford, S.; et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: Integrating morphologic, clinical, and genomic data. Blood 2022, 140, 1200–1228. [Google Scholar] [CrossRef] [PubMed]
- Gou, X.; Chen, Z.; Shangguan, Y. Global, regional, and national burden of myelodysplastic syndromes and myeloproliferative neoplasms, 1990–2021: An analysis from the global burden of disease study 2021. Front. Oncol. 2025, 15, 1559382. [Google Scholar] [CrossRef]
- Goksu, S.Y.; Ozer, M.; Goksu, B.B.; Wang, R.; Khatib, J.; Patel, P.A.; Vusirikala, M.; Cole, S.; Seyhanli, A.; Collins, R.H.; et al. The impact of race and ethnicity on outcomes of patients with myelodysplastic syndromes: A population-based analysis. Leuk. Lymphoma 2022, 63, 1651–1659. [Google Scholar] [CrossRef]
- Economic Commission for Latin America and the Caribbean (ECLAC). Demographic Observatory, 2024 (LC/PUB.2024/22-P), Santiago, 2024. Available online: https://repositorio.cepal.org/server/api/core/bitstreams/dc566b9e-b3ef-44e9-a167-995614696404/content (accessed on 7 April 2025).
- Crisp, R.; Grille, S.; Belli, C.; Diaz, L.; Undurraga, S.; Navarro, J.; Vidal, G.; Gusmao, B.; Reyes, J.; Huaman, F.; et al. Myelodysplastic syndromes in Latin America: State of the art. Blood Adv. 2018, 2 (Suppl. S1), 60–62. [Google Scholar] [CrossRef]
- Norris, E.T.; Wang, L.; Conley, A.B.; Rishishwar, L.; Marino-Ramirez, L.; Valderrama-Aguirre, A.; Jordan, I.K. Genetic ancestry, admixture and health determinants in Latin America. BMC Genom. 2018, 19 (Suppl. S8), 861. [Google Scholar] [CrossRef] [PubMed]
- Borges, D.P.; Dos Santos, R.; Velloso, E.R.P.; Ribeiro Junior, H.L.; Larripa, I.B.; Camacho, M.F.; Gonzalez, J.; Pratx, L.D.B.; Magalhaes, S.M.M.; Belli, C.B.; et al. Functional polymorphisms of DNA repair genes in Latin America reinforces the heterogeneity of Myelodysplastic Syndrome. Hematol. Transfus. Cell Ther. 2023, 45, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro Junior, H.L. Molecular monitoring of myelodysplastic neoplasm: Don’t just watch this space, consider the patient’s ancestry. Br. J. Haematol. 2024, 205, 759–760. [Google Scholar] [CrossRef]
- Belli, C.B.; Pinheiro, R.F.; Bestach, Y.; Larripa, I.B.; da Silva Tanizawa, R.S.; Alfonso, G.; Gonzalez, J.; Rosenhain, M.; Watman, N.; Cavalcante de Andrade Silva, M.; et al. Myelodysplastic syndromes in South America: A multinational study of 1080 patients. Am. J. Hematol. 2015, 90, 851–858. [Google Scholar] [CrossRef]
- Gonzalez, J.S.; Perusini, M.A.; Basquiera, A.L.; Alfonso, G.; Fantl, D.; Lima, W.M.; Nucifora, E.; Lazzarino, C.; Novoa, V.; de Andrade Silva, M.C.; et al. Prognostic assessment for chronic myelomonocytic leukemia in the context of the World Health Organization 2016 proposal: A multicenter study of 280 patients. Ann. Hematol. 2021, 100, 1439–1449. [Google Scholar] [CrossRef]
- Azevedo, R.S.; Belli, C.; Bassolli, L.; Ferri, L.; Perusini, M.A.; Enrico, A.; Pereira, T.; Junior, W.; Buccheri, V.; Pinheiro, R.F.; et al. Age, Blasts, Performance Status and Lenalidomide Therapy Influence the Outcome of Myelodysplastic Syndrome With Isolated Del(5q): A Study of 58 South American Patients. Clin. Lymphoma Myeloma Leuk. 2022, 22, e1–e6. [Google Scholar] [CrossRef]
- Lazzarino, C.; Arbelbide, J.; Schuster, S.; Crisp, R.; Gonzalez, J.; Tavares, R.; Perusini, M.A.; Di Stefano, M.; Pintos, N.; Espinosa, D.; et al. Severe thrombocytopenia as a predictor of survival and response to hypomethylating agents in myelodysplastic syndromes: A Latin-American cohort of 212 patients. Am. J. Hematol. 2020, 95, E323–E326. [Google Scholar] [CrossRef] [PubMed]
- Castelo, L.; da Silva, W.F.; Lincango, M.; Buccheri, V.; Perusini, A.; Arbelbide, J.; Iastrebner, M.; Gonzalez, J.; Pereyra, P.; Pereira, T.D.M.; et al. Alternative Dosing Schedules of Azacitidine: A Real-World Study Across South American Centers. Clin. Lymphoma Myeloma Leuk. 2024, 24, 407–411. [Google Scholar] [CrossRef]
- Moura, A.T.G.; Duarte, F.B.; Barbosa, M.C.; Santos, T.; Lemes, R.P.G. Prolonged response to recombinant human erythropoietin treatment in patients with myelodysplastic syndrome at a single referral centre in Brazil. Clinics 2019, 74, e771. [Google Scholar] [CrossRef]
- Basquiera, A.L.; Rivas, M.M.; Remaggi, G.; Klein, G.; Milovic, V.; Foncuberta, M.C.; Saba, S.; Milone, J.H.; Arbelbide, J.; Jaimovich, G.; et al. Allogeneic hematopoietic stem cell transplantation in adults with myelodysplastic syndrome: Experience of the Argentinean Group of Bone Marrow Transplantation (GATMO). Hematology 2016, 21, 162–169. [Google Scholar] [CrossRef]
- Duarte, F.B.; Moura, A.T.G.; Funke, V.A.M.; Colturato, V.A.R.; Hamerschlak, N.; Vilela, N.C.; Lopes, L.F.; de Almeida Macedo, M.C.M.; Vigorito, A.C.; de Almeida Soares, R.D.; et al. Impact of Treatment Prior to Allogeneic Transplantation of Hematopoietic Stem Cells in Patients with Myelodysplastic Syndrome: Results of the Latin American Bone Marrow Transplant Registry. Biol. Blood Marrow Transpl. 2020, 26, 1021–1024. [Google Scholar] [CrossRef] [PubMed]
- Bejar, R.; Stevenson, K.; Abdel-Wahab, O.; Galili, N.; Nilsson, B.; Garcia-Manero, G.; Kantarjian, H.; Raza, A.; Levine, R.L.; Neuberg, D.; et al. Clinical effect of point mutations in myelodysplastic syndromes. N. Engl. J. Med. 2011, 364, 2496–2506. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Sanada, M.; Shiraishi, Y.; Nowak, D.; Nagata, Y.; Yamamoto, R.; Sato, Y.; Sato-Otsubo, A.; Kon, A.; Nagasaki, M.; et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011, 478, 64–69. [Google Scholar] [CrossRef]
- Duployez, N.; Preudhomme, C. Monitoring molecular changes in the management of myelodysplastic syndromes. Br. J. Haematol. 2024, 205, 772–779. [Google Scholar] [CrossRef] [PubMed]
- Papaemmanuil, E.; Gerstung, M.; Malcovati, L.; Tauro, S.; Gundem, G.; Van Loo, P.; Yoon, C.J.; Ellis, P.; Wedge, D.C.; Pellagatti, A.; et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 2013, 122, 3616–3627. [Google Scholar] [CrossRef]
- Haferlach, T.; Nagata, Y.; Grossmann, V.; Okuno, Y.; Bacher, U.; Nagae, G.; Schnittger, S.; Sanada, M.; Kon, A.; Alpermann, T.; et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 2014, 28, 241–247. [Google Scholar] [CrossRef]
- Bernard, E.; Tuechler, H.; Greenberg, P.L.; Hasserjian, R.P.; Arango Ossa, J.E.; Nannya, Y.; Devlin, S.M.; Creignou, M.; Pinel, P.; Monnier, L.; et al. Molecular International Prognostic Scoring System for Myelodysplastic Syndromes. NEJM Evid. 2022, 1, EVIDoa2200008. [Google Scholar] [CrossRef]
- Lincango, M.; Andreoli, V.; Rivello, H.G.; Bender, A.; Catalan, A.I.; Rahhal, M.; Delamer, R.; Asinari, M.; Orgueira, A.M.; Castro, M.B.; et al. Assessing the Relevance of Non-molecular Prognostic Systems for Myelodysplastic Syndrome in the Era of Next-Generation Sequencing. Ann. Lab. Med. 2025, 45, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Catalán, A.I.; Boada, M.; Ottati, C.; Ranero, S.; Cayota, A.; Lens, D.; Grille, S. Integration of NGS and CNV Analysis in Prognostic Evaluation of MDS in a Developing Country: Insights from Uruguay. Blood Glob. Hematol. 2025, 100014. Available online: https://ashpublications.org/bloodglobal/article/doi/10.1016/j.bglo.2025.100014/537254/Integration-of-NGS-and-CNV-Analysis-in-Prognostic (accessed on 29 May 2025). [CrossRef]
- Tseng, C.C.; Obeng, E.A. RNA splicing as a therapeutic target in myelodysplastic syndromes. Semin. Hematol. 2024, 61, 431–441. [Google Scholar] [CrossRef]
- Reinig, E.; Yang, F.; Traer, E.; Arora, R.; Brown, S.; Rattray, R.; Braziel, R.; Fan, G.; Press, R.; Dunlap, J. Targeted Next-Generation Sequencing in Myelodysplastic Syndrome and Chronic Myelomonocytic Leukemia Aids Diagnosis in Challenging Cases and Identifies Frequent Spliceosome Mutations in Transformed Acute Myeloid Leukemia. Am. J. Clin. Pathol. 2016, 145, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Thol, F.; Kade, S.; Schlarmann, C.; Loffeld, P.; Morgan, M.; Krauter, J.; Wlodarski, M.W.; Kolking, B.; Wichmann, M.; Gorlich, K.; et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood 2012, 119, 3578–3584. [Google Scholar] [CrossRef]
- Tefferi, A.; Lasho, T.L.; Patnaik, M.M.; Saeed, L.; Mudireddy, M.; Idossa, D.; Finke, C.; Ketterling, R.P.; Pardanani, A.; Gangat, N. Targeted next-generation sequencing in myelodysplastic syndromes and prognostic interaction between mutations and IPSS-R. Am. J. Hematol. 2017, 92, 1311–1317. [Google Scholar] [CrossRef]
- Lindsley, R.C.; Mar, B.G.; Mazzola, E.; Grauman, P.V.; Shareef, S.; Allen, S.L.; Pigneux, A.; Wetzler, M.; Stuart, R.K.; Erba, H.P.; et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood 2015, 125, 1367–1376. [Google Scholar] [CrossRef] [PubMed]
- Nagehan, P.; Sabbir, M.; Song, J.; Mohammad, H. Impact of single versus multiple spliceosome mutations on myelodysplastic syndrome. J. Clin. Exp. Hematop. 2023, 63, 173–176. [Google Scholar] [CrossRef]
- Jiang, M.; Chen, M.; Liu, Q.; Jin, Z.; Yang, X.; Zhang, W. SF3B1 mutations in myelodysplastic syndromes: A potential therapeutic target for modulating the entire disease process. Front. Oncol. 2023, 13, 1116438. [Google Scholar] [CrossRef]
- Malcovati, L.; Stevenson, K.; Papaemmanuil, E.; Neuberg, D.; Bejar, R.; Boultwood, J.; Bowen, D.T.; Campbell, P.J.; Ebert, B.L.; Fenaux, P.; et al. SF3B1-mutant MDS as a distinct disease subtype: A proposal from the International Working Group for the Prognosis of MDS. Blood 2020, 136, 157–170. [Google Scholar] [CrossRef]
- Lincango, M.; Larripa, I.; Belli, C. Bioinformatic Evaluation of Differentially Expressed Genes in Myelodysplastic Syndromes with SF3B1 Variants. Medicina 2022, 82 (Suppl. SV), 98. [Google Scholar]
- Lincango, M.; González, J.; Toloza, M.J.; Pintos, N.; Fiorito, A.; Iastrebner, M.; Alberbide, J.; Ferro, H.; Engelberger, I.; Enrico, A.; et al. Validation of Novel Hub Genes Expression in Patients with Myelodysplastic Syndrome and SF3B1 Mutations. Medicina 2024, 84 (Suppl. SV), 108. [Google Scholar]
- Donaires, F.S.; Martelli, F.; Alves-Paiva, R.M.; Magalhaes, S.M.; Pinheiro, R.F.; Calado, R.T. Splicing factor SF3B1 mutations and ring sideroblasts in myelodysplastic syndromes: A Brazilian cohort screening study. Rev. Bras. Hematol. Hemoter. 2016, 38, 320–324. [Google Scholar] [CrossRef][Green Version]
- Sarmiento, M.; Rocca, G.S.; Rahhal, M.; Lincango Yupanki, M.; Zubieta, M.; Metrebian, F.; Narbaitz, M.; Larripa, I.B.; Belli, C.B. Efficacy of lenalidomide in a patient with systemic mastocytosis associated with SF3B1-mutant myelodysplastic syndrome. Leuk. Lymphoma 2021, 62, 3027–3030. [Google Scholar] [CrossRef] [PubMed]
- Badar, T.; Vanegas, Y.A.M.; Nanaa, A.; Foran, J.M.; Al-Kali, A.; Mangaonkar, A.; Murthy, H.; Alkhateeb, H.B.; Viswanatha, D.; He, R.; et al. U2AF1 pathogenic variants in myeloid neoplasms and precursor states: Distribution of co-mutations and prognostic heterogeneity. Blood Cancer J. 2023, 13, 149. [Google Scholar] [CrossRef] [PubMed]
- Bersanelli, M.; Travaglino, E.; Meggendorfer, M.; Matteuzzi, T.; Sala, C.; Mosca, E.; Chiereghin, C.; Di Nanni, N.; Gnocchi, M.; Zampini, M.; et al. Classification and Personalized Prognostic Assessment on the Basis of Clinical and Genomic Features in Myelodysplastic Syndromes. J. Clin. Oncol. 2021, 39, 1223–1233. [Google Scholar] [CrossRef] [PubMed]
- Kuendgen, A.; Muller-Thomas, C.; Lauseker, M.; Haferlach, T.; Urbaniak, P.; Schroeder, T.; Brings, C.; Wulfert, M.; Meggendorfer, M.; Hildebrandt, B.; et al. Efficacy of azacitidine is independent of molecular and clinical characteristics—An analysis of 128 patients with myelodysplastic syndromes or acute myeloid leukemia and a review of the literature. Oncotarget 2018, 9, 27882–27894. [Google Scholar] [CrossRef]
- Kim, E.; Ilagan, J.O.; Liang, Y.; Daubner, G.M.; Lee, S.C.; Ramakrishnan, A.; Li, Y.; Chung, Y.R.; Micol, J.B.; Murphy, M.E.; et al. SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer Cell 2015, 27, 617–630. [Google Scholar] [CrossRef]
- Makishima, H.; Yoshizato, T.; Yoshida, K.; Sekeres, M.A.; Radivoyevitch, T.; Suzuki, H.; Przychodzen, B.; Nagata, Y.; Meggendorfer, M.; Sanada, M.; et al. Dynamics of clonal evolution in myelodysplastic syndromes. Nat. Genet. 2017, 49, 204–212. [Google Scholar] [CrossRef]
- Thol, F.; Friesen, I.; Damm, F.; Yun, H.; Weissinger, E.M.; Krauter, J.; Wagner, K.; Chaturvedi, A.; Sharma, A.; Wichmann, M.; et al. Prognostic significance of ASXL1 mutations in patients with myelodysplastic syndromes. J. Clin. Oncol. 2011, 29, 2499–2506. [Google Scholar] [CrossRef]
- Yang, L.; Wei, X.; Gong, Y. Prognosis and risk factors for ASXL1 mutations in patients with newly diagnosed acute myeloid leukemia and myelodysplastic syndrome. Cancer Med. 2024, 13, e6871. [Google Scholar] [CrossRef]
- Lin, Y.; Zheng, Y.; Wang, Z.C.; Wang, S.Y. Prognostic significance of ASXL1 mutations in myelodysplastic syndromes and chronic myelomonocytic leukemia: A meta-analysis. Hematology 2016, 21, 454–461. [Google Scholar] [CrossRef]
- Rinke, J.; Chase, A.; Cross, N.C.P.; Hochhaus, A.; Ernst, T. EZH2 in Myeloid Malignancies. Cells 2020, 9, 1639. [Google Scholar] [CrossRef]
- Ball, S.; Aguirre, L.E.; Jain, A.G.; Ali, N.A.; Tinsley, S.M.; Chan, O.; Kuykendall, A.T.; Sweet, K.; Lancet, J.E.; Sallman, D.A.; et al. Clinical characteristics and outcomes of EZH2-mutant myelodysplastic syndrome: A large single institution analysis of 1774 patients. Leuk. Res. 2023, 124, 106999. [Google Scholar] [CrossRef] [PubMed]
- Damm, F.; Chesnais, V.; Nagata, Y.; Yoshida, K.; Scourzic, L.; Okuno, Y.; Itzykson, R.; Sanada, M.; Shiraishi, Y.; Gelsi-Boyer, V.; et al. BCOR and BCORL1 mutations in myelodysplastic syndromes and related disorders. Blood 2013, 122, 3169–3177. [Google Scholar] [CrossRef] [PubMed]
- Sportoletti, P.; Sorcini, D.; Falini, B. BCOR gene alterations in hematologic diseases. Blood 2021, 138, 2455–2468. [Google Scholar] [CrossRef]
- Baranwal, A.; Gurney, M.; Basmaci, R.; Katamesh, B.; He, R.; Viswanatha, D.S.; Greipp, P.; Foran, J.; Badar, T.; Murthy, H.; et al. Genetic landscape and clinical outcomes of patients with BCOR mutated myeloid neoplasms. Haematologica 2024, 109, 1779–1791. [Google Scholar] [CrossRef]
- Liu, Y.; Cheng, H.; Cheng, M.; Sun, M.; Ma, J.; Gong, T. Next generation sequencing reveals the mutation landscape of Chinese MDS patients and the association between mutations and AML transformations. Hematology 2024, 29, 2392469. [Google Scholar] [CrossRef] [PubMed]
- Meyer, C.; Larghero, P.; Almeida Lopes, B.; Burmeister, T.; Groger, D.; Sutton, R.; Venn, N.C.; Cazzaniga, G.; Corral Abascal, L.; Tsaur, G.; et al. The KMT2A recombinome of acute leukemias in 2023. Leukemia 2023, 37, 988–1005. [Google Scholar] [CrossRef]
- Wei, Q.; Hu, S.; Xu, J.; Loghavi, S.; Daver, N.; Toruner, G.A.; Wang, W.; Medeiros, L.J.; Tang, G. Detection of KMT2A Partial Tandem Duplication by Optical Genome Mapping in Myeloid Neoplasms: Associated Cytogenetics, Gene Mutations, Treatment Responses, and Patient Outcomes. Cancers 2024, 16, 4193. [Google Scholar] [CrossRef]
- Lee, W.H.; Tsai, M.T.; Tsai, C.H.; Tien, F.M.; Lo, M.Y.; Tseng, M.H.; Kuo, Y.Y.; Liu, M.C.; Yang, Y.T.; Chen, J.C.; et al. Validation of the molecular international prognostic scoring system in patients with myelodysplastic syndromes defined by international consensus classification. Blood Cancer J. 2023, 13, 120. [Google Scholar] [CrossRef]
- Fathima, S.; Alsugair, A.; He, R.; Mangaonkar, A.A.; Begna, K.H.; Pardanani, A.; Zepeda Mendoza, C.J.; Reichard, K.K.; Gangat, N.; Tefferi, A. Myeloid neoplasms with PHF6 mutations: Context-dependent genomic and prognostic characterization in 176 informative cases. Blood Cancer J. 2025, 15, 28. [Google Scholar] [CrossRef]
- Zuo, Z.; Medeiros, L.J.; Garces, S.; Routbort, M.J.; Ok, C.Y.; Loghavi, S.; Kanagal-Shamanna, R.; Jelloul, F.Z.; Garcia-Manero, G.; Chien, K.S.; et al. Concurrent Mutations in SF3B1 and PHF6 in Myeloid Neoplasms. Biology 2022, 12, 13. [Google Scholar] [CrossRef]
- Sudunagunta, V.S.; Viny, A.D. Untangling the loops of STAG2 mutations in myelodysplastic syndrome. Leuk. Lymphoma 2025, 66, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Kon, A.; Shih, L.Y.; Minamino, M.; Sanada, M.; Shiraishi, Y.; Nagata, Y.; Yoshida, K.; Okuno, Y.; Bando, M.; Nakato, R.; et al. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat. Genet. 2013, 45, 1232–1237. [Google Scholar] [CrossRef] [PubMed]
- Katamesh, B.; Nanaa, A.; He, R.; Viswanatha, D.; Nguyen, P.; Greipp, P.; Bessonen, K.; Gangat, N.; Begna, K.; Mangaonkar, A.; et al. Clinical and prognostic impact of STAG2 mutations in myeloid neoplasms: The Mayo Clinic experience. Blood Adv. 2023, 7, 1351–1355. [Google Scholar] [CrossRef] [PubMed]
- Deb, P.Q.; Xiao, W. “Ring-form” megakaryocytic dysplasia in STAG2-mutated myelodysplastic neoplasm. Blood 2024, 143, 2218. [Google Scholar] [CrossRef]
- Kawashima, N.; Kubota, Y.; Bravo-Perez, C.; Guarnera, L.; Williams, N.D.; Durmaz, A.; Witt, M.; Ahmed, A.; Gurnari, C.; Maciejewski, J.P.; et al. Landscape of biallelic DNMT3A mutant myeloid neoplasms. J. Hematol. Oncol. 2024, 17, 87. [Google Scholar] [CrossRef]
- Yang, L.; Rau, R.; Goodell, M.A. DNMT3A in haematological malignancies. Nat. Rev. Cancer 2015, 15, 152–165. [Google Scholar] [CrossRef]
- Challen, G.A.; Sun, D.; Jeong, M.; Luo, M.; Jelinek, J.; Berg, J.S.; Bock, C.; Vasanthakumar, A.; Gu, H.; Xi, Y.; et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat. Genet. 2011, 44, 23–31. [Google Scholar] [CrossRef]
- Jaiswal, S.; Fontanillas, P.; Flannick, J.; Manning, A.; Grauman, P.V.; Mar, B.G.; Lindsley, R.C.; Mermel, C.H.; Burtt, N.; Chavez, A.; et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014, 371, 2488–2498. [Google Scholar] [CrossRef]
- Lee, W.H.; Lin, C.C.; Tsai, C.H.; Tien, F.M.; Lo, M.Y.; Ni, S.C.; Yao, M.; Tseng, M.H.; Kuo, Y.Y.; Liu, M.C.; et al. Clinico-genetic and prognostic analyses of 716 patients with primary myelodysplastic syndrome and myelodysplastic syndrome/acute myeloid leukemia based on the 2022 International Consensus Classification. Am. J. Hematol. 2023, 98, 398–407. [Google Scholar] [CrossRef]
- Maggioni, G.; Bersanelli, M.; Travaglino, E.; Piérola, A.A.; Kasprzak, A.; Montserrat, A.S.; Sauta, E.; Sala, C.; Matteuzzi, T.; Meggendorfer, M.; et al. A sex-informed approach to improve the personalised decision making process in myelodysplastic syndromes: A multicentre, observational cohort study. Lancet Haematol. 2023, 10, e117–e128. [Google Scholar] [CrossRef]
- Jawad, M.; Afkhami, M.; Ding, Y.; Zhang, X.; Li, P.; Young, K.; Xu, M.L.; Cui, W.; Zhao, Y.; Halene, S.; et al. DNMT3A R882 Mutations Confer Unique Clinicopathologic Features in MDS Including a High Risk of AML Transformation. Front. Oncol. 2022, 12, 849376. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, Y.; Wang, C.; Wang, X. TET (Ten-eleven translocation) family proteins: Structure, biological functions and applications. Signal Transduct. Target. Ther. 2023, 8, 297. [Google Scholar] [CrossRef]
- Hawking, Z.L.; Allan, J.M. Landscape of TET2 Mutations: From Hematological Malignancies to Solid Tumors. Cancer Med. 2025, 14, e70792. [Google Scholar] [CrossRef]
- Smith, A.E.; Mohamedali, A.M.; Kulasekararaj, A.; Lim, Z.; Gaken, J.; Lea, N.C.; Przychodzen, B.; Mian, S.A.; Nasser, E.E.; Shooter, C.; et al. Next-generation sequencing of the TET2 gene in 355 MDS and CMML patients reveals low-abundance mutant clones with early origins, but indicates no definite prognostic value. Blood 2010, 116, 3923–3932. [Google Scholar] [CrossRef]
- Danishevich, A.; Chegodar, A.; Bodunova, N.; Konovalov, F.; Nefedova, M.; Kremneva, N.; Kurbanov, N.; Bilyalov, A.; Nikolaev, S.; Khatkov, I.; et al. Myelodysplastic Syndrome: Clinical Characteristics and Significance of Preclinically Detecting Biallelic Mutations in the TET2 Gene. Life 2024, 14, 637. [Google Scholar] [CrossRef]
- Awada, H.; Nagata, Y.; Goyal, A.; Asad, M.F.; Patel, B.; Hirsch, C.M.; Kuzmanovic, T.; Guan, Y.; Przychodzen, B.P.; Aly, M.; et al. Invariant phenotype and molecular association of biallelic TET2 mutant myeloid neoplasia. Blood Adv. 2019, 3, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Bezerra, M.F.; Larrazabal, B.R.; Lima, A.S.; Mello, M.R.; Pimentel, R.F.; Weinhauser, I.; Costa, F.F.; Fertrin, K.Y.; Araujo, A.S.; Machado, C.G.; et al. Screening for myeloid mutations in patients with myelodysplastic syndromes and AML with myelodysplasia-related changes. Hematol. Transfus. Cell Ther. 2022, 44, 328–331. [Google Scholar] [CrossRef]
- Komrokji, R.; Al Ali, N.; Chan, O.; Sweet, K.; Kuykendall, A.; Lancet, J.; Padron, E.; Sallman, D.A. IDH mutations are enriched in myelodysplastic syndrome patients with severe neutropenia and can be a potential for targeted therapy. Haematologica 2023, 108, 1168–1172. [Google Scholar] [CrossRef] [PubMed]
- Patnaik, M.M.; Hanson, C.A.; Hodnefield, J.M.; Lasho, T.L.; Finke, C.M.; Knudson, R.A.; Ketterling, R.P.; Pardanani, A.; Tefferi, A. Differential prognostic effect of IDH1 versus IDH2 mutations in myelodysplastic syndromes: A Mayo Clinic study of 277 patients. Leukemia 2012, 26, 101–105. [Google Scholar] [CrossRef]
- Wang, N.; Wang, F.; Shan, N.; Sui, X.; Xu, H. IDH1 Mutation Is an Independent Inferior Prognostic Indicator for Patients with Myelodysplastic Syndromes. Acta Haematol. 2017, 138, 143–151. [Google Scholar] [CrossRef]
- Jain, A.G.; Ball, S.; Aguirre, L.; Al Ali, N.; Kaldas, D.; Tinsley-Vance, S.; Kuykendall, A.; Chan, O.; Sweet, K.; Lancet, J.E.; et al. Patterns of lower risk myelodysplastic syndrome progression: Factors predicting progression to high-risk myelodysplastic syndrome and acute myeloid leukemia. Haematologica 2024, 109, 2157–2164. [Google Scholar] [CrossRef] [PubMed]
- Bellissimo, D.C.; Speck, N.A. RUNX1 Mutations in Inherited and Sporadic Leukemia. Front. Cell Dev. Biol. 2017, 5, 111. [Google Scholar] [CrossRef] [PubMed]
- Sutandyo, N.; Mulyasari, R.; Kosasih, A.; Rinaldi, I.; Louisa, M.; Kevinsyah, A.P.; Winston, K. Association of Somatic Gene Mutations with Risk of Transformation into Acute Myeloid Leukemia in Patients with Myelodysplastic Syndrome: A Systematic Review and Meta-Analysis. Asian Pac. J. Cancer Prev. 2022, 23, 1107–1116. [Google Scholar] [CrossRef]
- He, W.; Zhao, C.; Hu, H. Prognostic effect of RUNX1 mutations in myelodysplastic syndromes: A meta-analysis. Hematology 2020, 25, 494–501. [Google Scholar] [CrossRef]
- Wang, Y.H.; Yao, C.Y.; Lin, C.C.; Chen, C.L.; Hsu, C.L.; Tsai, C.H.; Hou, H.A.; Chou, W.C.; Tien, H.F. Higher RUNX1 expression levels are associated with worse overall and leukaemia-free survival in myelodysplastic syndrome patients. EJHaem 2022, 3, 1209–1219. [Google Scholar] [CrossRef]
- Watad, A.; Kacar, M.; Bragazzi, N.L.; Zhou, Q.; Jassam, M.; Taylor, J.; Roman, E.; Smith, A.; Jones, R.A.; Amital, H.; et al. Somatic Mutations and the Risk of Undifferentiated Autoinflammatory Disease in MDS: An Under-Recognized but Prognostically Important Complication. Front. Immunol. 2021, 12, 610019. [Google Scholar] [CrossRef]
- Okano, T.; Nishimura, A.; Inoue, K.; Naruto, T.; Tokoro, S.; Tomoda, T.; Kamiya, T.; Simbo, A.; Akutsu, Y.; Okamoto, K.; et al. Somatic mutation in RUNX1 underlies mucocutaneus inflammatory manifestations. Rheumatology 2021, 60, e429–e431. [Google Scholar] [CrossRef]
- Dermawan, J.K.; Wensel, C.; Visconte, V.; Maciejewski, J.P.; Cook, J.R.; Bosler, D.S. Clinically Significant CUX1 Mutations Are Frequently Subclonal and Common in Myeloid Disorders With a High Number of Co-mutated Genes and Dysplastic Features. Am. J. Clin. Pathol. 2022, 157, 586–594. [Google Scholar] [CrossRef] [PubMed]
- Hulea, L.; Nepveu, A. CUX1 transcription factors: From biochemical activities and cell-based assays to mouse models and human diseases. Gene 2012, 497, 18–26. [Google Scholar] [CrossRef]
- Aly, M.; Ramdzan, Z.M.; Nagata, Y.; Balasubramanian, S.K.; Hosono, N.; Makishima, H.; Visconte, V.; Kuzmanovic, T.; Adema, V.; Nazha, A.; et al. Distinct clinical and biological implications of CUX1 in myeloid neoplasms. Blood Adv. 2019, 3, 2164–2178. [Google Scholar] [CrossRef]
- Wang, Q.; Dong, S.; Yao, H.; Wen, L.; Qiu, H.; Qin, L.; Ma, L.; Chen, S. ETV6 mutation in a cohort of 970 patients with hematologic malignancies. Haematologica 2014, 99, e176–e178. [Google Scholar] [CrossRef] [PubMed]
- Gurney, M.; Chekkaf, I.; Baranwal, A.; Basmaci, R.; Katamesh, B.; Greipp, P.; Foran, J.M.; Badar, T.; Mangaonkar, A.A.; Begna, K.H.; et al. The clinical and molecular spectrum of ETV6 mutated myeloid neoplasms. Br. J. Haematol. 2023, 202, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Olivier, M.; Hollstein, M.; Hainaut, P. TP53 mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2010, 2, a001008. [Google Scholar] [CrossRef] [PubMed]
- Bernard, E.; Nannya, Y.; Hasserjian, R.P.; Devlin, S.M.; Tuechler, H.; Medina-Martinez, J.S.; Yoshizato, T.; Shiozawa, Y.; Saiki, R.; Malcovati, L.; et al. Implications of TP53 allelic state for genome stability, clinical presentation and outcomes in myelodysplastic syndromes. Nat. Med. 2020, 26, 1549–1556. [Google Scholar] [CrossRef]
- Siddon, A.J.; Weinberg, O.K. Diagnosis and Classification of Myelodysplastic Syndromes with Mutated TP53. Clin. Lab. Med. 2023, 43, 607–614. [Google Scholar] [CrossRef]
- Boada, M.; Arbelbide, J.; Basquiera, A.; Velloso, E.; Iastrebner, M.; Catalán, A.; Grille, S. TP53 Myelodysplastic Syndromes in Latin America. Real World Data from Latin American MDS Group (GLAM). Leuk. Res. 2023, 128S, 107158. [Google Scholar] [CrossRef]
- Hsu, J.I.; Dayaram, T.; Tovy, A.; De Braekeleer, E.; Jeong, M.; Wang, F.; Zhang, J.; Heffernan, T.P.; Gera, S.; Kovacs, J.J.; et al. PPM1D Mutations Drive Clonal Hematopoiesis in Response to Cytotoxic Chemotherapy. Cell Stem Cell 2018, 23, 700–713. [Google Scholar] [CrossRef]
- Fandrei, D.; Pegliasco, J.; Pasquier, F.; Ibrahim, N.; Kfoury, M.; Berthon, C.; Heiblig, M.; Lebon, D.; Marcais, A.; Meunier, M.; et al. Clonal Evolution of PPM1D Mutations in the Spectrum of Myeloid Disorders. Clin. Cancer Res. 2025, OF1–OF13. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Wu, L.Y.; Guo, J.; He, Q.; Zhang, Z.; Li, X. Somatic mutations of activating signalling, transcription factor, and tumour suppressor are a precondition for leukaemia transformation in myelodysplastic syndromes. J. Cell. Mol. Med. 2022, 26, 5901–5916. [Google Scholar] [CrossRef]
- Menssen, A.J.; Khanna, A.; Miller, C.A.; Nonavinkere Srivatsan, S.; Chang, G.S.; Shao, J.; Robinson, J.; O’Laughlin, M.; Fronick, C.C.; Fulton, R.S.; et al. Convergent Clonal Evolution of Signaling Gene Mutations Is a Hallmark of Myelodysplastic Syndrome Progression. Blood Cancer Discov. 2022, 3, 330–345. [Google Scholar] [CrossRef]
- Sumiyoshi, R.; Tashiro, H.; Shirasaki, R.; Matsuo, T.; Yamamoto, T.; Matsumoto, K.; Ooi, J.; Shirafuji, N. The FLT3 internal tandem duplication mutation at disease diagnosis is a negative prognostic factor in myelodysplastic syndrome patients. Leuk. Res. 2022, 113, 106790. [Google Scholar] [CrossRef] [PubMed]
- da Silva-Coelho, P.; Kroeze, L.I.; Yoshida, K.; Koorenhof-Scheele, T.N.; Knops, R.; van de Locht, L.T.; de Graaf, A.O.; Massop, M.; Sandmann, S.; Dugas, M.; et al. Clonal evolution in myelodysplastic syndromes. Nat. Commun. 2017, 8, 15099. [Google Scholar] [CrossRef]
- Ren, Y.; Lang, W.; Mei, C.; Luo, Y.; Ye, L.; Wang, L.; Zhou, X.; Xu, G.; Ma, L.; Jin, J.; et al. Co-mutation landscape and clinical significance of RAS pathway related gene mutations in patients with myelodysplastic syndrome. Hematol. Oncol. 2023, 41, 159–166. [Google Scholar] [CrossRef]
- Pinheiro, R.F.; Goes, J.V.C.; Sampaio, L.R.; Germano de Oliveira, R.T.; Lima, S.C.S.; Furtado, C.L.M.; de Paula Borges, D.; Costa, M.B.; da Silva Monte, C.; Minete, N.F.; et al. The Ataxia-telangiectasia mutated (ATM) is the most important gene for repairing the DNA in Myelodysplastic Neoplasm. DNA Repair 2025, 146, 103803. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.J.; Seto, E. HATs and HDACs: From structure, function and regulation to novel strategies for therapy and prevention. Oncogene 2007, 26, 5310–5318. [Google Scholar] [CrossRef]
- Dai, Y.; Faller, D.V. Transcription Regulation by Class III Histone Deacetylases (HDACs)-Sirtuins. Transl. Oncogenomics 2008, 3, 53–65. [Google Scholar] [CrossRef]
- Goes, J.V.C.; Viana, M.A.; Sampaio, L.R.; Cavalcante, C.B.A.; Melo, M.M.L.; de Oliveira, R.T.G.; Borges, D.P.; Goncalves, P.G.; Pinheiro, R.F.; Ribeiro-Junior, H.L. Gene expression patterns of Sirtuin family members (SIRT1 TO SIRT7): Insights into pathogenesis and prognostic of Myelodysplastic neoplasm. Gene 2024, 915, 148428. [Google Scholar] [CrossRef]
- Greenberg, P.L.; Tuechler, H.; Schanz, J.; Sanz, G.; Garcia-Manero, G.; Sole, F.; Bennett, J.M.; Bowen, D.; Fenaux, P.; Dreyfus, F.; et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood 2012, 120, 2454–2465. [Google Scholar] [CrossRef] [PubMed]
- Baliakas, P.; Tesi, B.; Cammenga, J.; Stray-Pedersen, A.; Jahnukainen, K.; Andersen, M.K.; Agerstam, H.; Creignou, M.; Dybedal, I.; Raaschou-Jensen, K.; et al. How to manage patients with germline DDX41 variants: Recommendations from the Nordic working group on germline predisposition for myeloid neoplasms. Hemasphere 2024, 8, e145. [Google Scholar] [CrossRef]
- Nazha, A.; Komrokji, R.; Meggendorfer, M.; Jia, X.; Radakovich, N.; Shreve, J.; Hilton, C.B.; Nagata, Y.; Hamilton, B.K.; Mukherjee, S.; et al. Personalized Prediction Model to Risk Stratify Patients with Myelodysplastic Syndromes. J. Clin. Oncol. 2021, 39, 3737–3746. [Google Scholar] [CrossRef]
- Mosquera Orgueira, A.; Perez Encinas, M.M.; Diaz Varela, N.A.; Mora, E.; Diaz-Beya, M.; Montoro, M.J.; Pomares, H.; Ramos, F.; Tormo, M.; Jerez, A.; et al. Machine Learning Improves Risk Stratification in Myelodysplastic Neoplasms: An Analysis of the Spanish Group of Myelodysplastic Syndromes. Hemasphere 2023, 7, e961. [Google Scholar] [CrossRef] [PubMed]
- Huber, S.; Haferlach, T.; Muller, H.; Meggendorfer, M.; Hutter, S.; Hoermann, G.; Baer, C.; Kern, W.; Haferlach, C. MDS subclassification-do we still have to count blasts? Leukemia 2023, 37, 942–945. [Google Scholar] [CrossRef] [PubMed]
- Kewan, T.; Durmaz, A.; Bahaj, W.; Gurnari, C.; Terkawi, L.; Awada, H.; Ogbue, O.D.; Ahmed, R.; Pagliuca, S.; Awada, H.; et al. Molecular patterns identify distinct subclasses of myeloid neoplasia. Nat. Commun. 2023, 14, 3136. [Google Scholar] [CrossRef]
- Bernard, E.; Hasserjian, R.P.; Greenberg, P.L.; Arango Ossa, J.E.; Creignou, M.; Tuechler, H.; Gutierrez-Abril, J.; Domenico, D.; Medina-Martinez, J.S.; Levine, M.; et al. Molecular taxonomy of myelodysplastic syndromes and its clinical implications. Blood 2024, 144, 1617–1632. [Google Scholar] [CrossRef] [PubMed]
- Loghavi, S.; Kanagal-Shamanna, R.; Khoury, J.D.; Medeiros, L.J.; Naresh, K.N.; Nejati, R.; Patnaik, M.M.; Project, W.H.O.t.E.C. Fifth Edition of the World Health Classification of Tumors of the Hematopoietic and Lymphoid Tissue: Myeloid Neoplasms. Mod. Pathol. 2024, 37, 100397. [Google Scholar] [CrossRef]
- Vaughan, L.; Pimanda, J.E. Seeing MDS through the lens of genomics. Blood 2024, 144, 1552–1554. [Google Scholar] [CrossRef]
- Mendonca, P.D.S.; Pinheiro, R.F.; Magalhaes, S.M.M. Myelodysplastic Syndrome Over Time: A Comparative Analysis of Overall Outcome. Mayo Clin. Proc. 2019, 94, 2593–2594. [Google Scholar] [CrossRef]
- Al-Kali, A.; Zblewski, D.; Foran, J.M.; Patnaik, M.S.; Larrabee, B.R.; Gangat, N.; Begna, K.H.; Elliott, M.A.; Hogan, W.J.; Tefferi, A.; et al. Outcome of Myelodysplastic Syndromes Over Time in the United States: A National Cancer Data Base Study from 2004–2013. Mayo Clin. Proc. 2019, 94, 1467–1474. [Google Scholar] [CrossRef]
- Burke, S.; Chowdhury, O.; Rouault-Pierre, K. Low-risk MDS-A spotlight on precision medicine for SF3B1-mutated patients. Hemasphere 2025, 9, e70103. [Google Scholar] [CrossRef]
- DiNardo, C.D.; Roboz, G.J.; Watts, J.M.; Madanat, Y.F.; Prince, G.T.; Baratam, P.; de Botton, S.; Stein, A.; Foran, J.M.; Arellano, M.L.; et al. Final phase 1 substudy results of ivosidenib for patients with mutant IDH1 relapsed/refractory myelodysplastic syndrome. Blood Adv. 2024, 8, 4209–4220. [Google Scholar] [CrossRef]
- DiNardo, C.D.; Venugopal, S.; Lachowiez, C.; Takahashi, K.; Loghavi, S.; Montalban-Bravo, G.; Wang, X.; Carraway, H.; Sekeres, M.; Sukkur, A.; et al. Targeted therapy with the mutant IDH2 inhibitor enasidenib for high-risk IDH2-mutant myelodysplastic syndrome. Blood Adv. 2023, 7, 2378–2387. [Google Scholar] [CrossRef] [PubMed]
- Othman, J.; Tiong, I.S.; O’Nions, J.; Dennis, M.; Mokretar, K.; Ivey, A.; Austin, M.; Latif, A.L.; Amer, M.; Chan, W.Y.; et al. Molecular MRD is strongly prognostic in patients with NPM1-mutated AML receiving venetoclax-based nonintensive therapy. Blood 2024, 143, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Senapati, J.; Urrutia, S.; Loghavi, S.; Short, N.J.; Issa, G.C.; Maiti, A.; Abbas, H.A.; Daver, N.G.; Pemmaraju, N.; Pierce, S.; et al. Venetoclax abrogates the prognostic impact of splicing factor gene mutations in newly diagnosed acute myeloid leukemia. Blood 2023, 142, 1647–1657. [Google Scholar] [CrossRef] [PubMed]
- Lachowiez, C.A.; Reville, P.K.; Kantarjian, H.; Jabbour, E.; Borthakur, G.; Daver, N.; Issa, G.; Furudate, K.; Tanaka, T.; Pierce, S.; et al. Contemporary outcomes in IDH-mutated acute myeloid leukemia: The impact of co-occurring NPM1 mutations and venetoclax-based treatment. Am. J. Hematol. 2022, 97, 1443–1452. [Google Scholar] [CrossRef]
- Nanaa, A.; He, R.; Foran, J.M.; Badar, T.; Gangat, N.; Pardanani, A.; Hogan, W.J.; Litzow, M.R.; Patnaik, M.; Al-Kali, A.; et al. Venetoclax plus hypomethylating agents in DDX41-mutated acute myeloid leukaemia and myelodysplastic syndrome: Mayo Clinic series on 12 patients. Br. J. Haematol. 2024, 204, 171–176. [Google Scholar] [CrossRef]
- Itzykson, R.; Kosmider, O.; Cluzeau, T.; Mansat-De Mas, V.; Dreyfus, F.; Beyne-Rauzy, O.; Quesnel, B.; Vey, N.; Gelsi-Boyer, V.; Raynaud, S.; et al. Impact of TET2 mutations on response rate to azacitidine in myelodysplastic syndromes and low blast count acute myeloid leukemias. Leukemia 2011, 25, 1147–1152. [Google Scholar] [CrossRef] [PubMed]
- Traina, F.; Visconte, V.; Elson, P.; Tabarroki, A.; Jankowska, A.M.; Hasrouni, E.; Sugimoto, Y.; Szpurka, H.; Makishima, H.; O’Keefe, C.L.; et al. Impact of molecular mutations on treatment response to DNMT inhibitors in myelodysplasia and related neoplasms. Leukemia 2014, 28, 78–87. [Google Scholar] [CrossRef]
- Hu, C.; Wang, X. Predictive and prognostic value of gene mutations in myelodysplastic syndrome treated with hypomethylating agents: A meta-analysis. Leuk. Lymphoma 2022, 63, 2336–2351. [Google Scholar] [CrossRef]
- Tentori, C.A.; Gregorio, C.; Robin, M.; Gagelmann, N.; Gurnari, C.; Ball, S.; Caballero Berrocal, J.C.; Lanino, L.; D’Amico, S.; Spreafico, M.; et al. Clinical and Genomic-Based Decision Support System to Define the Optimal Timing of Allogeneic Hematopoietic Stem-Cell Transplantation in Patients With Myelodysplastic Syndromes. J. Clin. Oncol. 2024, 42, 2873–2886. [Google Scholar] [CrossRef]
- Gurnari, C.; Robin, M.; Ades, L.; Aljurf, M.; Almeida, A.; Duarte, F.B.; Bernard, E.; Cutler, C.; Della Porta, M.G.; De Witte, T.; et al. Clinical-genomic profiling of MDS to inform allo-HCT: Recommendations from an international panel on behalf of the EBMT. Blood 2025, 145, 1987–2001. [Google Scholar] [CrossRef]
- Zhang, J.; Walsh, M.F.; Wu, G.; Edmonson, M.N.; Gruber, T.A.; Easton, J.; Hedges, D.; Ma, X.; Zhou, X.; Yergeau, D.A.; et al. Germline Mutations in Predisposition Genes in Pediatric Cancer. N. Engl. J. Med. 2015, 373, 2336–2346. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, J.R.; Ma, J.; Lamprecht, T.; Walsh, M.; Wang, S.; Bryant, V.; Song, G.; Wu, G.; Easton, J.; Kesserwan, C.; et al. The genomic landscape of pediatric myelodysplastic syndromes. Nat. Commun. 2017, 8, 1557. [Google Scholar] [CrossRef]
- Feurstein, S.; Churpek, J.E.; Walsh, T.; Keel, S.; Hakkarainen, M.; Schroeder, T.; Germing, U.; Geyh, S.; Heuser, M.; Thol, F.; et al. Germline variants drive myelodysplastic syndrome in young adults. Leukemia 2021, 35, 2439–2444. [Google Scholar] [CrossRef]
- Sebert, M.; Passet, M.; Raimbault, A.; Rahme, R.; Raffoux, E.; Sicre de Fontbrune, F.; Cerrano, M.; Quentin, S.; Vasquez, N.; Da Costa, M.; et al. Germline DDX41 mutations define a significant entity within adult MDS/AML patients. Blood 2019, 134, 1441–1444. [Google Scholar] [CrossRef] [PubMed]
- Churpek, J.E.; Pyrtel, K.; Kanchi, K.L.; Shao, J.; Koboldt, D.; Miller, C.A.; Shen, D.; Fulton, R.; O’Laughlin, M.; Fronick, C.; et al. Genomic analysis of germ line and somatic variants in familial myelodysplasia/acute myeloid leukemia. Blood 2015, 126, 2484–2490. [Google Scholar] [CrossRef] [PubMed]
- DiNardo, C.D.; Bannon, S.A.; Routbort, M.; Franklin, A.; Mork, M.; Armanios, M.; Mace, E.M.; Orange, J.S.; Jeff-Eke, M.; Churpek, J.E.; et al. Evaluation of Patients and Families With Concern for Predispositions to Hematologic Malignancies Within the Hereditary Hematologic Malignancy Clinic (HHMC). Clin. Lymphoma Myeloma Leuk. 2016, 16, 417–428. [Google Scholar] [CrossRef]
- Feurstein, S.; Trottier, A.M.; Estrada-Merly, N.; Pozsgai, M.; McNeely, K.; Drazer, M.W.; Ruhle, B.; Sadera, K.; Koppayi, A.L.; Scott, B.L.; et al. Germ line predisposition variants occur in myelodysplastic syndrome patients of all ages. Blood 2022, 140, 2533–2548. [Google Scholar] [CrossRef]
- Trottier, A.M.; Feurstein, S.; Godley, L.A. Germline predisposition to myeloid neoplasms: Characteristics and management of high versus variable penetrance disorders. Best. Pract. Res. Clin. Haematol. 2024, 37, 101537. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef]
- Owen, C.; Barnett, M.; Fitzgibbon, J. Familial myelodysplasia and acute myeloid leukaemia—A review. Br. J. Haematol. 2008, 140, 123–132. [Google Scholar] [CrossRef]
- Smith, M.L.; Cavenagh, J.D.; Lister, T.A.; Fitzgibbon, J. Mutation of CEBPA in familial acute myeloid leukemia. N. Engl. J. Med. 2004, 351, 2403–2407. [Google Scholar] [CrossRef] [PubMed]
- Tawana, K.; Wang, J.; Renneville, A.; Bodor, C.; Hills, R.; Loveday, C.; Savic, A.; Van Delft, F.W.; Treleaven, J.; Georgiades, P.; et al. Disease evolution and outcomes in familial AML with germline CEBPA mutations. Blood 2015, 126, 1214–1223. [Google Scholar] [CrossRef]
- Taskesen, E.; Bullinger, L.; Corbacioglu, A.; Sanders, M.A.; Erpelinck, C.A.; Wouters, B.J.; van der Poel-van de Luytgaarde, S.C.; Damm, F.; Krauter, J.; Ganser, A.; et al. Prognostic impact, concurrent genetic mutations, and gene expression features of AML with CEBPA mutations in a cohort of 1182 cytogenetically normal AML patients: Further evidence for CEBPA double mutant AML as a distinctive disease entity. Blood 2011, 117, 2469–2475. [Google Scholar] [CrossRef]
- Polprasert, C.; Schulze, I.; Sekeres, M.A.; Makishima, H.; Przychodzen, B.; Hosono, N.; Singh, J.; Padgett, R.A.; Gu, X.; Phillips, J.G.; et al. Inherited and Somatic Defects in DDX41 in Myeloid Neoplasms. Cancer Cell 2015, 27, 658–670. [Google Scholar] [CrossRef] [PubMed]
- Quesada, A.E.; Routbort, M.J.; DiNardo, C.D.; Bueso-Ramos, C.E.; Kanagal-Shamanna, R.; Khoury, J.D.; Thakral, B.; Zuo, Z.; Yin, C.C.; Loghavi, S.; et al. DDX41 mutations in myeloid neoplasms are associated with male gender, TP53 mutations and high-risk disease. Am. J. Hematol. 2019, 94, 757–766. [Google Scholar] [CrossRef] [PubMed]
- Makishima, H.; Saiki, R.; Nannya, Y.; Korotev, S.; Gurnari, C.; Takeda, J.; Momozawa, Y.; Best, S.; Krishnamurthy, P.; Yoshizato, T.; et al. Germ line DDX41 mutations define a unique subtype of myeloid neoplasms. Blood 2023, 141, 534–549. [Google Scholar] [CrossRef]
- Auger, N.; Douet-Guilbert, N.; Quessada, J.; Theisen, O.; Lafage-Pochitaloff, M.; Troadec, M.B. Cytogenetics in the management of myelodysplastic neoplasms (myelodysplastic syndromes, MDS): Guidelines from the groupe francophone de cytogenetique hematologique (GFCH). Curr. Res. Transl. Med. 2023, 71, 103409. [Google Scholar] [CrossRef]
- Homan, C.C.; Drazer, M.W.; Yu, K.; Lawrence, D.M.; Feng, J.; Arriola-Martinez, L.; Pozsgai, M.J.; McNeely, K.E.; Ha, T.; Venugopal, P.; et al. Somatic mutational landscape of hereditary hematopoietic malignancies caused by germline variants in RUNX1, GATA2, and DDX41. Blood Adv. 2023, 7, 6092–6107. [Google Scholar] [CrossRef]
- Saygin, C.; Roloff, G.; Hahn, C.N.; Chhetri, R.; Gill, S.; Elmariah, H.; Talati, C.; Nunley, E.; Gao, G.; Kim, A.; et al. Allogeneic hematopoietic stem cell transplant outcomes in adults with inherited myeloid malignancies. Blood Adv. 2023, 7, 549–554. [Google Scholar] [CrossRef]
- Owen, C.J.; Toze, C.L.; Koochin, A.; Forrest, D.L.; Smith, C.A.; Stevens, J.M.; Jackson, S.C.; Poon, M.C.; Sinclair, G.D.; Leber, B.; et al. Five new pedigrees with inherited RUNX1 mutations causing familial platelet disorder with propensity to myeloid malignancy. Blood 2008, 112, 4639–4645. [Google Scholar] [CrossRef]
- Song, W.J.; Sullivan, M.G.; Legare, R.D.; Hutchings, S.; Tan, X.; Kufrin, D.; Ratajczak, J.; Resende, I.C.; Haworth, C.; Hock, R.; et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat. Genet. 1999, 23, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Arepally, G.; Rebbeck, T.R.; Song, W.; Gilliland, G.; Maris, J.M.; Poncz, M. Evidence for genetic homogeneity in a familial platelet disorder with predisposition to acute myelogenous leukemia (FPD/AML). Blood 1998, 92, 2600–2602. [Google Scholar] [CrossRef] [PubMed]
- Beri-Dexheimer, M.; Latger-Cannard, V.; Philippe, C.; Bonnet, C.; Chambon, P.; Roth, V.; Gregoire, M.J.; Bordigoni, P.; Lecompte, T.; Leheup, B.; et al. Clinical phenotype of germline RUNX1 haploinsufficiency: From point mutations to large genomic deletions. Eur. J. Hum. Genet. 2008, 16, 1014–1018. [Google Scholar] [CrossRef] [PubMed]
- Churpek, J.E.; Garcia, J.S.; Madzo, J.; Jackson, S.A.; Onel, K.; Godley, L.A. Identification and molecular characterization of a novel 3′ mutation in RUNX1 in a family with familial platelet disorder. Leuk. Lymphoma 2010, 51, 1931–1935. [Google Scholar] [CrossRef]
- Michaud, J.; Wu, F.; Osato, M.; Cottles, G.M.; Yanagida, M.; Asou, N.; Shigesada, K.; Ito, Y.; Benson, K.F.; Raskind, W.H.; et al. In vitro analyses of known and novel RUNX1/AML1 mutations in dominant familial platelet disorder with predisposition to acute myelogenous leukemia: Implications for mechanisms of pathogenesis. Blood 2002, 99, 1364–1372. [Google Scholar] [CrossRef]
- Noris, P.; Pecci, A. Hereditary thrombocytopenias: A growing list of disorders. Hematol. Am. Soc. Hematol. Educ. Program. 2017, 2017, 385–399. [Google Scholar] [CrossRef]
- Noris, P.; Perrotta, S.; Seri, M.; Pecci, A.; Gnan, C.; Loffredo, G.; Pujol-Moix, N.; Zecca, M.; Scognamiglio, F.; De Rocco, D.; et al. Mutations in ANKRD26 are responsible for a frequent form of inherited thrombocytopenia: Analysis of 78 patients from 21 families. Blood 2011, 117, 6673–6680. [Google Scholar] [CrossRef]
- Noris, P.; Favier, R.; Alessi, M.C.; Geddis, A.E.; Kunishima, S.; Heller, P.G.; Giordano, P.; Niederhoffer, K.Y.; Bussel, J.B.; Podda, G.M.; et al. ANKRD26-related thrombocytopenia and myeloid malignancies. Blood 2013, 122, 1987–1989. [Google Scholar] [CrossRef]
- Marconi, C.; Canobbio, I.; Bozzi, V.; Pippucci, T.; Simonetti, G.; Melazzini, F.; Angori, S.; Martinelli, G.; Saglio, G.; Torti, M.; et al. 5′UTR point substitutions and N-terminal truncating mutations of ANKRD26 in acute myeloid leukemia. J. Hematol. Oncol. 2017, 10, 18. [Google Scholar] [CrossRef]
- Noetzli, L.; Lo, R.W.; Lee-Sherick, A.B.; Callaghan, M.; Noris, P.; Savoia, A.; Rajpurkar, M.; Jones, K.; Gowan, K.; Balduini, C.; et al. Germline mutations in ETV6 are associated with thrombocytopenia, red cell macrocytosis and predisposition to lymphoblastic leukemia. Nat. Genet. 2015, 47, 535–538. [Google Scholar] [CrossRef]
- Rampersaud, E.; Ziegler, D.S.; Iacobucci, I.; Payne-Turner, D.; Churchman, M.L.; Schrader, K.A.; Joseph, V.; Offit, K.; Tucker, K.; Sutton, R.; et al. Germline deletion of ETV6 in familial acute lymphoblastic leukemia. Blood Adv. 2019, 3, 1039–1046. [Google Scholar] [CrossRef]
- Zhang, M.Y.; Churpek, J.E.; Keel, S.B.; Walsh, T.; Lee, M.K.; Loeb, K.R.; Gulsuner, S.; Pritchard, C.C.; Sanchez-Bonilla, M.; Delrow, J.J.; et al. Germline ETV6 mutations in familial thrombocytopenia and hematologic malignancy. Nat. Genet. 2015, 47, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Wlodarski, M.W.; Hirabayashi, S.; Pastor, V.; Stary, J.; Hasle, H.; Masetti, R.; Dworzak, M.; Schmugge, M.; van den Heuvel-Eibrink, M.; Ussowicz, M.; et al. Prevalence, clinical characteristics, and prognosis of GATA2-related myelodysplastic syndromes in children and adolescents. Blood 2016, 127, 1387–1397, quiz 1518. [Google Scholar] [CrossRef]
- Hahn, C.N.; Chong, C.E.; Carmichael, C.L.; Wilkins, E.J.; Brautigan, P.J.; Li, X.C.; Babic, M.; Lin, M.; Carmagnac, A.; Lee, Y.K.; et al. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat. Genet. 2011, 43, 1012–1017. [Google Scholar] [CrossRef] [PubMed]
- Donadieu, J.; Lamant, M.; Fieschi, C.; de Fontbrune, F.S.; Caye, A.; Ouachee, M.; Beaupain, B.; Bustamante, J.; Poirel, H.A.; Isidor, B.; et al. Natural history of GATA2 deficiency in a survey of 79 French and Belgian patients. Haematologica 2018, 103, 1278–1287. [Google Scholar] [CrossRef]
- Spinner, M.A.; Sanchez, L.A.; Hsu, A.P.; Shaw, P.A.; Zerbe, C.S.; Calvo, K.R.; Arthur, D.C.; Gu, W.; Gould, C.M.; Brewer, C.C.; et al. GATA2 deficiency: A protean disorder of hematopoiesis, lymphatics, and immunity. Blood 2014, 123, 809–821. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, S.S.; Kozyra, E.J.; Wlodarski, M.W. Germline predisposition in myeloid neoplasms: Unique genetic and clinical features of GATA2 deficiency and SAMD9/SAMD9L syndromes. Best. Pract. Res. Clin. Haematol. 2020, 33, 101197. [Google Scholar] [CrossRef]
- Bluteau, O.; Sebert, M.; Leblanc, T.; Peffault de Latour, R.; Quentin, S.; Lainey, E.; Hernandez, L.; Dalle, J.H.; Sicre de Fontbrune, F.; Lengline, E.; et al. A landscape of germ line mutations in a cohort of inherited bone marrow failure patients. Blood 2018, 131, 717–732. [Google Scholar] [CrossRef]
- Narumi, S.; Amano, N.; Ishii, T.; Katsumata, N.; Muroya, K.; Adachi, M.; Toyoshima, K.; Tanaka, Y.; Fukuzawa, R.; Miyako, K.; et al. SAMD9 mutations cause a novel multisystem disorder, MIRAGE syndrome, and are associated with loss of chromosome 7. Nat. Genet. 2016, 48, 792–797. [Google Scholar] [CrossRef]
- Gonzalez, K.D.; Noltner, K.A.; Buzin, C.H.; Gu, D.; Wen-Fong, C.Y.; Nguyen, V.Q.; Han, J.H.; Lowstuter, K.; Longmate, J.; Sommer, S.S.; et al. Beyond Li Fraumeni Syndrome: Clinical characteristics of families with p53 germline mutations. J. Clin. Oncol. 2009, 27, 1250–1256. [Google Scholar] [CrossRef]
- Luo, M.; Wong, D.; Zelley, K.; Wu, J.; Schubert, J.; Denenberg, E.H.; Fanning, E.A.; Chen, J.; Gallo, D.; Golenberg, N.; et al. Identification of TP53 germline variants in pediatric patients undergoing tumor testing: Strategy and prevalence. J. Natl. Cancer Inst. 2024, 116, 1356–1365. [Google Scholar] [CrossRef] [PubMed]
- Palmero, E.I.; Schuler-Faccini, L.; Caleffi, M.; Achatz, M.I.; Olivier, M.; Martel-Planche, G.; Marcel, V.; Aguiar, E.; Giacomazzi, J.; Ewald, I.P.; et al. Detection of R337H, a germline TP53 mutation predisposing to multiple cancers, in asymptomatic women participating in a breast cancer screening program in Southern Brazil. Cancer Lett. 2008, 261, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Garritano, S.; Gemignani, F.; Palmero, E.I.; Olivier, M.; Martel-Planche, G.; Le Calvez-Kelm, F.; Brugieres, L.; Vargas, F.R.; Brentani, R.R.; Ashton-Prolla, P.; et al. Detailed haplotype analysis at the TP53 locus in p.R337H mutation carriers in the population of Southern Brazil: Evidence for a founder effect. Hum. Mutat. 2010, 31, 143–150. [Google Scholar] [CrossRef]
- Galante, P.A.F.; Guardia, G.D.A.; Pisani, J.; Sandoval, R.L.; Barros-Filho, M.C.; Gifoni, A.; Patrao, D.F.C.; Ashton-Prolla, P.; de Vasconcellos, V.F.; Freycon, C.; et al. Personalized screening strategies for TP53 R337H carriers: A retrospective cohort study of tumor spectrum in Li-Fraumeni syndrome adult carriers. Lancet Reg. Health. Am. 2025, 42, 100982. [Google Scholar] [CrossRef]
- Custodio, G.; Parise, G.A.; Kiesel Filho, N.; Komechen, H.; Sabbaga, C.C.; Rosati, R.; Grisa, L.; Parise, I.Z.; Pianovski, M.A.; Fiori, C.M.; et al. Impact of neonatal screening and surveillance for the TP53 R337H mutation on early detection of childhood adrenocortical tumors. J. Clin. Oncol. 2013, 31, 2619–2626. [Google Scholar] [CrossRef] [PubMed]
- Vieira, I.A.; Pezzi, E.H.; Bandeira, I.C.; Reis, L.B.; de Araujo Rocha, Y.M.; Fernandes, B.V.; Siebert, M.; Miyamoto, K.N.; Siqueira, M.B.; Achatz, M.I.; et al. Functional pri-miR-34b/c rs4938723 and KRAS 3′UTR rs61764370 SNPs: Novel phenotype modifiers in Li-Fraumeni Syndrome? Gene 2024, 898, 148069. [Google Scholar] [CrossRef]
- Baliakas, P.; Tesi, B.; Wartiovaara-Kautto, U.; Stray-Pedersen, A.; Friis, L.S.; Dybedal, I.; Hovland, R.; Jahnukainen, K.; Raaschou-Jensen, K.; Ljungman, P.; et al. Nordic Guidelines for Germline Predisposition to Myeloid Neoplasms in Adults: Recommendations for Genetic Diagnosis, Clinical Management and Follow-up. Hemasphere 2019, 3, e321. [Google Scholar] [CrossRef]
- Hemoterapia, S.E.d.H.y. Manual para el Diagnóstico y Atención Clínica en la Predisposición Germinal a Neoplasias Hematológicas. 2024. Available online: https://sehh.es/publicaciones/manuales-publicaciones/126044-manual-para-el-diagnostico-y-atencion-clinica-en-la-predisposicion-germinal-a-neoplasias-hematologicas (accessed on 7 April 2025).
- Dokal, I.; Tummala, H.; Vulliamy, T. Inherited bone marrow failure in the pediatric patient. Blood 2022, 140, 556–570. [Google Scholar] [CrossRef]
- Roloff, G.W.; Godley, L.A.; Drazer, M.W. Assessment of technical heterogeneity among diagnostic tests to detect germline risk variants for hematopoietic malignancies. Genet. Med. 2021, 23, 211–214. [Google Scholar] [CrossRef]
- DeRoin, L.; Cavalcante de Andrade Silva, M.; Petras, K.; Arndt, K.; Phillips, N.; Wanjari, P.; Subramanian, H.P.; Montes, D.; McElherne, J.; Theissen, M.; et al. Feasibility and limitations of cultured skin fibroblasts for germline genetic testing in hematologic disorders. Hum. Mutat. 2022, 43, 950–962. [Google Scholar] [CrossRef]
- Drazer, M.W.; Homan, C.C.; Yu, K.; Cavalcante de Andrade Silva, M.; McNeely, K.E.; Pozsgai, M.J.; Acevedo-Mendez, M.G.; Segal, J.P.; Wang, P.; Feng, J.; et al. Clonal hematopoiesis in patients with ANKRD26 or ETV6 germline mutations. Blood Adv. 2022, 6, 4357–4359. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Rodrigues, F.; Munger, E.; Ma, X.; Groarke, E.M.; Tang, Y.; Patel, B.A.; Catto, L.F.B.; Cle, D.V.; Niewisch, M.R.; Alves-Paiva, R.M.; et al. Differential diagnosis of bone marrow failure syndromes guided by machine learning. Blood 2023, 141, 2100–2113. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, L.J.; Barozzi, S.; Isidori, F.; Ganiewich, D.; De Luca, G.; Bozzi, V.; Marta, R.F.; Melazzini, F.; Pippucci, T.; Heller, P.G.; et al. Two novel families with RUNX1 variants indicate glycine 168 as a new mutational hotspot: Implications for FPD/AML diagnosis. Br. J. Haematol. 2024, 205, 2315–2320. [Google Scholar] [CrossRef]
- Cunningham, L.; Merguerian, M.; Calvo, K.R.; Davis, J.; Deuitch, N.T.; Dulau-Florea, A.; Patel, N.; Yu, K.; Sacco, K.; Bhattacharya, S.; et al. Natural history study of patients with familial platelet disorder with associated myeloid malignancy. Blood 2023, 142, 2146–2158. [Google Scholar] [CrossRef]
- Bove, V.; Spangenberg, M.N.; Ottati, C.; Vazquez, L.; Catalan, A.I.; Grille, S. Myelodysplastic syndrome with dual germline RUNX1 and DDX41 variants: A rare genetic predisposition case. Fam. Cancer 2025, 24, 20. [Google Scholar] [CrossRef]
- Spangenberg, M.N.; Boada, M.; Ottati, C.; Vazquez, L.; Catalan, A.; Grille, S. Unusual Co-Occurrence of Multiple Myeloma and AML in a Patient With Germline CEBPA Variant. Expanding the Spectrum of Hereditary Hematologic Malignancies. Clin. Genet. 2025, 107, 576–578. [Google Scholar] [CrossRef]
- Boada, M.; Catalan, A.I.; Ottati, C.; Bentancour, F.; Lens, D.; Guillermo, C.; Grille, S. Germline CEBPA mutation in familial acute myeloid leukemia. Hematol. Rep. 2021, 13, 9114. [Google Scholar] [CrossRef]
- Belli, C.; Bender, A.; Jauk, F.; Alonso, C.; Asinari, M.; Rahhal, M.; De Paula Sabino, A.; Duarte, F.; Calado, R.; Campregher, P.; et al. P008-Topic: AS01-Diagnosis/AS01c-Molecular aberrations (cytogenetic, genetic, gene expression): Real-world laboratory practice patterns on next-generation-sequencing (NGS) technology in Latin America. Leuk. Res. 2023, 128, 107143. [Google Scholar] [CrossRef]
- Crisp, R.; Bestach, Y.; Kornblihtt, L.; Lazzarino, C.; Iastrebner, M.; Gonzalez, J.; Arbelbide, J.; Belli, C.B.; Grupo de Estudio de Sindromes Mielodisplasicos, S.A.d.H. Preferences and limitations of hematologists to address the complexity of myelodysplastic syndromes. Medicina 2019, 79, 174–184. [Google Scholar]
- Grille Montauban, S.; Hernandez-Perez, C.R.; Velloso, E.; Novoa, V.; Lorand-Metze, I.; Gonzalez, J.; Solari, L.; Cismondi, V.; Serrano, J.C.; Burgnini, A.; et al. Flow cytometry “Ogata score” for the diagnosis of myelodysplastic syndromes in a real-life setting. A Latin American experience. Int. J. Lab. Hematol. 2019, 41, 536–541. [Google Scholar] [CrossRef]
- Grille, S.; Velloso, E.D.; Boada, M.; Apodaca Chavez, E.; Gomez-De Leon, A.; Rodríguez-Zuñiga, A.C.; Martínez Moreno, E.; Perez-Jacobo, F.; Alfonso, G.; Serrano, J.C.; et al. Myelodysplastic Syndromes in Latin-America—Results from a Novel International Registry: Re-Glam. Blood 2022, 140 (Suppl. S1), 4096–4097. [Google Scholar] [CrossRef]
- Cazzola, M.; Malcovati, L. Genome sequencing in the management of myelodysplastic syndromes and related disorders. Haematologica 2025, 110, 312–329. [Google Scholar] [CrossRef] [PubMed]
| Author/Year | N/Diagnosis | Region | Methods | Median Follow-Up | Main Findings |
|---|---|---|---|---|---|
| Kewan et al./2023 [115] | 3588/MDS and sAML | USA and Europe | NGS (40 genes) + ML | NI | MDS classification, prognostication, and prediction of treatment response, based solely on genetic factors (based on karyotype and mutation status). Fourteen molecularly distinct clusters. Correlation with OS (independent of IPSS-M) and response to treatment. |
| Huber et al./2023 [114] | 735/De novo MDS | Europe (Germany) | WGS | 9.3 years | MDS classification and prognostication based solely on genetic. Nine genetically defined, mutually exclusive hierarchical subgroups (based on karyotype and mutation status). Correlation with OS and IPSS-M risk groups. |
| Mosquera et al./2023 [113] | 7202/MDS and sAML | Spain | ML | 4.9 years | Enhanced MDS prognostication based on non-molecular variables (traditional clinical and laboratory). The model, AIPSS-MDS, performed better than IPSS-R and similar to IPSS-M. Correlation with OS and LFS. |
| Bernard et al./2024 [116] | 3233/MDS or related disorders | USA, Europe, and Japan | NGS (152-gene panel) | NI | Classification and prognostication based on gene mutations, copy-number alterations, and copy neutral loss of heterozygocity. Sixteen molecular groups and two residual groups with negative findings, with different OS. Prognosis of t-MDS and myelodysplastic/myeloproliferative neoplasms depended on genetic subtypes. |
| Lincango et al./2024 [41] | 182/MDS or related disorders | Latin America (AR and UY) | NGS (various gene panels) | 1.9 years | AIPSS-MDS validation for prognosis, showing similar prognostic discrimination to IPSS-M. AIPSS-MDS useful in resource-limited centers without molecular testing. |
| Group | Entity | % | Reported Features |
|---|---|---|---|
| Validated five established subgroups | DDX41 | 3.3 | 56% of patients with mutated DDX41 had both a putative germline DDX41 variant (defined here as >30% VAF) and a somatic DDX41 mutation, 37% had only a putative germline DDX41 variant, and 7% had only somatic DDX41 mutations. |
| AML-like | 2.0 | NPM1 mutations or at least 2 events from WT1, FLT3, MLLPTD, or MYC mutations. | |
| TP53 complex | 10.0 | Multi-hit TP53 mutations were present in 74% of cases, of which 91% had a complex karyotype. | |
| del(5q) | 6.9 | Presence of del(5q) as the sole cytogenetic abnormality or with 1 additional abnormality excluding −7/7q. Monoallelic TP53 mutations were significantly enriched in this group. | |
| SF3B1 | 14.0 | Indolent clinical course. | |
| Confirmed three previously reported subgroups | Bi-allelic TET2 | 13.0 | Early biallelic TET2 mutations with splicing factor mutations in 80% of patients, most commonly affecting SRSF2, SF3B1, or ZRSR2. Modulation of phenotype by ASXL1 and RAS mutations driving monocytosis and JAK2 driving thrombocytosis. |
| der(1;7) | 0.5 | ETNK1 mutations were enriched in this group. | |
| CCUS-like | 6.9 | 46% had a single mutated gene (TET2 or DNMT3A), 8% had loss of Y without gene mutations, and 6% had only ≥2 DTA mutations. | |
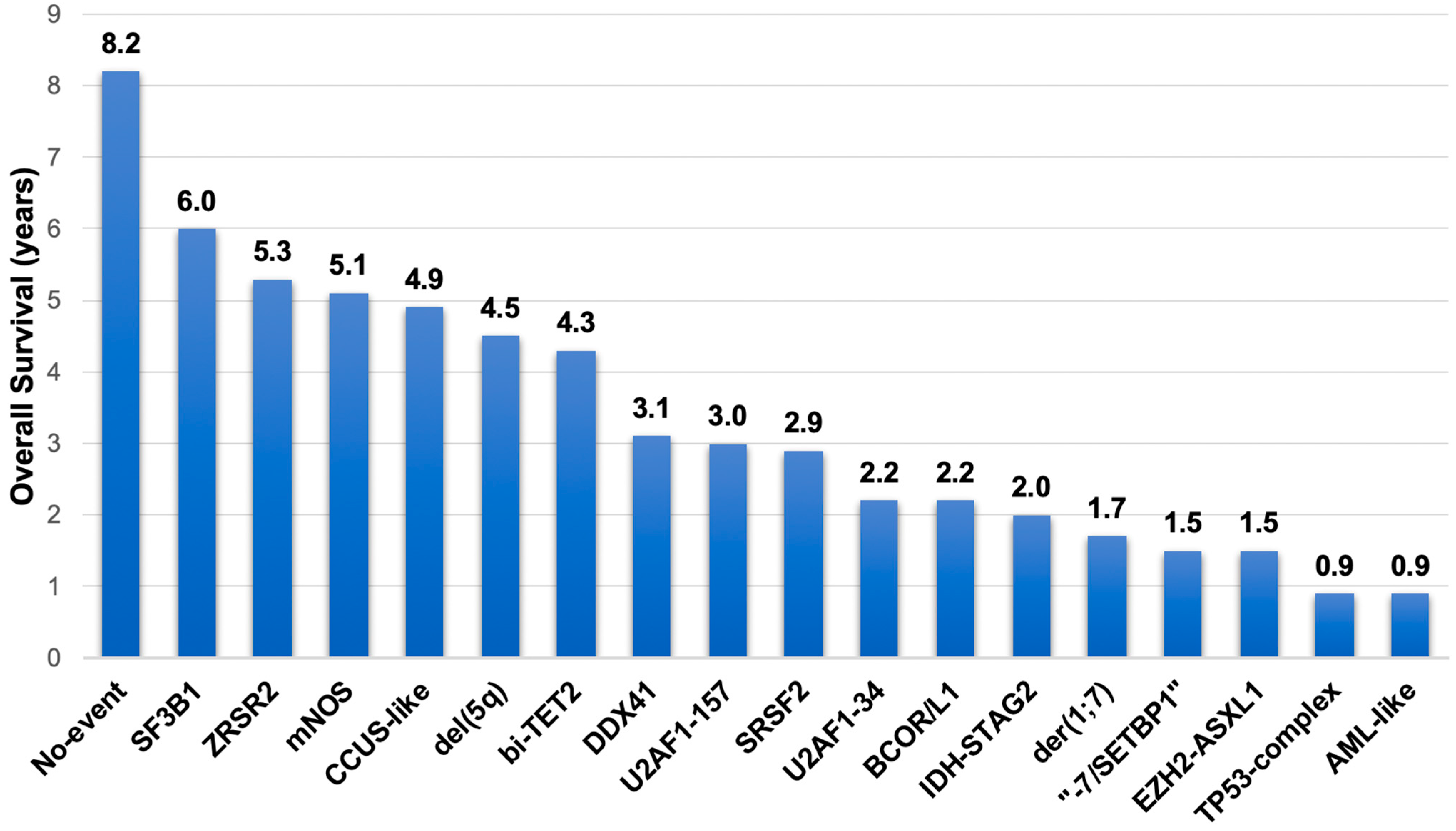
| Eight novel subgroups | “−7/SETBP1” | 4.9 | SETBP1 mutations and/or −7 in the absence of complex karyotype. GATA2 variants were prevalent. |
| EZH2-ASXL1 | 4.0 | ASXL1 and EZH2 mutation co-occurrence. High molecular complexity (75% of patients with ≥5 mutated genes). | |
| IDH-STAG2 | 8.9 | Mutations at the IDH2 R140 hot spot, IDH1, and/or STAG2 co-occurring with either SRSF2 or ASXL1 mutations | |
| BCOR/L1 | 3.5 | 83% of patients had mutations in BCOR, 33% in BCORL1, and 17% in both genes. | |
| U2AF1 | 4.3 | 58% had a Q157 mutation, 41% had a S34 mutation, and 1% had both. | |
| SRSF2 | 2.2 | Aggressive disease. | |
| ZRSR2 | 1.3 | Indolent clinical course. | |
| Two subgroups without defining genetic events | Not otherwise specified | 7.9 | Presence of other cytogenetic abnormalities and/or mutations in 51 other recurrently mutated genes. |
| No event | 6.5 | Absence of any recurrent drivers evaluated. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Basquiera, A.L.; Andreoli, V.; Grille, S.; Belli, C.B. Progress in the Genetics of Myelodysplastic Syndromes with a Latin American Perspective. Genes 2025, 16, 687. https://doi.org/10.3390/genes16060687
Basquiera AL, Andreoli V, Grille S, Belli CB. Progress in the Genetics of Myelodysplastic Syndromes with a Latin American Perspective. Genes. 2025; 16(6):687. https://doi.org/10.3390/genes16060687
Chicago/Turabian StyleBasquiera, Ana Lisa, Verónica Andreoli, Sofía Grille, and Carolina Bárbara Belli. 2025. "Progress in the Genetics of Myelodysplastic Syndromes with a Latin American Perspective" Genes 16, no. 6: 687. https://doi.org/10.3390/genes16060687
APA StyleBasquiera, A. L., Andreoli, V., Grille, S., & Belli, C. B. (2025). Progress in the Genetics of Myelodysplastic Syndromes with a Latin American Perspective. Genes, 16(6), 687. https://doi.org/10.3390/genes16060687





