Abstract
Background: The order Suliformes exhibits significant karyotype diversity, with Sula species showing a Z1Z1Z2Z2/Z1Z2W multiple-sex chromosome system, an uncommon occurrence in avians. Satellite DNAs (satDNAs), which consist of tandemly repeated sequences, often vary considerably even among closely related species, making them valuable markers for studying karyotypic evolution, particularly that of sex chromosome evolution. This study aims to characterize and investigate the potential role of these sequences in the karyotypic evolution of the group, with special attention to the sex chromosomes. Methods: Through characterizing satDNAs in two Suliformes species (Sula leucogaster and Nannopterum brasilianum) using BGISEQ-500 platform and bioinformatics analysis. Their chromosomal distribution was mapped by fluorescence in situ hybridization (FISH) within their own karyotypes and in three additional Suliformes species (S. sula, S. dactylatra, and Fregata magnificens). Results: Five satDNAs were identified in S. leucogaster and eight in N. brasilianum. Within the genus Sula, three species shared specific satDNA sequences, although with different hybridization patterns. In contrast, the satDNAs of N. brasilianum were species-specific. Additionally, the Z chromosome, including Z2 in Sula species, showed reduced accumulation of repetitive DNAs. Conclusions: These results suggest that differential accumulation of repetitive sequences may have contributed to the diversification of karyotypes in this group, particularly influencing the structure and differentiation of sex chromosomes.
1. Introduction
Satellite DNA (satDNA) can constitute up to 50% of eukaryotic genomes, typically arranged in tandemly repeated monomers within the genome [1,2,3,4,5]. The term satellitome denotes the entire set of satDNAs within a species’ genome [6]. Characterizing the satellitome elucidates genome evolution through the examination of the presence and absence of specific satDNAs, their abundance, and their chromosomal distribution among various related species [7,8,9]. The Library Hypothesis, proposed by [10], affirms that satDNA can vary in copy number, frequency in the genome, and chromosomal localization among related species. The long-term conservation of satDNAs has been increasingly demonstrated across various groups, including plants [8], reptiles [11], nematodes [12], anurans [13], insects [9,14,15], fishes [16,17], and mammals [18,19,20]. In addition, [21] suggests that species that diverged over 50 million years ago (Mya) typically exhibit no similarities in their satellite DNAs (satDNAs).
Satellite DNAs (satDNAs), once considered “junk” due to their association with heterochromatin and low gene content [22], are now recognized as playing essential roles in genome function and organization. They are involved in cellular processes such as kinetochore assembly, X chromosome recognition, and meiotic segregation [23,24,25]. For instance, in Drosophila melanogaster, the 1.688 satDNA family is crucial for centromere function, heterochromatin formation, and dosage compensation [26,27]. Their rapid sequence evolution across species may contribute to hybrid incompatibilities and speciation [21,28,29].
In sex chromosomes, limited recombination results in independent evolution and accumulation of repetitive DNAs, particularly in W and Y chromosomes due to their haploid condition [7,30,31]. The W chromosome can harbor over 50% of repetitive DNAs in birds, whereas the entire genome comprises roughly 10% [32,33]. For example, in the Wattled Jacana Jacana jacana, four out of eleven satDNAs are present in the W chromosome, while none are found in the Z chromosome [34].
Thus far, the satellitome has been studied in a limited number of avian species, and, in contrast to other taxa, avian satDNAs are predominantly GC-rich and found in microchromosomes [34,35,36,37,38]. In Turdus leucomelas, two satDNAs were W-specific, TleSat06 and TleSat08, but a W-autosomal translocation with a small autosome, identified by comparative genomic hybridization, incorporated these satDNA sequences in a microchromosome [37].
In the order Suliformes, approximately 16% of species have undergone karyotyping, with the majority exhibiting diploid numbers (2n) lower than the hypothesized ancestral avian karyotype (2n = 80) [39,40,41,42,43,44,45,46,47]. The genus Sula also hosts rare cases of a multiple Z1Z1Z2Z2/Z1Z2W sex chromosome system in birds, where males display 2n = 76, characterized by Z1Z1Z2Z2 sex chromosomes, whereas females exhibit 2n = 75, possessing Z1Z2W sex chromosomes. A Robertsonian translocation between the W ancestral chromosome and a microchromosome was suggested as the most plausible hypothesis for the emergence of this system [47].
This study aims to investigate the role of the satellitome in the evolution of the karyotype of Suliformes using bioinformatics and molecular cytogenetic techniques, focusing on the sex chromosomes of the genus Sula. To achieve this, the satellitome of S. leucogaster and N. brasilianum was identified and subsequently hybridized into three closely related species (S. dactylatra, S. sula, and F. magnificens).
2. Materials and Methods
2.1. Sampling and Chromosomal Obtainment
The fibroblast cell culture was established using feather pulp from female and male individuals of N. brasilianum, S. leucogaster, S. dactylatra, S. sula, and F. magnificens (Table 1), following [48]. The individuals were captured in their natural environment and were promptly returned after sampling. Metaphase chromosome spreads were obtained by treatment with colchicine (0.05% at 37 °C) for 1 h, hypotonic solution (0.075 M KCl at 37 °C) for 8 min, and fixation with 3:1 methanol/acetic acid. Additionally, for N. brasilianum and S. leucogaster, 1 mL of blood was collected from the cutaneous ulnar vein for posterior DNA extraction. The sample collection was approved by the Sistema de Autorização e Informação em Biodiversidade (SISBIO) under protocol number 64234-5. Furthermore, the ethics committee from the Universidade Federal do Rio Grande do Sul (CEUA) approved the experiments involving animals under protocol number 42827.

Table 1.
The locality and family of the five species of the Suliformes order analyzed in this study.
2.2. DNA Extraction, Genome Sequencing, and Bioinformatic Analysis
The genomic DNA (gDNA) from one male and one female of S. leucogaster and N. brasilianum was extracted with the PureLink™ Genomic DNA Mini Kit (Invitrogen, Carlsbad, CA, USA), following the protocol provided by the manufacturer. Using the BGISEQ-500 platform at BGI (BGI Shenzhen Corporation, Shenzhen, China), these four libraries yielded 150 base pairs (bp) paired-end sequences, including 2 Gb for each of the female and male whole genomes.
Genomic libraries were quality-trimmed with Trimmomatic [49]. The satellitome was characterized by a female specimen of both N. brasilianum and S. leucogaster. Satellite DNA identification for each species was performed using multiple iterations of the TAREAN tool [50]. Initially, 2 × 500,000 reads were input into TAREAN to identify satellite sequences. The satDNAs detected were subsequently filtered using DeconSeq [51] until TAREAN no longer identified additional satDNA sequences. Other tandemly repeated sequences, such as multigene families, were also identified and removed. Next, a similarity search was conducted using RepeatMasker software 4.1.5 (https://github.com/fjruizruano/satminer/blob/master/rm_homology.py, accessed on 30 October 2023) to eliminate redundancies and categorize the identified satDNAs into three groups based on the classification proposed by [6]: (i) superfamilies (50–80% similarity), (ii) variants of a single satDNA family (80–95% similarity), and (iii) different copies of the same satDNA variant (more than 95% similarity). Using RepeatMasker [52], we calculated the divergence and abundance of each satDNA with a Python script (https://github.com/fjruizruano/ngs-protocols/blob/master/repeat_masker_run_big.py, accessed on 30 October 2023).
To estimate the relative abundance of each satDNA, we randomly selected 2 × 5,000,000 reads and mapped them against their satellitomes. The number of mapped reads was then divided by the number of analyzed nucleotides. The satDNA families were named incorporating the species abbreviation (Nbr for N. brasilianum and Sle for S. leucogaster), the term “Sat,” the catalog number, and the monomer size (in base pairs) in decreasing order of abundance. We also performed a comparative analysis between male and female specimens by using the satellites found and running RepeatMasker in males to find the differential abundance of satDNAs. A comparative analysis was also performed between female and male individuals to assess differential satDNA abundance. We considered a sex-exclusive satDNA when the presence was only in one sex or the ratio was <0.5 or >1.5. Additionally, we conducted a comparative analysis between N. brasilianum and S. leucogaster using the homology.v2 script (https://github.com/fjruizruano/satminer/blob/master/rm_homology.py, accessed on 25 April 2025) and de novo assembly in Geneious 7.1.3.
2.3. SatDNA Probes and Fluorescence In Situ Hybridization (FISH)
Primers were designed for three out of five described satDNAs for S. leucogaster and seven out of eight described satDNAs for N. brasilianum (Table S1). The satDNAs with the shorter monomer length for both species (SleSat03, SleSat04, and NbrSat07) were labeled with biotin at the 5′ end during synthesis by ThermoFisher (ThermoFisher Scientific). The satDNAs of both species were amplified by polymerase chain reaction (PCR) with a start denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 45 s, annealing at 62–70.6 °C for 20–60 s, extension at 72 °C for 1 min, and final extension at 72 °C for 10 min (Table S2). The DNA concentrations used were 10 ng, 1 ng, 0.1 ng, 0.01 ng, 0.0001 ng, and 0.00001 ng. Ideal DNA template concentrations and amplification temperatures for each satDNA are described in Table S2. The PCR products were examined on a 2% electrophoresis gel and quantified in a NanoDrop LITE spectrophotometer (Thermo Scientific™, Wilmington, DE, USA) to ensure the amplification and integrity of satDNAs. The satDNAs amplified by PCR were labeled with the BioNick™ DNA Labeling System (Invitrogen, Carlsbad, CA, USA), following the manufacturer’s instructions.
The probes were hybridized following the protocol described by [53]. After hybridization, the probes were detected with Streptavidin-Cy3 dye, and the chromosomes were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) solution.
2.4. Image Processing
The chromosomal spreads were analyzed in a Zeiss Axio Imager A2 (ZEISS, Oberkochen, Germany) microscope with a 64-bit AxioCam MRc (ZEISS) attached and processed using Axio Vision SE 64 REL 4.8.3 (ZEISS) software. At least 10 metaphase chromosomal spreads were analyzed to confirm the FISH results.
3. Results
The diploid number and chromosome morphologies of all five species in this study align with the previous description of their karyotypes [46,47]. N. brasilianum presented 2n = 74, S. leucogaster and S. dactylatra presented 2n = 75 for females and 2n = 76 for male individuals, S. sula presented 2n = 76 for males, and F. magnificens presented 2n = 76 (for both sexes).
3.1. Abundance and Characteristics of the satDNAs
Five satDNAs were identified in S. leucogaster, while N. brasilianum exhibited eight distinct satDNAs (Table 2). In S. leucogaster, the monomer lengths varied from 17 to 190 base pairs (bp), with three designated as long satDNAs, each surpassing 100 bp in length (SleSat01, SleSat02, and SleSat05). Conversely, N. brasilianum exhibited a broader diversity in monomer length, spanning from 20 to 2559 bp. Of these, four satDNA were categorized as short, each measuring less than 100 bp (NbrSat04, NbrSat04, NbrSat07, and NbrSat08), while the others were classed as lengthy (NbrSat01, NbrSat02, NbrSat05, and NbrSat06). NbrSat02 and NbrSat05 demonstrated lengths beyond 2 kilobases (Kb).

Table 2.
General characteristics of satDNAs found in S. leucogaster (SleSat) and N. brasilianum (NbrSat).
Differences between male and female libraries were observed for only one satDNA in each species (Figures S1 and S2, Table 2). In S. leucogaster, SleSat04 exhibited significantly higher abundance in the female compared to the male library, whereas in N. brasilianum, NbrSat07 was detected exclusively in the female library. No satDNA sequences were shared between the two species (N. brasilianum and S. leucogaster). Most satDNAs on the analyzed species showed a higher percentage of CG. In S. leucogaster, only SleSat04 deviated from this trend, displaying a lower C+G content (47.1%), while the other four ranged from 54.7% to 63%. In N. brasilianum, NbrSat04 (42.2%) and NbrSat06 (38.4%) showed lower C+G content, whereas the remaining ranged from 52.1% to 71.4%.
3.2. Chromosomal Location of S. leucogaster satDNAs (SleSatDNAs)
All SleSatDNAs exhibited distinct hybridization patterns on their chromosomes, with most hybridization signals detected in the microchromosomes (Table 3, Figure 1). Furthermore, all SleSatDNAs, except for SleSat04, exhibited hybridization signals on the chromosomes of S. dactylatra (Table 3, Figure 2), while only SleSat03 and SleSat05 were present on the chromosomes of S. sula (Table 3, Figure 3). It is important to highlight that there was no evidence of hybridization signals for SleSat01 on the sex chromosomes of S. leucogaster or SleSat03 on Z2 of S. dactylatra. Moreover, SleSat04 exclusively showed signs on the W chromosome of S. leucogaster. In contrast, SleSat05 exhibited the same hybridization pattern across all species within the genus Sula, with only one microchromosome pair showing hybridization signals.

Table 3.
Hybridization results of SatDNA families found in S. leucogaster (SleSat) and N. brasilianum (NbrSat) and in three closely related species (S. dactylatra, S. sula, and F. magnificens).
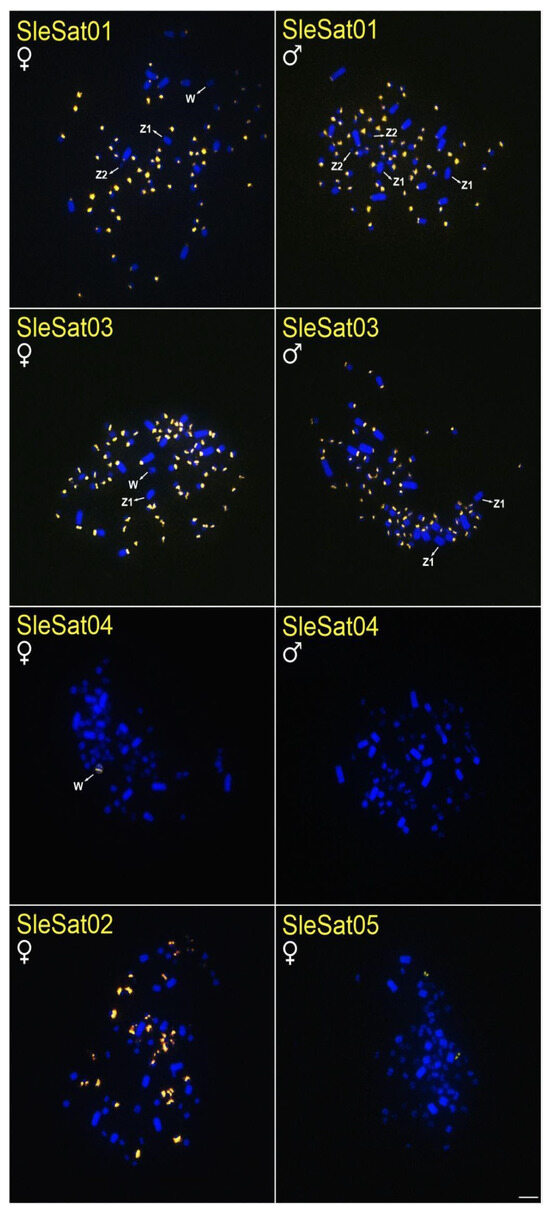
Figure 1.
Metaphase plates of females and males of S. leucogaster hybridized with different SleSatDNAs. Bar = 5 µm.
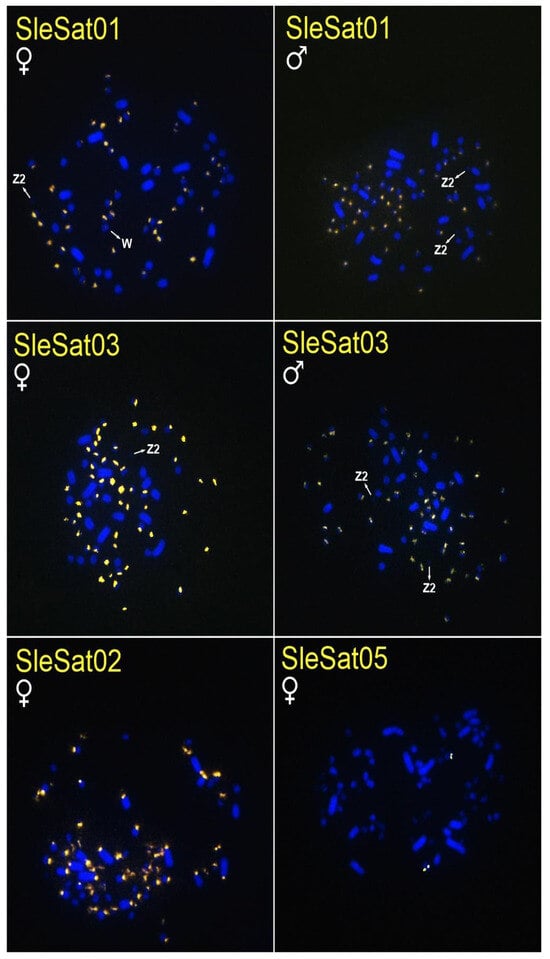
Figure 2.
Metaphase plates of females and males of S. dactylatra hybridized with different SleSatDNAs. Bar = 5 µm.
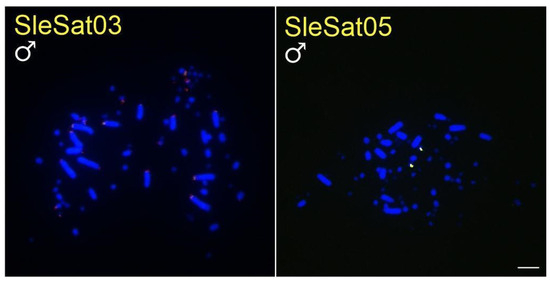
Figure 3.
Metaphase plates of males of S. sula hybridized with different SleSatDNAs. Bar = 5 µm.
SleSat01 probes produced no hybridization signals on the W chromosome of S. leucogaster, whereas weak signals were detected on the W chromosome of S. dactylatra (Table 3, Figure 1). Additionally, SleSat04, which is specific to the female library, exhibited hybridization signals exclusively on the W chromosomes of S. leucogaster (Table 3, Figure 1). Neither SleSat01 nor SleSat03 showed any signals on the Z chromosome; however, SleSat03 was detected on the Z2 chromosome of S. leucogaster (Table 3, Figure 1), and both SatDNAs failed to produce hybridization signals on the Z2 chromosome of S. dactylatra (Table 3, Figure 2).
The SleSatDNAs showed no hybridization signals in N. brasilianum and F. magnificens chromosomes, indicating the absence of these satDNAs in these species. It is important to consider the possibility of their existence in low abundance and/or scattered distribution, which would not be detected due to the limitations of the FISH technique.
3.3. Chromosomal Location of N. brasilianum satDNAs (NbrSatDNAs)
The NbrSatDNAs hybridization results revealed that, except for NbrSat07 and NbrSat08, all these sequences were localized on microchromosomes of N. brasilianum (Table 3, Figure 4). NbrSat01 and NbrSat03 had a similar hybridization pattern; however, NbrSat01 showed weaker signals (Table 3, Figure 4). NbrSat07 exclusively hybridized to the W sex chromosome (Table 3, Figure 4), whereas no signals were seen for NbrSat08. The Z sex chromosome showed no hybridization signals of any NbrSatDNAs (Table 3, Figure 4).
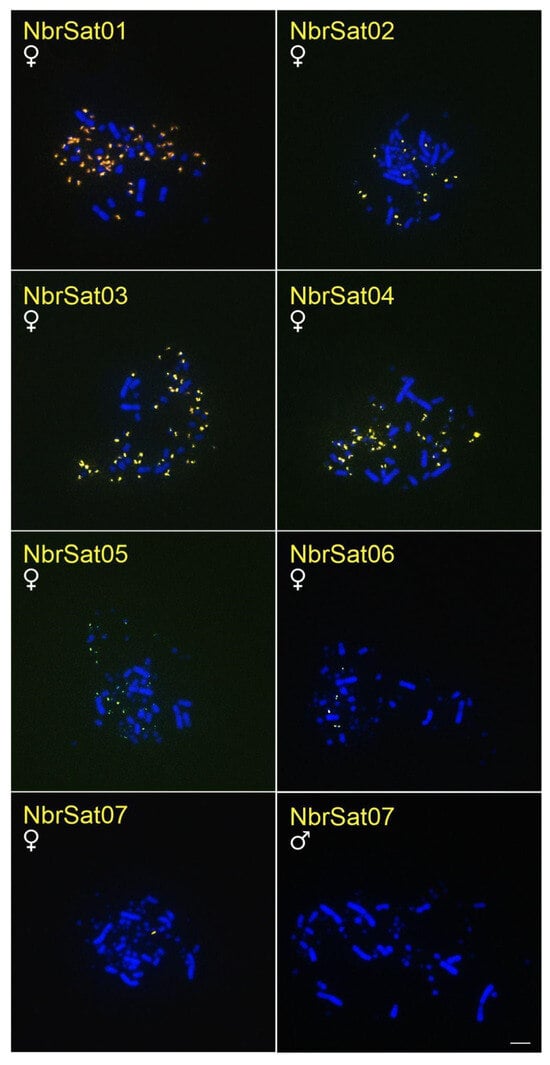
Figure 4.
Metaphase plates of males and females of N. brasilianum hybridized with different NbrSatDNAs. Bar = 5 µm.
No FISH signals corresponding to any NbrSatDNAs were detected in F. magnificens and the three Sula species examined.
4. Discussion
In the present study, we characterized and compared, for the first time, the complete set of satellite DNAs (satellitome) in two Suliformes species, S. leucogaster and N. brasilianum. Additionally, we used fluorescence in situ hybridization (FISH) to investigate the presence of the identified satDNAs in three other species: S. dactylatra, S. sula, and F. magnificens. Our results revealed limited conservation of satDNA subsets across these species, with a higher number of sequences shared between closely related taxa, such as S. leucogaster and S. dactylatra. Furthermore, we demonstrated that the recently emerged Z1 and Z2 chromosomes in the genus Sula tend to accumulate fewer satDNA sequences compared to the autosomes and the W chromosome, a pattern consistent with previous findings for the typical Z chromosomes in birds.
4.1. Overview of Satellite DNA in S. leucogaster and N. brasilianum
We identified five satDNAs in S. leucogaster and eight in N. brasilianum. Among birds, the number of satDNA families ranges from four in C. cristata to twenty-eight in Corvus woodfordi and C. splendens [38,54]. The variety of satDNA numbers among various species is markedly variable, with a minimum of one in the moth Cydalima perspectalis and a maximum of 248 in the crayfish Pontastacus leptodactylus [14,55]. In closely related species, such as the root-knot nematode genus Meloidogyne, the amount of satDNAs may vary, ranging from 38 in M. floridensis to 81 in M. arenaria [12]. These differences in the number of satDNAs highlight the remarkable diversity of satDNA across species.
The AT content in bird satDNAs is typically lower compared to other taxa, a trend that extends to insects [56], amphibians [13], fishes [57], reptiles [58], and mammals [18,20]. The higher GC content in birds is widely documented [34,35,36,37,38,54,59,60] and may correlate with the comparatively GC-rich structure of their microchromosomes, as shown in Gallus gallus and Meleagris gallopavo [61,62]. Another potential reason is the pronounced preferential fixing of GC over AT in avian species [63].
DNA monomers surpassing 2 Kb in length have been observed in N. brasilianum and 12 other avian species, constituting around 37.5% of the 32 species examined, with the majority exhibiting one or two such monomers. The longest monomer was identified in the wattled J. jacana, reaching 6514 bp [34,35,36,37,38,54]. Longer satDNA sequences are variants of shorter ones that have undergone mutations [64]. The notably long satDNA monomers are extensively recorded in several species, indicating that they are a prevalent feature in birds as well [11,18,65,66,67].
4.2. Comparative Chromosomal Mapping of Satellite DNAs in Suliformes
Four satDNAs identified in S. leucogaster generated FISH signals in S. dactylatra, while only two were successfully hybridized in S. sula chromosomes. This pattern likely indicates the recent divergence of these species, with S. sula splitting from their common ancestor roughly 5.9 million years ago (Mya) and S. leucogaster and S. dactylatra diverging around 3.6 Mya [68]. The satDNAs of N. brasilianum hybridized only with its chromosomes. This pattern is likely due to its substantial divergence from the genus Sula, estimated at approximately 50 Mya [69]. Similarly, no evidence of hybridization signals was found in F. magnificens, which may be attributed to its divergence from the other four species around 59 Mya [69]. These results corroborate the findings of [21], who reported that plant and animal species with divergence times exceeding 50 Mya typically show no similarity in their repetitive DNA sequences. Furthermore, shared satDNA among the three Sula species supports the Library Hypothesis [10]. This hypothesis proposes that closely related species retain common satDNAs, though their genomic abundance may vary. The idea is exemplified by the five satDNAs of S. leucogaster, which persist within the genus Sula but differ in distribution.
Except for SleSat04 in S. leucogaster and NbrSat07 in N. brasilianum, all other satDNAs hybridized to centromeric regions and covered some microchromosomes (SleSat01, SleSat02, SleSat03, NbrSat01, NbrSat03, and NbrSat04). This pattern is similar to the satDNAs TleSat1 and TleSat3 of T. leucomelas and VchSat1 of V. chilensis, which also hybridized into centromeric regions [35,37]. According to [70], such distributions provide evidence that these satDNAs may have a significant role in the function of the centromere by either aiding the binding of proteins or serving mainly as structural components of the centromere.
4.3. Satellite DNA Mapping in Suliformes Sheds Light on Sex Chromosome Evolution
The absence of recombination in the W chromosome makes it highly susceptible to accumulating repetitive DNA [7]. Additionally, the size and repetitive DNA content of the W chromosome vary significantly among bird species [7,71,72]. In Suliformes, W morphology differs across all studied species [47]. This may result from the buildup of various repeating DNAs, including satellite DNA and transposable elements. Indeed, the SleSat04 hybridized exclusively to the W chromosome of S. leucogaster, with no signals detected in the other two Sula species (Figure 1). Similarly, NbrSat07 was restricted to the W chromosome of N. brasilianum, showing no hybridization signals in different species tested (Figure 4). This pattern of W-linked satDNA accumulation has been documented in five other avian species, where hybridization signals suggest a significant buildup of repetitive DNA on the W chromosome [34,35,36,37,38].
In most avian species, the Z chromosome shows little constitutive heterochromatin, mainly in the centromere region, though it may be entirely absent [41,60,73,74]. Corroborating these findings, previous studies have shown that the Z chromosome in N. brasilianum and the Z1 and Z2 chromosomes in the genus Sula lack constitutive heterochromatin [47]. In this study, we extend these findings by demonstrating that these chromosomes also exhibit few satDNA hybridization signals. This indicates that although satDNA exists on the Z chromosome, specifically SleSat03 on the Z2 chromosome of S. leucogaster (Figure 1), is comparatively scarce. Similar findings have been observed in V. chilensis, where only VchSat01 is located in the centromere of all chromosomes, including the Z chromosome [34]. Conversely, in the red-legged seriema Cariama cristata, the Z chromosome is the largest element in the karyotype, attributed in part to the accumulation of the satDNA CcrSat02-1104 [38]. Additionally, in the pale-breasted Thrush T. leucomelas, two satDNA sequences hybridize to the centromeric region of the Z chromosome, which is likewise heterochromatic [37].
5. Conclusions
Here, we describe the satDNA of S. leucogaster and N. brasilianum. Eleven of the thirteen satDNAs characterized have a higher content of GC, a general feature for bird satellites, since this observation was also made in other birds. Also, most satDNA were localized in the same sites as constitutive heterochromatin and centromere sites. Ref. [21] stated that species that diverged more than 50 Mya showed no satDNA resemblance. This statement is confirmed here since only the genus Sula, with 5.9 Mya of divergence, showed shared satDNAs [69]. Finally, the Z chromosome of N. brasilianum and S. leucogaster showed none to little satDNA hybridization signals, indicating that the Z chromosome has little tendency to accumulate satDNA, including the Z2 of genus Sula.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/genes16060633/s1, Table S1: Primers design for amplification of satellitomes from Sula leucogaster (SleSat) and Nannopterum brasialianum (NbrSat); Table S2: Termocycle program for amplification of satellitomes from Sula leucogaster (SleSat) and Nannopterum brasialianum (NbrSat); Figure S1: Landscape from female (A) and male (B) of Nannopterum brasilianum with satDNA families in order of abundance; Figure S2: Landscape from female (A) and male (B) of Sula leucogaster with satDNA families in order of abundance.
Author Contributions
Conceptualization, L.C.P., R.K. and T.R.O.d.F.; methodology, L.C.P., N.d.S., R.U., F.P.-F. and R.K.; software, L.C.P. and N.d.S.; validation, L.C.P.; formal analysis, L.C.P. and N.d.S.; investigation, L.C.P., N.d.S., R.U., F.P.-F., M.d.B.C. and R.K.; resources, R.K. and T.R.O.d.F.; data curation, L.C.P.; writing—original draft preparation, L.C.P., N.d.S., R.U., F.P.-F., M.d.B.C., R.K. and T.R.O.d.F.; writing—review and editing, L.C.P., N.d.S., R.U., F.P.-F., M.d.B.C., R.K. and T.R.O.d.F.; visualization, L.C.P., N.d.S., R.U., F.P.-F., M.d.B.C., R.K. and T.R.O.d.F.; supervision, R.K. and T.R.O.d.F.; project administration, R.K. and T.R.O.d.F.; funding acquisition, R.K. and T.R.O.d.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Conselho Nacional de Desenvolvimento Científico e Tecnológico (Proc. 406747/2023-7 and Proc. 130070/2022-0) and Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (Proc. 24/2551-0001269-9).
Institutional Review Board Statement
The animal study protocol was approved by the Sistema de Autorização e Informação em Biodiversidade (SISBIO, No. 64234-5) and the Ethics Committee from the Universidade Federal do Rio Grande do Sul (CEUA, No. 42827).
Data Availability Statement
The sequencing reads have been deposited in the Sequence Read Archive (SRA-NCBI) and are available under the following accession numbers: Nannopterum brasialianum—SRR33660189 (male) and SRR33660190 (female); Sula leucogaster—SRR33660187 (male) and SRR33660188 (female). The original contributions presented in this study are included in the article/Supplementary Material.
Acknowledgments
The authors would like to thank Parque Nacional Marinho dos Abrolhos, Cynthia Campolina, Guilherme Tavares Nunes, Júlia Jacoby, Márcio Reppening, Maria Bernadete Silva Barbosa, Leandro Bugoni, and Victor Libardi Silva for kindly providing the samples of the specimens used in this study.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Charlesworth, B.; Sniegowski, P.; Stephan, W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 1994, 371, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T. Genomes, genes and junk: The large-scale organization of plant chromosomes. Trends Plant Sci. 1998, 3, 195–199. [Google Scholar] [CrossRef]
- Richard, G.-F.; Kerrest, A.; Dujon, B. Comparative Genomics and Molecular Dynamics of DNA Repeats in Eukaryotes. Microbiol. Mol. Biol. Rev. 2008, 72, 686–727. [Google Scholar] [CrossRef] [PubMed]
- Plohl, M.; Meštrović, N.; Mravinac, B. Satellite DNA Evolution. In Repetitive DNA; Garrido-Ramos, M.A., Ed.; S. Karger AG: Basel, Switzerland, 2012; Volume 7, pp. 126–152. ISBN 978-3-318-02149-3. [Google Scholar]
- Garrido-Ramos, M. Satellite DNA: An Evolving Topic. Genes 2017, 8, 230. [Google Scholar] [CrossRef]
- Ruiz-Ruano, F.J.; López-León, M.D.; Cabrero, J.; Camacho, J.P.M. High-throughput analysis of the satellitome illuminates satellite DNA evolution. Sci. Rep. 2016, 6, 28333. [Google Scholar] [CrossRef]
- Peona, V.; Palacios-Gimenez, O.M.; Blommaert, J.; Liu, J.; Haryoko, T.; Jønsson, K.A.; Irestedt, M.; Zhou, Q.; Jern, P.; Suh, A. The avian W chromosome is a refugium for endogenous retroviruses with likely effects on female-biased mutational load and genetic incompatibilities. Phil. Trans. R. Soc. B 2021, 376, 20200186. [Google Scholar] [CrossRef]
- Wei, L.; Liu, B.; Zhang, C.; Yu, Y.; Yang, X.; Dou, Q.; Dong, Q. Identification and characterization of satellite DNAs in Poa L. Mol. Cytogenet. 2020, 13, 47. [Google Scholar] [CrossRef]
- De Lima, L.G.; Ruiz-Ruano, F.J. In-Depth Satellitome Analyses of 37 Drosophila Species Illuminate Repetitive DNA Evolution in the Drosophila Genus. Genome Biol. Evol. 2022, 14, evac064. [Google Scholar] [CrossRef]
- Fry, K.; Salser, W. Nucleotide sequences of HS-α satellite DNA from kangaroo rat dipodomys ordii and characterization of similar sequences in other rodents. Cell 1977, 12, 1069–1084. [Google Scholar] [CrossRef]
- Yurchenko, A.; Pšenička, T.; Mora, P.; Ortega, J.A.M.; Baca, A.S.; Rovatsos, M. Cytogenetic Analysis of Satellitome of Madagascar Leaf-Tailed Geckos. Genes 2024, 15, 429. [Google Scholar] [CrossRef]
- Despot-Slade, E.; Širca, S.; Mravinac, B.; Castagnone-Sereno, P.; Plohl, M.; Meštrović, N. Satellitome analyses in nematodes illuminate complex species history and show conserved features in satellite DNAs. BMC Biol. 2022, 20, 259. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, M.J.; Fogarin Destro, R.; Gazoni, T.; Narimatsu, H.; Pereira Dos Santos, P.S.; Haddad, C.F.B.; Parise-Maltempi, P.P. Great Abundance of Satellite DNA in Proceratophrys (Anura, Odontophrynidae) Revealed by Genome Sequencing. Cytogenet. Genome Res. 2020, 160, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Cabral-de-Mello, D.C.; Zrzavá, M.; Kubíčková, S.; Rendón, P.; Marec, F. The Role of Satellite DNAs in Genome Architecture and Sex Chromosome Evolution in Crambidae Moths. Front. Genet. 2021, 12, 661417. [Google Scholar] [CrossRef] [PubMed]
- Pereira, J.A.; Milani, D.; Ferretti, A.B.S.M.; Bardella, V.B.; Cabral-de-Mello, D.C.; Lopes, D.M. The extensive amplification of heterochromatin in Melipona bees revealed by high throughput genomic and chromosomal analysis. Chromosoma 2021, 130, 251–262. [Google Scholar] [CrossRef]
- Voleníková, A.; Lukšíková, K.; Mora, P.; Pavlica, T.; Altmanová, M.; Štundlová, J.; Pelikánová, Š.; Simanovsky, S.A.; Jankásek, M.; Reichard, M.; et al. Fast satellite DNA evolution in Nothobranchius annual killifishes. Chromosome Res. 2023, 31, 33. [Google Scholar] [CrossRef]
- Crepaldi, C.; Martí, E.; Gonçalves, É.M.; Martí, D.A.; Parise-Maltempi, P.P. Genomic Differences Between the Sexes in a Fish Species Seen Through Satellite DNAs. Front. Genet. 2021, 12, 728670. [Google Scholar] [CrossRef]
- Sena, R.S.; Heringer, P.; Valeri, M.P.; Pereira, V.S.; Kuhn, G.C.S.; Svartman, M. Identification and characterization of satellite DNAs in two-toed sloths of the genus Choloepus (Megalonychidae, Xenarthra). Sci. Rep. 2020, 10, 19202. [Google Scholar] [CrossRef]
- Vozdova, M.; Kubickova, S.; Martínková, N.; Galindo, D.J.; Bernegossi, A.M.; Cernohorska, H.; Kadlcikova, D.; Musilová, P.; Duarte, J.M.; Rubes, J. Satellite DNA in Neotropical Deer Species. Genes 2021, 12, 123. [Google Scholar] [CrossRef]
- Gutiérrez, J.; Aleix-Mata, G.; Montiel, E.E.; Cabral-de-Mello, D.C.; Marchal, J.A.; Sánchez, A. Satellitome Analysis on Talpa aquitania Genome and Inferences about the satDNAs Evolution on Some Talpidae. Genes 2022, 14, 117. [Google Scholar] [CrossRef]
- Melters, D.P.; Bradnam, K.R.; Young, H.A.; Telis, N.; May, M.R.; Ruby, J.G.; Sebra, R.; Peluso, P.; Eid, J.; Rank, D.; et al. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 2013, 14, R10. [Google Scholar] [CrossRef]
- Ohno, S. So much “junk” DNA in our genome. Brookhaven Symp. Biol. 1972, 23, 366–370. [Google Scholar] [PubMed]
- Dernburg, A.F.; Sedat, J.W.; Hawley, R.S. Direct Evidence of a Role for Heterochromatin in Meiotic Chromosome Segregation. Cell 1996, 86, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, G.C.S. Satellite DNA transcripts have diverse biological roles in Drosophila. Heredity 2015, 115, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Shatskikh, A.S.; Kotov, A.A.; Adashev, V.E.; Bazylev, S.S.; Olenina, L.V. Functional Significance of Satellite DNAs: Insights From Drosophila. Front. Cell. Dev. Biol. 2020, 8, 312. [Google Scholar] [CrossRef]
- Cattani, M.V.; Presgraves, D.C. Incompatibility between X chromosome factor and pericentric heterochromatic region causes lethality in hybrids between Drosophila melanogaster and its sibling species. Genetics 2012, 191, 549–559. [Google Scholar] [CrossRef]
- Rošić, S.; Köhler, F.; Erhardt, S. Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J. Cell. Biol. 2014, 207, 335–349. [Google Scholar] [CrossRef]
- Bayes, J.J.; Malik, H.S. Altered Heterochromatin Binding by a Hybrid Sterility Protein in Drosophila Sibling Species. Science 2009, 326, 1538–1541. [Google Scholar] [CrossRef]
- Jagannathan, M.; Yamashita, Y.M. Defective Satellite DNA Clustering into Chromocenters Underlies Hybrid Incompatibility in Drosophila. Mol. Biol. Evol. 2021, 38, 4977–4986. [Google Scholar] [CrossRef]
- Ferretti, A.B.S.M.; Milani, D.; Palacios-Gimenez, O.M.; Ruiz-Ruano, F.J.; Cabral-de-Mello, D.C. High dynamism for neo-sex chromosomes: Satellite DNAs reveal complex evolution in a grasshopper. Heredity 2020, 125, 124–137. [Google Scholar] [CrossRef]
- Gatto, K.P.; Busin, C.S.; Lourenço, L.B. Unraveling the Sex Chromosome Heteromorphism of the Paradoxical Frog Pseudis tocantins. PLoS ONE 2016, 11, e0156176. [Google Scholar] [CrossRef]
- Smeds, L.; Warmuth, V.; Bolivar, P.; Uebbing, S.; Burri, R.; Suh, A.; Nater, A.; Bureš, S.; Garamszegi, L.Z.; Hogner, S.; et al. Evolutionary analysis of the female-specific avian W chromosome. Nat. Commun. 2015, 6, 7330. [Google Scholar] [CrossRef] [PubMed]
- Peona, V.; Blom, M.P.K.; Xu, L.; Burri, R.; Sullivan, S.; Bunikis, I.; Liachko, I.; Haryoko, T.; Jønsson, K.A.; Zhou, Q.; et al. Identifying the causes and consequences of assembly gaps using a multiplatform genome assembly of a bird-of-paradise. Mol. Ecol. Resour. 2021, 21, 263–286. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, A.M.; Souza, G.M.; Toma, G.A.; Dos Santos, N.; Dos Santos, R.Z.; Goes, C.A.G.; Deon, G.A.; Setti, P.G.; Porto-Foresti, F.; Utsunomia, R.; et al. Satellite DNAs, heterochromatin, and sex chromosomes of the wattled jacana (Charadriiformes; Jacanidae): A species with highly rearranged karyotype. Genome 2024, 67, 109–118. [Google Scholar] [CrossRef]
- Kretschmer, R.; Toma, G.A.; Deon, G.A.; Dos Santos, N.; Dos Santos, R.Z.; Utsunomia, R.; Porto-Foresti, F.; Gunski, R.J.; Garnero, A.D.V.; Liehr, T.; et al. Satellitome Analysis in the Southern Lapwing (Vanellus chilensis) Genome: Implications for SatDNA Evolution in Charadriiform Birds. Genes 2024, 15, 258. [Google Scholar] [CrossRef] [PubMed]
- Setti, P.G.; Deon, G.A.; Zeni Dos Santos, R.; Goes, C.A.G.; Garnero, A.D.V.; Gunski, R.J.; De Oliveira, E.H.C.; Porto-Foresti, F.; De Freitas, T.R.O.; Silva, F.A.O.; et al. Evolution of bird sex chromosomes: A cytogenomic approach in Palaeognathae species. BMC Ecol. Evo. 2024, 24, 51. [Google Scholar] [CrossRef]
- Souza, G.M.; Kretschmer, R.; Toma, G.A.; De Oliveira, A.M.; Deon, G.A.; Setti, P.G.; Zeni Dos Santos, R.; Goes, C.A.G.; Del Valle Garnero, A.; Gunski, R.J.; et al. Satellitome analysis on the pale-breasted thrush Turdus leucomelas (Passeriformes; Turdidae) uncovers the putative co-evolution of sex chromosomes and satellite DNAs. Sci. Rep. 2024, 14, 20656. [Google Scholar] [CrossRef]
- Souza, G.M.; Vidal, J.A.D.; Utsunomia, R.; Deon, G.A.; De Oliveira, E.H.C.; Franca, R.T.; Porto-Foresti, F.; Liehr, T.; De Souza, F.H.S.; Kretschmer, R.; et al. Cytogenomic analysis in Seriemas (Cariamidae): Insights into an Atypical Avian Karyotype. J. Hered. 2025, esaf012. [Google Scholar] [CrossRef]
- Theodorescu, R.C. The Karyotypic Evolution in Two Pelecaniformes Species (Aves). Caryologia 1975, 28, 459–466. [Google Scholar] [CrossRef]
- Patnaik, A.K.; Samanta, M.; Prasad, R. Chromosome complement and banding patterns in a pelecaniform bird, Phalacrocorax niger. J. Hered. 1981, 72, 447–449. [Google Scholar] [CrossRef]
- Belterman, R.H.R.; De Boer, L.E.M. A karyological study of 55 species of birds, including karyotypes of 39 species new to cytology. Genetica 1984, 65, 39–82. [Google Scholar] [CrossRef]
- Bhunya, S.P.; Mohanty, M.K. Localization of constitutive heterochromatin (C-band) and nucleolus organizers (NORs) in the somatic chromosomes of a pelecaniform bird Phalacrocorax niger (Viellot). Chrom. Inform. Serv. 1985, 39, 17–19. [Google Scholar]
- Ebied, A.M.; Hassan, H.A.; Abu Almaaty, A.H.; Yaseen, A.E. Karyotypic Characterization of Ten Species of Birds. Cytologia 2005, 70, 181–194. [Google Scholar] [CrossRef]
- Ledesma, M.A.; Cardozo, D.E.; Montalti, D.; Leotta, G.A.; Gunski, R. Estudios citogenéticos en tres especies de Aves Antárticas. Rev. De Cienc. Y Tecnol. 2005, 7. [Google Scholar]
- Griffin, D.K.; Robertson, L.B.W.; Tempest, H.G.; Skinner, B.M. The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenet. Genome Res. 2007, 117, 64–77. [Google Scholar] [CrossRef]
- Kretschmer, R.; De Souza, M.S.; Furo, I.D.O.; Romanov, M.N.; Gunski, R.J.; Garnero, A.D.V.; De Freitas, T.R.O.; De Oliveira, E.H.C.; O’Connor, R.E.; Griffin, D.K. Interspecies Chromosome Mapping in Caprimulgiformes, Piciformes, Suliformes, and Trogoniformes (Aves): Cytogenomic Insight into Microchromosome Organization and Karyotype Evolution in Birds. Cells 2021, 10, 826. [Google Scholar] [CrossRef] [PubMed]
- Pozzobon, L.C.; Toma, G.A.; Cioffi, M.D.B.; De Oliveira, E.H.C.; Kretschmer, R.; De Freitas, T.R.O. Karyotype evolution of suliformes and description of a ♂Z1Z1Z2Z2/♀Z1Z2W multiple sex chromosome system in boobies (Sula spp.). Genome 2025, 68, 1–11. [Google Scholar] [CrossRef]
- Sasaki, M.; Hitotsumachi, S.; Makino, S.; Terao, T. A Comparative Study of the Chromosomes in the Chum Salmon, the Kokanee Salmon and their Hybrids. Caryologia 1968, 21, 389–394. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Novák, P.; Neumann, P.; Macas, J. Global analysis of repetitive DNA from unassembled sequence reads using RepeatExplorer2. Nat. Protoc. 2020, 15, 3745–3776. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Fast Identification and Removal of Sequence Contamination from Genomic and Metagenomic Datasets. PLoS ONE 2011, 6, e17288. [Google Scholar] [CrossRef]
- Smit, A.F.A.; Hubley, R.; Green, P. RepeatMasker Open-4.1.5. 2013–2015. Available online: http://www.repeatmasker.org. (accessed on 30 October 2023).
- Kretschmer, R.; dos Santos, M.S.; Furo, I.D.O.; Cioffi, M.D.B. FISH—In Birds. In Cytogenetics and Molecular Cytogenetics; Liehr, T., Ed.; CRC Press: Boca Raton, FL, USA, 2022; Volume 1, pp. 263–280. ISBN 978-1-003-22365-8. [Google Scholar]
- Peona, V.; Kutschera, V.E.; Blom, M.P.K.; Irestedt, M.; Suh, A. Satellite DNA evolution in Corvoidea inferred from short and long reads. Mol. Ecol. 2023, 32, 1288–1305. [Google Scholar] [CrossRef] [PubMed]
- Boštjančić, L.L.; Bonassin, L.; Anušić, L.; Lovrenčić, L.; Besendorfer, V.; Maguire, I.; Grandjean, F.; Austin, C.M.; Greve, C.; Hamadou, A.B.; et al. The Pontastacus leptodactylus (Astacidae) Repeatome Provides Insight Into Genome Evolution and Reveals Remarkable Diversity of Satellite DNA. Front. Genet. 2021, 11, 611745. [Google Scholar] [CrossRef] [PubMed]
- Lorite, P.; Carrillo, J.A.; Aguilar, J.A.; Palomeque, T. Isolation and characterization of two families of satellite DNA with repetitive units of 135 bp and 2.5 kb in the ant Monomorium subopacum (Hymenoptera, Formicidae). Cytogenet. Genome Res. 2004, 105, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Crepaldi, C.; Parise-Maltempi, P.P. Heteromorphic Sex Chromosomes and Their DNA Content in Fish: An Insight through Satellite DNA Accumulation in Megaleporinus elongatus. Cytogenet. Genome Res. 2020, 160, 38–46. [Google Scholar] [CrossRef]
- Rojo, V.; Martínez-Lage, A.; Giovannotti, M.; González-Tizón, A.M.; Cerioni, P.N.; Barucchi, V.C.; Galán, P.; Olmo, E.; Naveira, H. Evolutionary dynamics of two satellite DNA families in rock lizards of the genus Iberolacerta (Squamata, Lacertidae): Different histories but common traits. Chromosome Res. 2015, 23, 441–461. [Google Scholar] [CrossRef]
- Yamada, K.; Yamada, K.; Nishida-Umehara, C.; Matsuda, Y. Characterization and chromosomal distribution of novel satellite DNA sequences of the lesser rhea (Pterocnemia pennata) and the greater rhea (Rhea americana). Chromosome Res. 2002, 10, 513–523. [Google Scholar] [CrossRef]
- Yamada, K.; Nishida-Umehara, C.; Matsuda, Y. A new family of satellite DNA sequences as a major component of centromeric heterochromatin in owls (Strigiformes). Chromosoma 2004, 112, 277–287. [Google Scholar] [CrossRef]
- Axelsson, E.; Webster, M.T.; Smith, N.G.C.; Burt, D.W.; Ellegren, H. Comparison of the chicken and turkey genomes reveals a higher rate of nucleotide divergence on microchromosomes than macrochromosomes. Genome Res. 2005, 15, 120–125. [Google Scholar] [CrossRef]
- Warren, W.C.; Hillier, L.W.; Tomlinson, C.; Minx, P.; Kremitzki, M.; Graves, T.; Markovic, C.; Bouk, N.; Pruitt, K.D.; Thibaud-Nissen, F.; et al. A New Chicken Genome Assembly Provides Insight into Avian Genome Structure. G3 Genes. Genomes Genet. 2017, 7, 109–117. [Google Scholar] [CrossRef]
- Bolívar, P.; Guéguen, L.; Duret, L.; Ellegren, H.; Mugal, C.F. GC-biased gene conversion conceals the prediction of the nearly neutral theory in avian genomes. Genome Biol. 2019, 20, 5. [Google Scholar] [CrossRef]
- Ruiz-Ruano, F.J.; Navarro-Domínguez, B.; Camacho, J.P.M.; Garrido-Ramos, M.A. Characterization of the satellitome in lower vascular plants: The case of the endangered fern Vandenboschia speciosa. Ann. Bot. 2019, 123, 587–599. [Google Scholar] [CrossRef] [PubMed]
- Goes, C.A.G.; Dos Santos, N.; Rodrigues, P.H.D.M.; Stornioli, J.H.F.; Silva, A.B.D.; Dos Santos, R.Z.; Vidal, J.A.D.; Silva, D.M.Z.D.A.; Artoni, R.F.; Foresti, F.; et al. The Satellite DNA Catalogues of Two Serrasalmidae (Teleostei, Characiformes): Conservation of General satDNA Features over 30 Million Years. Genes 2022, 14, 91. [Google Scholar] [CrossRef] [PubMed]
- Rico-Porras, J.M.; Mora, P.; Palomeque, T.; Montiel, E.E.; Cabral-de-Mello, D.C.; Lorite, P. Heterochromatin Is Not the Only Place for satDNAs: The High Diversity of satDNAs in the Euchromatin of the Beetle Chrysolina americana (Coleoptera, Chrysomelidae). Genes 2024, 15, 395. [Google Scholar] [CrossRef] [PubMed]
- Gálvez-Galván, A.; Barea, L.; Garrido-Ramos, M.A.; Prieto, P. Highly divergent satellitomes of two barley species of agronomic importance, Hordeum chilense and H. vulgare. Plant Mol. Biol. 2024, 114, 108. [Google Scholar] [CrossRef]
- Patterson, S.A.; Morris-Pocock, J.A.; Friesen, V.L. A multilocus phylogeny of the Sulidae (Aves: Pelecaniformes). Mol. Phylogenetics Evol. 2011, 58, 181–191. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Hedges, S.B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evo. 2017, 34, 1812–1819. [Google Scholar] [CrossRef]
- Talbert, P.B.; Henikoff, S. What makes a centromere? Exp. Cell. Res. 2020, 389, 111895. [Google Scholar] [CrossRef]
- Rutkowska, J.; Lagisz, M.; Nakagawa, S. The long and the short of avian W chromosomes: No evidence for gradual W shortening. Biol. Lett. 2012, 8, 636–638. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhang, J.; Bachtrog, D.; An, N.; Huang, Q.; Jarvis, E.D.; Gilbert, M.T.P.; Zhang, G. Complex evolutionary trajectories of sex chromosomes across bird taxa. Science 2014, 346, 1246338. [Google Scholar] [CrossRef]
- Schmid, M.; Steinlein, C. The Hypermethylated Regions in Avian Chromosomes. Cytogenet. Genome Res. 2017, 151, 216–227. [Google Scholar] [CrossRef]
- Wójcik, E.; Smalec, E. Constitutive heterochromatin in chromosomes of duck hybrids and goose hybrids. Poult. Sci. 2017, 96, 18–26. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).