Genome-Wide Association Studies and Candidate Genes for Egg Production Traits in Layers from an F2 Crossbred Population Produced Using Two Divergently Selected Chicken Breeds, Russian White and Cornish White
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Birds
2.2. Performance Data
2.3. Sampling and DNA Isolation
2.4. Genotyping and Quality Control of SNPs
2.5. Principal Component Analysis
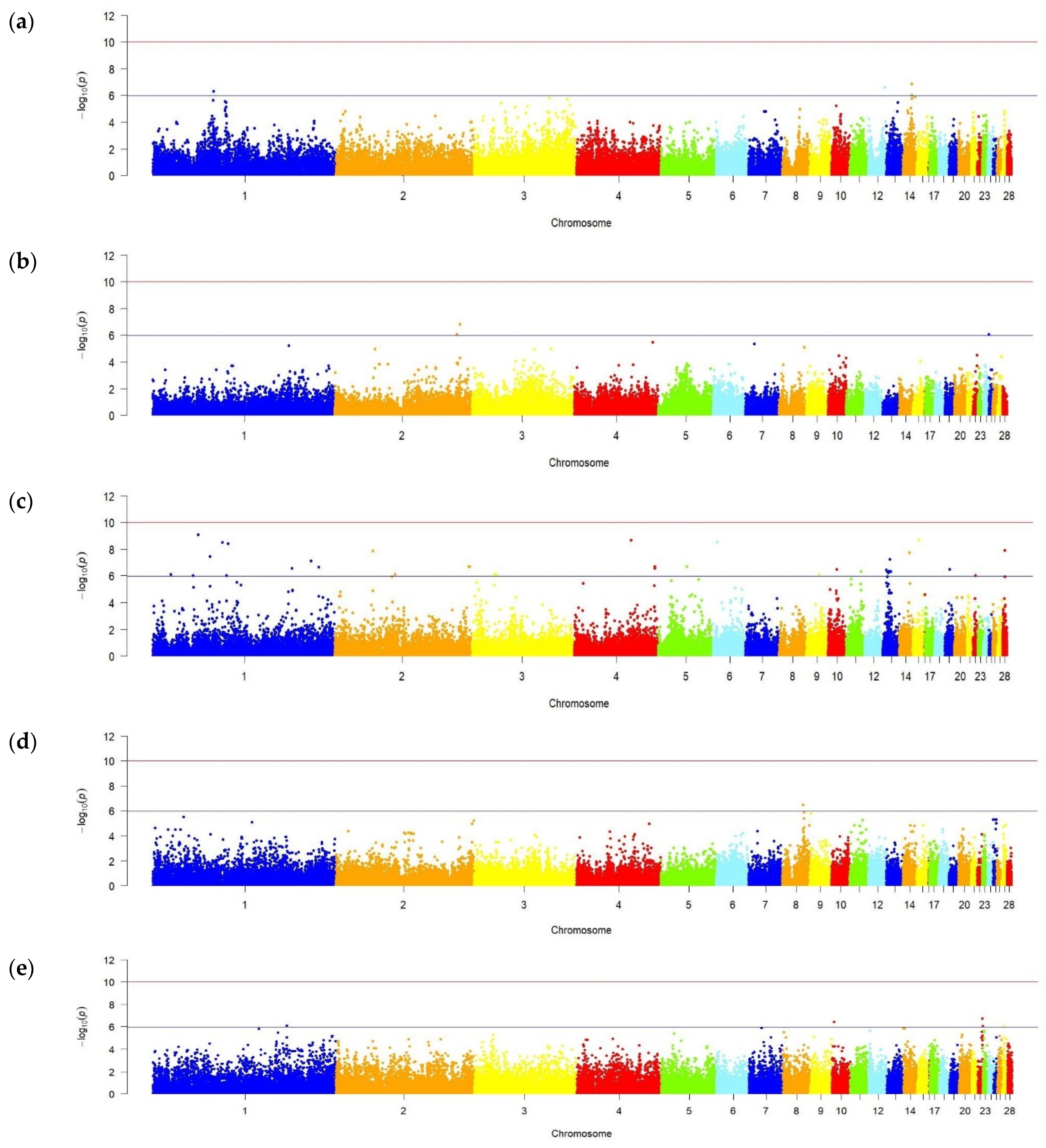
2.6. Genome-Wide Association Studies
3. Results
3.1. Phenotypic Data and Population Stratification
3.2. Genome-Wide Association Analysis Output
3.3. Candidate Genes
4. Discussion
4.1. Egg Number and Egg Weight
4.2. Age at First Egg and Duration of Egg Laying
4.3. Egg Laying Cycle and Egg Laying Interval
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Meuwissen, T.; Hayes, B.; MacLeod, I.; Goddard, M. Identification of genomic variants causing variation in quantitative traits: A review. Agriculture 2022, 12, 1713. [Google Scholar] [CrossRef]
- Plemyashov, K.V.; Smaragdov, M.G.; Romanov, M.N. Molekulyarno-geneticheskiy polimorfizm v populyatsiyakh zhivotnykh i yego primeneniye v intensivnoy selektsii molochnogo skota: Obzor [Molecular Genetic Polymorphism in Animal Populations and Its Application in Intensive Breeding of Dairy Cattle—A Review]. In Molecular Genetic Technologies for Analysis of Gene Expression Related to Animal Productivity and Disease Resistance, Proceedings of the Materials of the 3rd International Scientific and Practical Conference, Moscow, Russia, 30 September 2021; Sel’skokhozyaistvennye Tekhnologii: Moscow, Russia, 2021; pp. 368–378, (In Russian with English summary). [Google Scholar]
- Meuwissen, T.; Hayes, B.; Goddard, M. Accelerating improvement of livestock with genomic selection. Annu. Rev. Anim. Biosci. 2013, 1, 221–237. [Google Scholar] [CrossRef]
- Georges, M.; Charlier, C.; Hayes, B. Harnessing genomic information for livestock improvement. Nat. Rev. Genet. 2019, 20, 135–156. [Google Scholar] [CrossRef]
- Plemyashov, K.V.; Smaragdov, M.G.; Romanov, M.N. Genomnaya otsenka plemennykh bykov [Genomic Assessment of Breeding Bulls]. In Molecular Genetic Technologies for Analysis of Gene Expression Related to Animal Productivity and Disease Resistance, Proceedings of the Materials of the 3rd International Scientific and Practical Conference, Moscow, Russia, 30 September 2021; Pozyabin, S.V., Kochish, I.I., Romanov, M.N., Eds.; Sel’skokhozyaistvennye Tekhnologii: Moscow, Russia, 2021; pp. 363–367. Available online: https://elib.spbstu.ru/dl/2/z21-43.pdf/info (accessed on 21 April 2025).
- Romanov, M.N.; Kochish, I.I.; Sharafetdinov, G.R.; Myasnikova, O.V.; Nikonov, I.N.; Selina, M.V.; Surai, P.F. Napravleniya sovremennykh biotekhnologicheskikh razrabotok dlya realizatsii geneticheskogo potentsiala yaichnoy ptitsy [Towards Advanced Biotechnological Developments to Realize the Genetic Potential of Egg-type Poultry]. In Molecular Genetic Technologies for Analysis of Gene Expression Related to Animal Productivity and Disease Resistance, Proceedings of the Materials of the 3rd International Scientific and Practical Conference, Moscow, Russia, 30 September 2021; Pozyabin, S.V., Kochish, I.I., Romanov, M.N., Eds.; Sel’skokhozyaistvennye Tekhnologii: Moscow, Russia, 2021; pp. 40–51, (In Russian with English summary). [Google Scholar]
- Kutnyuk, P.I.; Bondarenko, Y.V.; Ivanova, T.V. Kariometrychnyy metod vyznachennya henetychnoho potentsialu nesuchosti kurey [Karyometric method of determining the genetic potential of laying hens]. Ptakhivnytstvo [Poult. Farming] 2001, 50, 75–81. (In Ukrainian) [Google Scholar]
- Chomchuen, K.; Tuntiyasawasdikul, V.; Chankitisakul, V.; Boonkum, W. Genetic Evaluation of body weights and egg production traits using a multi-trait animal model and selection index in Thai native synthetic chickens (Kaimook e-san2). Animals 2022, 12, 335. [Google Scholar] [CrossRef]
- Shtele, A.L. Problem of egg productivity in hens and its early prediction. Sel’skokhozyaistvennaya Biol. [Agric. Biol.] 2014, 6, 26–35. [Google Scholar] [CrossRef]
- Groen, A.F. Breeding objectives and selection strategies for layer production. In Poultry Genetic, Breeding and Biotechnology; Muir, W.M., Aggrey, S.E., Eds.; CABI Publishing: Wallingford, UK, 2003; pp. 101–112. Available online: https://books.google.com/books?id=IPSSDwAAQBAJ&pg=PA101 (accessed on 21 April 2025).
- Narushin, V.G.; Volkova, N.A.; Vetokh, A.N.; Dzhagaev, A.Y.; Volkova, L.A.; Griffin, D.K.; Romanov, M.N.; Zinovieva, N.A. Metabolic rate and egg production in Japanese quails can be predicted by assessing growth parameters of laying hens. Animals 2024, 14, 258. [Google Scholar] [CrossRef]
- Lukianova, V.D.; Kovalenko, V.P.; Kosenko, N.F.; Bondarenko, Y.V. Immunogenetic Method in Poultry Line Selection for Heterosis Effect on Egg Production. In Proceedings of the XVI World’s Poultry Congress, Proceedings & Abstracts, Rio de Janeiro, Brazil, 17–21 September 1978; World’s Poultry Science Association Brazil Branch: Sao Paulo, Brazil, 1978; Volume 12, pp. 2116–2122. Available online: https://scholar.google.com/citations?view_op=view_citation&citation_for_view=c1PnDYEAAAAJ:K3LRdlH-MEoC (accessed on 21 April 2025).
- Podstreshny, A.P.; Bondarenko, Y.V.; Rozhkovskiy, A.V.; Gintovt, V.E. Ispolzovanie markernyih priznakov pri sozdanii perspektivnyih kombinatsiy yaichnyih kur [The use of marker traits in the creation of promising combinations of egg-laying hens]. Ptitsevodstvo [Poult. Farming] 1984, 37, 9–14. (In Russian) [Google Scholar]
- Khvostik, V.P.; Bondarenko, Y.V.; Paskevych, G.A. Prohnozuvannya nesuchosti kurey riznoho henetychnoho pokhodzhennya [Prediction of laying hens of different genetic origins]. Nauk. Visn. Lʹvivsʹkoho Nats. un-tu vet. Medytsyny ta Biotekhnolohiy im. S. Z. Gzhytsʹkoho Ser. Silʹsʹkohospodarsʹki Nauky [Sci. Messin. LNU Vet. Med. Biotech. Ser. Agric. Sci.] 2023, 25, 60–65, (In Ukrainian with English summary). [Google Scholar] [CrossRef]
- Altahat, E.; Al-Sharafat, A.; Altarawneh, M. Factors affecting profitability of layer hens enterprises. Am. J. Agric. Biol. Sci. 2012, 7, 106–113. [Google Scholar] [CrossRef]
- Romanov, M.N. Qualitative and Quantitative Egg Characteristics in Laying Hens of Different Genotype. In Egg and Egg Products Quality, Proceedings of the 6th European Symposium on Quality of Eggs and Eggs Products, Zaragoza, Spain, 25–29 September 1995; World’s Poultry Science Association Spanish Branch: Madrid, Spain; pp. 203–206. Available online: https://scholar.google.com/citations?view_op=view_citation&citation_for_view=1fz40BYAAAAJ:KYgttONoxcsC (accessed on 21 April 2025).
- Mesías, F.J.; Martínez-Carrasco, F.; Martínez, J.M.; Gaspar, P. Functional and organic eggs as an alternative to conventional production: A conjoint analysis of consumers’ preferences. J. Sci. Food Agric. 2011, 91, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Li, D.; Liu, J.; Chen, S.; Qu, L.; Zheng, J.; Xu, G.; Yang, N. A genome-wide SNP scan reveals novel loci for egg production and quality traits in White Leghorn and brown-egg dwarf layers. PLoS ONE 2011, 6, e28600. [Google Scholar] [CrossRef] [PubMed]
- Shcherbatov, V.I.; Shkuro, A.G. Cycles and intervals in hen egg laying. E3S Web Conf. 2021, 285, 04009. [Google Scholar] [CrossRef]
- Tan, Y.G.; Xu, X.L.; Cao, H.Y.; Zhou, W.; Yin, Z.Z. Effect of age at first egg on reproduction performance and characterization of the hypothalamo-pituitary-gonadal axis in chickens. Poult. Sci. 2021, 100, 101325. [Google Scholar] [CrossRef]
- Xu, H.; Zeng, H.; Luo, C.; Zhang, D.; Wang, Q.; Sun, L.; Yang, L.; Zhou, M.; Nie, Q.; Zhang, X. Genetic effects of polymorphisms in candidate genes and the QTL region on chicken age at first egg. BMC Genet. 2011, 12, 33. [Google Scholar] [CrossRef]
- He, Z.; Ouyang, Q.; Chen, Q.; Song, Y.; Hu, J.; Hu, S.; He, H.; Li, L.; Liu, H.; Wang, J. Molecular mechanisms of hypothalamic-pituitary-ovarian/thyroid axis regulating age at first egg in geese. Poult. Sci. 2024, 103, 103478. [Google Scholar] [CrossRef]
- Dzhagaev, A.Y.; Volkova, N.A.; Zinovieva, N.A. Search for genes associated with the age at first egg in laying hens (Gallus gallus L.). Sel’skokhozyaistvennaya Biol. [Agric. Biol.] 2024, 59, 658–665. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Z.; Cao, D.; Liu, J.; Li, F.; Han, H.; Han, H.; Lei, Q.; Liu, W.; Li, D.; et al. Elucidation of the genetic determination of clutch traits in Chinese local chickens of the Laiwu Black breed. BMC Genom. 2023, 24, 686. [Google Scholar] [CrossRef]
- Vargas, D.; Galíndez, R.; De Basilio, V.; Martínez, G. Age at the first egg in Japanese quail (Coturnix coturnix japonica) under experimental conditions. Rev. Cient. Fac. Cienc. Vet. Univ. Zulia 2009, 19, 181–186. [Google Scholar]
- Balcha, K.A.; Mengesha, Y.T.; Senbeta, E.K.; Zeleke, N.A. Evaluation of different traits from day-old to age at first eggs of Fayoumi and White leghorn chickens and their reciprocal crossbreeds. J. Adv. Vet. Anim. Res. 2021, 8, 1–6. [Google Scholar] [CrossRef]
- Mesele, T.L. Reproduction and production performance of improved chickens, their production constraints, and opportunities under Ethiopian conditions. Trop. Anim. Health Prod. 2023, 55, 245. [Google Scholar] [CrossRef] [PubMed]
- Romanov, M.N. Impact of genotype, nutrition and management systems on goose production efficiency. In Proceedings of the 11th European Symposium on Waterfowl, Nantes, France, 8–10 September 1997; Association Groupe Francais de la WPSA (French Branch): Paris, France; World Society for the Protection of Animals French Branch: Nantes, France, 1997; pp. 33–41. Available online: https://kar.kent.ac.uk/46409/ (accessed on 21 April 2025).
- De Juan, A.F.; Scappaticcio, R.; Aguirre, L.; Mateos, G.G.; Camara, L. Effects of the composition of the pre-peak diet fed from 18 to 29 wk of age on egg production, egg quality, and the development of the gastrointestinal tract of brown-egg laying hens from 18 to 61 wk. J. Appl. Poult. Res. 2024, 33, 100415. [Google Scholar] [CrossRef]
- Bi, R.; Yang, M.; Liu, X.; Guo, F.; Hu, F.; Huang, J.; Abbas, W.; Xu, T.; Liu, W.; Wang, Z. Effects of chlorogenic acid on productive and reproductive performances, egg quality, antioxidant functions, and intestinal microenvironment in aged breeder laying hens. Poult. Sci. 2024, 103, 104060. [Google Scholar] [CrossRef]
- Kochish, I.I.; Surai, P.F.; Romanov, M.N.; Smolensky, V.I.; Nikonov, I.N.; Selina, M.V.; Myasnikova, O.V.; Kolesnikova, R.R. Metodicheskiye Rekomendatsii po Primeneniyu Osnov Tekhnologii Kormleniya Yaichnykh kur, Obespechivayushchey Vysokiy Protsent Realizatsii ikh Geneticheskogo Potentsiala Produktivnosti [Methodical Recommendations on the Application of the Foundations of the Technology of Feeding Egg-Type Chickens, Providing a High Percentage of Realization of Their Genetic Potential for Productivity]; Sel’skokhozyaistvennye Tekhnologii: Moscow, Russia, 2019; ISBN 978-5-6043642-6-0. Available online: https://www.researchgate.net/publication/371531267_Methodical_recommendations_on_the_application_of_the_foundations_of_the_technology_of_feeding_egg-type_chickens_providing_a_high_percentage_of_realization_of_their_genetic_potential_for_productivity_M (accessed on 21 April 2025). (In Russian)
- Romanov, M.N.; Grozina, A.A.; Ilina, L.A.; Laptev, G.Y.; Yildirim, E.A.; Filippova, V.A.; Tyurina, D.G.; Fisinin, V.I.; Kochish, I.I.; Griffin, D.K.; et al. From feed regulation to regulated feeding: Intestinal microbiome and performance optimization in broiler chickens in response to antibiotic and probiotic treatment. In Proceedings of the Life of Genomes 2022, Abstracts of the International Conference, Kazan, Russia, 23–24 November 2022; Research Center “Regulatory Genomics”, Institute of Fundamental Medicine and Biology, Kazan (Volga Region) Federal University: Kazan, Russia, 2022; pp. 44–45. Available online: http://lifeofgenomes.r-genomics.com/wp-content/uploads/2022/12/Absctracts.pdf (accessed on 21 April 2025).
- Bratyshko, N.I.; Prytulenko, O.V.; Lemesheva, M.M.; Tereshchenko, O.V. Rekomendatsiyi z Normuvannya Hodivli Silʹsʹkohospodarsʹkoyi Ptytsi [Recommendations on Rationing of Feeding of Agricultural Poultry], 3rd ed.; Tereshchenko, O.V., Ed.; Poultry Research Institute, NAAS, NTMT: Birky, Ukraine, 2010; Available online: https://scholar.google.com/citations?view_op=view_citation&citation_for_view=deB5xCwAAAAJ:RHpTSmoSYBkC (accessed on 21 April 2025). (In Ukrainian)
- Katerynych, O.O.; Pankova, S.M.; Tereshchenko, O.V.; Ruda, S.V.; Havilei, O.V.; Riabinina, O.V.; Muzyka, N.M.; Ionov, I.A. Vyroshchuvannia, Utrymannia ta Hodivlia Yaiechnykh ta Miaso-Yaiechnykh Kurei [Rearing, Maintenance and Feeding of Egg and Meat-Egg Hens: Scientific and Practical Guide]; Poultry Research Institute, DDSP NAAS: Birky, Ukraine, 2017; Available online: https://scholar.google.com/scholar?cluster=10199343115289849189 (accessed on 21 April 2025). (In Ukrainian)
- Shkurko, M.I.; Bondarenko, U.V.; Ostapenko, V.I. Produktyvnist’ molodnyaka kachok riznykh henotypiv v umovakh prysadybnoho hospodarstva [Productivity of young ducks of different genotypes under conditions of farming household]. Visn. Sums’kogo Nac. Agrar. Un-tu Ser. Tvarynnytstvo [Bull. Sumy Natl. Agrar. Univ. Ser. Livest.] 2015, 6, 75–78, (In Ukrainian with English summary). [Google Scholar]
- Shkurko, M.I.; Bondarenko, Y.V. Porivnyal’na kharakterystyka molodnyaka kachok riznykh henotypiv v umovakh prysadybnoho hospodarstva [Comparative Characteristics of Young Ducks of Different Genotypes in Homestead Farming Conditions]. In Proceedings of the Materials of the Scientific Practical Conference of Teachers, Postgraduates and Students of the Sumy National Agrarian University, Sumy, Ukraine, 20–24 April 2015; Sumy National Agrarian University: Sumy, Ukraine, 2015; Volume 2, p. 103. Available online: https://scholar.google.com/citations?view_op=view_citation&citation_for_view=c1PnDYEAAAAJ:nrtMV_XWKgEC (accessed on 21 April 2025). (In Ukrainian).
- Podstreshnyi, O.; Tereshchenko, O. Utrymannya doroslykh perepeliv [Maintenance of adult quails]. Ahrar. Krayina [Agrar. Country] 2012, 6, 8–9. (In Ukrainian) [Google Scholar]
- Wang, S.D.; Jan, D.F.; Yeh, L.T.; Wu, G.C.; Chen, L.R. Effect of exposure to long photoperiod during the rearing period on the age at first egg and the subsequent reproductive performance in geese. Anim. Reprod. Sci. 2002, 73, 227–234. [Google Scholar] [CrossRef]
- Kavtarashvili, A.S.; Fisinin, V.I.; Buyarov, V.S.; Kolokolnikova, T.N. The effects of lighting regimes on the oviposition time and egg quality in laying hens (review). Sel’skokhozyaistvennaya Biol. [Agric. Biol.] 2019, 54, 1095–1109. [Google Scholar] [CrossRef]
- Siopes, T.D. Initiation of egg production by turkey breeder hens: Sexual maturation and age at lighting. Poult. Sci. 2010, 89, 1490–1496. [Google Scholar] [CrossRef]
- Zhao, X.; Yue, S.; Zhang, Y.; Sun, J.; Peng, F.; Guo, Z. The TRHDE and TSHR genes regulate laying traits in domesticated Zi geese. Curr. Issues Mol. Biol. 2025, 47, 331. [Google Scholar] [CrossRef]
- Zhao, X.; Yue, S.; Sun, J.; Zhang, Y.; Peng, F.; Ma, Z.; He, H.; Li, M.; Guo, Z. RNA-seq reveals the effects of light on reproductive traits in domesticated geese. Braz. J. Poult. Sci. 2025, 27, eRBCA-2024-2016. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, N.; Yan, Y.; Li, G.; Liu, A.; Wu, G.; Sun, C. Genome-wide association analysis of egg production performance in chickens across the whole laying period. BMC Genet. 2019, 20, 67. [Google Scholar] [CrossRef]
- Chen, A.; Zhao, X.; Wen, J.; Zhao, X.; Wang, G.; Zhang, X.; Ren, X.; Zhang, Y.; Cheng, X.; Yu, X.; et al. Genetic parameter estimation and molecular foundation of chicken egg-laying trait. Poult. Sci. 2024, 103, 103627. [Google Scholar] [CrossRef]
- Ou, J.T.; Tang, S.Q.; Sun, D.X.; Zhang, Y. Polymorphisms of three neuroendocrine-correlated genes associated with growth and reproductive traits in the chicken. Poult. Sci. 2009, 88, 722–727. [Google Scholar] [CrossRef]
- Gorodnaja, A.V.; Moiseeva, I.G.; Glazko, V.I.; Sevastjanova, A.A. Polimorfizm nekotorykh genetiko-biokhimicheskikh sistem u kur [Polymorphism of some genetic-biochemical systems in chicken]. Dokl. Ross. Akad. s-kh. Nauk [Proc. Russ. Acad. Agric. Sci.] 1997, 2, 31–33, (In Russian with English summary). [Google Scholar]
- Wolc, A.; Arango, J.; Jankowski, T.; Dunn, I.; Settar, P.; Fulton, J.E.; O’Sullivan, N.P.; Preisinger, R.; Fernando, R.L.; Garrick, D.J.; et al. Genome-wide association study for egg production and quality in layer chickens. J. Anim. Breed. Genet. 2014, 131, 173–182. [Google Scholar] [CrossRef]
- Dementeva, N.V.; Romanov, M.N.; Kudinov, A.A.; Mitrofanova, O.V.; Stanishevskaya, O.I.; Terletsky, V.P.; Fedorova, E.S.; Nikitkina, E.V.; Plemyashov, K.V. Studying the structure of a gene pool population of the Russian White chicken breed by genome-wide SNP scan. Sel’skokhozyaistvennaya Biol. [Agric. Biol.] 2017, 52, 1166–1174. [Google Scholar] [CrossRef]
- Zhang, L.; Zheng, M.Q.; Liu, R.R.; Wen, J.; Wu, D.; Hu, Y.D.; Sun, Y.F.; Li, P.; Liu, L.; Zhao, G.P. Genome-wide association of thymus and spleen mass in chicken. Sci. Agric. Sin. 2012, 45, 3165–3175. [Google Scholar] [CrossRef]
- Dementeva, N.V.; Kudinov, A.A.; Mitrofanova, O.V.; Stanishevskaya, O.I.; Fedorova, E.S.; Romanov, M.N. Genome-wide association study of reproductive traits in a gene pool breed of the Russian White chickens. Reprod. Domest. Anim. 2018, 53 (Suppl. S2), 123–124. [Google Scholar]
- Volkova, N.A.; Romanov, M.N.; Vetokh, A.N.; Larionova, P.V.; Volkova, L.A.; Abdelmanova, A.S.; Sermyagin, A.A.; Griffin, D.K.; Zinovieva, N.A. Genome-wide association study reveals the genetic architecture of growth and meat production traits in a chicken F2 resource population. Genes 2024, 15, 1246. [Google Scholar] [CrossRef]
- Tan, L.; Xu, N.; Liu, Z.; Cai, X.; Kong, Y.; Wen, Z.; Wang, Y.; Zhao, Y. Genome-wide association studies on longitudinal phenotypes reveal genetic mechanisms of egg production in chickens. Poult. Sci. 2025, 105280, in press. [Google Scholar] [CrossRef]
- Tu, T.-C.; Lin, C.-J.; Liu, M.-C.; Hsu, Z.-T.; Chen, C.-F. Genomic prediction and genome-wide association study for growth-related traits in Taiwan country chicken. Animals 2025, 15, 376. [Google Scholar] [CrossRef]
- Gao, G.; Gao, D.; Zhao, X.; Xu, S.; Zhang, K.; Wu, R.; Yin, C.; Li, J.; Xie, Y.; Hu, S.; et al. Genome-wide association study-based identification of SNPs and haplotypes associated with goose reproductive performance and egg quality. Front. Genet. 2021, 12, 602583. [Google Scholar] [CrossRef]
- Volkova, N.A.; Romanov, M.N.; Abdelmanova, A.S.; Larionova, P.V.; German, N.Y.; Vetokh, A.N.; Shakhin, A.V.; Volkova, L.A.; Sermyagin, A.A.; Anshakov, D.V.; et al. Genome-wide association study revealed putative SNPs and candidate genes associated with growth and meat traits in Japanese quail. Genes 2024, 15, 294. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, Y.; Wu, Q.; Lin, R.; Chen, H.; Zhang, M.; Lin, J.; Xu, E.; Li, M.; Cai, Y.; et al. Whole-genome sequencing identifies potential candidate genes for egg production traits in laying ducks (Anas platyrhynchos). Sci. Rep. 2023, 13, 1821. [Google Scholar] [CrossRef]
- Lu, Y.; Wei, B.; Yang, Q.; Han, X.; He, X.; Tao, Q.; Jiang, S.; Xu, M.; Bai, Y.; Zhang, T.; et al. Identification of candidate genes affecting the tibia quality in Nonghua duck. Poult. Sci. 2024, 103, 103515. [Google Scholar] [CrossRef]
- Cai, K.; Liu, R.; Wei, L.; Wang, X.; Cui, H.; Luo, N.; Wen, J.; Chang, Y.; Zhao, G. Genome-wide association analysis identify candidate genes for feed efficiency and growth traits in Wenchang chickens. BMC Genom. 2024, 25, 645. [Google Scholar] [CrossRef]
- Shah, T.M.; Patel, N.V.; Patel, A.B.; Upadhyay, M.R.; Mohapatra, A.; Singh, K.M.; Deshpande, S.D.; Joshi, C.G. A genome-wide approach to screen for genetic variants in broilers (Gallus gallus) with divergent feed conversion ratio. Mol. Genet. Genom. 2016, 291, 1715–1725. [Google Scholar] [CrossRef]
- Li, H.; Li, S.; Zhang, H.; Gu, J.; Dai, Y.; Wu, R.; Wang, Y.; Han, R.; Sun, G.; Zhang, Y.; et al. Integrated GWAS and transcriptome analysis reveals key genes associated with muscle fibre and fat traits in Gushi chicken. Br. Poult. Sci. 2025, 66, 31–41. [Google Scholar] [CrossRef]
- Fan, S.; Yuan, P.; Li, S.; Li, H.; Zhai, B.; Li, Y.; Zhang, H.; Gu, J.; Li, H.; Tian, Y.; et al. Genetic architecture and key regulatory genes of fatty acid composition in Gushi chicken breast muscle determined by GWAS and WGCNA. BMC Genom. 2023, 24, 434. [Google Scholar] [CrossRef]
- Luo, N.; An, B.; Wei, L.; Wen, J.; Zhao, G. Identification of molecular markers associated with body size traits through genome-wide association analysis in Wenchang chickens. Sci. Agric. Sin. 2024, 57, 2046–2060. [Google Scholar] [CrossRef]
- Zhang, G.M.; Liu, P.H.; Chen, L.; Zheng, J.M.; Zhao, G.P.; Xing, W.H.; Wen, J.; Li, Q.H. Genome-wide association study identifies variants associated with semen volume in white-feathered broilers. Anim. Genet. 2023, 54, 803–807. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.; Sun, Y.; Liu, N.; Xue, F.; Li, Y.; Xu, S.; Ye, J.; Zhang, L.; Chen, Y.; Chen, J. Single SNP- and pathway-based genome-wide association studies for beak deformity in chickens using high-density 600K SNP arrays. BMC Genom. 2018, 19, 501. [Google Scholar] [CrossRef] [PubMed]
- Walugembe, M.; Amuzu-Aweh, E.N.; Botchway, P.K.; Naazie, A.; Aning, G.; Wang, Y.; Saelao, P.; Kelly, T.; Gallardo, R.A.; Zhou, H.; et al. Genetic basis of response of Ghanaian local chickens to infection with a lentogenic Newcastle disease virus. Front. Genet. 2020, 11, 739. [Google Scholar] [CrossRef] [PubMed]
- Parveen, A.; Jackson, C.D.; Dey, S.; Tarrant, K.; Anthony, N.; Rhoads, D.D. Identification and validation of quantitative trait loci for ascites syndrome in broiler chickens using whole genome resequencing. BMC Genet. 2020, 21, 54. [Google Scholar] [CrossRef]
- Abdelmanova, A.S.; Dotsev, A.V.; Romanov, M.N.; Stanishevskaya, O.I.; Gladyr, E.A.; Rodionov, A.N.; Vetokh, A.N.; Volkova, N.A.; Fedorova, E.S.; Gusev, I.V.; et al. Unveiling comparative genomic trajectories of selection and key candidate genes in egg-type Russian White and meat-type White Cornish chickens. Biology 2021, 10, 876. [Google Scholar] [CrossRef]
- Moiseeva, I.G. Izmenchivost’ i nasleduyemost’ nekotorykh pokazateley kachestva yaits u kur russkoy beloy porody [Variability and heritability of some features of egg quality in Russkaya Belaya chickens]. Tr. In-ta Genet. AN SSSR [Trudy Inst. Genet.] 1964, 31, 302–308. (In Russian) [Google Scholar]
- Moiseeva, I.G. Soderzhaniye lipidov i kholesterina v yaytsakh kur russkoy beloy porody [Content of lipids and cholesterol in eggs of Russian White chickens]. Collect. Works Young Sci. All-Union Res. Tech. Poult. Inst. 1966, 8, 225–235. (In Russian) [Google Scholar]
- Romanov, M.N.; Shakhin, A.V.; Abdelmanova, A.S.; Volkova, N.A.; Efimov, D.N.; Fisinin, V.I.; Korshunova, L.G.; Anshakov, D.V.; Dotsev, A.V.; Griffin, D.K.; et al. Dissecting selective signatures and candidate genes in grandparent lines subject to high selection pressure for broiler production and in a local Russian chicken breed of Ushanka. Genes 2024, 15, 524. [Google Scholar] [CrossRef]
- Bondarenko, Y.V.; Sergheyeva, V.D.; Kuranova, E.N.; Krasnozhon, S.A.; Romanov, M.N. Autoseksnaya materinskaya forma myasnykh kur [Autosexing maternal form of meat-type chickens]. Ptitsevodstvo [Poult. Farming] 1987, 40, 6–11. (In Russian) [Google Scholar]
- Melnyk, V.O.; Ivko, I.I.; Tereshchenko, O.V. Resursozberihayuchi Tekhnolohiyi Vyroshchuvannya, Utrymannya ta Hodivli Yayechnykh i M’yaso-Yayechnykh Kurey: Metodychni Rekomendatsiyi [Resource-Saving Technologies for Raising, Keeping and Feeding Egg and Meat-Egg Chickens: Methodological Recommendations]; Poultry Research Institute, NAAS: Birky, Ukraine, 2011; Available online: https://scholar.google.com/citations?view_op=view_citation&citation_for_view=deB5xCwAAAAJ:FAceZFleit8C (accessed on 21 April 2025). (In Ukrainian)
- Tereshchenko, A.V.; Artemenko, A.B.; Pudov, V.Y. Skrytyj istocnik uvelicenia proizvodstva cyplat-brojlerov [A hidden source of increasing the production of broiler chickens]. Eksklyuziv Agro [Exclus. Agro] 2007, 4, 64–65. (In Russian) [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing: Vienna, Austria, 2018. Available online: https://www.r-project.org/ (accessed on 21 April 2025).
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S. PLINK 1.9.; Center for Human Genetic Research: Boston, MA, USA; Massachusetts General Hospital: Boston, MA, USA; Broad Institute of Harvard & MIT: Cambridge, MA, USA, 2017; Available online: https://zzz.bwh.harvard.edu/plink/index.shtml (accessed on 21 April 2025).
- DataCamp. Principal Component Analysis in R Tutorial. In Tutorials, R Programming; DataCamp, Inc.: New York, NY, USA, 2023; Available online: https://www.datacamp.com/tutorial/pca-analysis-r (accessed on 21 April 2025).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009; ISBN 978-0-387-98141-3. [Google Scholar] [CrossRef]
- GitHub. ggplot2. Tidyverse, GitHub, Inc.: San Francisco, CA, USA, 2023. Available online: https://github.com/tidyverse/ggplot2 (accessed on 21 April 2025).
- R Core Team. R-4, R Foundation for Statistical Computing: Vienna, Austria, 2020. Available online: https://cran.r-project.org/src/base/R-4/ (accessed on 21 April 2025).
- GitHub. qqman, GitHub, Inc.: San Francisco, CA, USA, 2024. Available online: https://github.com/qqman (accessed on 21 April 2025).
- Turner, S.D. qqman: An R package for visualizing GWAS results using Q-Q and Manhattan plots. J. Open Source Softw. 2018, 3, 731. [Google Scholar] [CrossRef]
- NCBI. Gallus gallus Genome Assembly GRCg6a. Genome; National Library of Medicine: Bethesda, MD, USA, 2018. Available online: https://ncbi.nlm.nih.gov/datasets/genome/GCF_000002315.6/ (accessed on 21 April 2025).
- NCBI. Gallus gallus (Chicken). Genome; National Library of Medicine: Bethesda, MD, USA, 2024. Available online: https://www.ncbi.nlm.nih.gov/datasets/genome/?taxon=9031 (accessed on 21 April 2025).
- Kinsella, R.J.; Kähäri, A.; Haider, S.; Zamora, J.; Proctor, G.; Spudich, G.; Almeida-King, J.; Staines, D.; Derwent, P.; Kerhornou, A.; et al. Ensembl BioMarts: A hub for data retrieval across taxonomic space. Database 2011, 2011, bar030. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Ren, J.; Gao, Z.; Lu, Y.; Li, M.; Hong, J.; Wu, J.; Wu, D.; Deng, W.; Xi, D.; Chong, Y. Application of GWAS and mGWAS in livestock and poultry breeding. Animals 2024, 14, 2382. [Google Scholar] [CrossRef]
- Sharma, A.; Lee, J.S.; Dang, C.G.; Sudrajad, P.; Kim, H.C.; Yeon, S.H.; Kang, H.S.; Lee, S.-H. Stories and challenges of genome wide association studies in livestock—A review. Asian-Australas. J. Anim. Sci. 2015, 28, 1371–1379. [Google Scholar] [CrossRef]
- International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 2004, 432, 695–716. [Google Scholar] [CrossRef]
- Dodgson, J.B.; Romanov, M.N. The Chicken Genome: From Maps to Sequence. In Proceedings of the 8th International Symposium on Avian Endocrinology: Symposium Talk and Plenary Lecture Abstracts, Scottsdale, AZ, USA, 6–11 June 2004; Arizona State University: Scottsdale, AZ, USA, 2004. Abstract T26. Available online: https://kar.kent.ac.uk/46516/ (accessed on 21 April 2025).
- Haqani, M.I.; Nakano, M.; Nagano, A.J.; Nakamura, Y.; Tsudzuki, M. Association analysis of production traits of Japanese quail (Coturnix japonica) using restriction-site associated DNA sequencing. Sci. Rep. 2023, 13, 21307. [Google Scholar] [CrossRef]
- Haqani, M.I.; Nomura, S.; Nakano, M.; Goto, T.; Nagano, A.J.; Takenouchi, A.; Nakamura, Y.; Ishikawa, A.; Tsudzuki, M. Mapping of quantitative trait loci controlling egg-quality and -production traits in Japanese quail (Coturnix japonica) using restriction-site associated DNA sequencing. Genes 2021, 12, 735. [Google Scholar] [CrossRef]
- Lithgow, P.E.; O’Connor, R.; Smith, D.; Fonseka, G.; Rathje, C.; Frodsham, R.; O’Brien, P.C.; Ferguson-Smith, M.A.; Skinner, B.M.; Griffin, D.K.; et al. Novel Tools for Characterising Inter- and Intra-chromosomal Rearrangements in Avian Microchromosomes. In Proceedings of the 2014 Meeting on Avian Model Systems, Cold Spring Harbor, New York, NY, USA, 5–8 March 2014; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2014; p. 56. Available online: https://kar.kent.ac.uk/46692/ (accessed on 21 April 2025).
- Sazanov, A.A.; Sazanova, A.L.; Nefedov, M.D.; Griffin, D.K.; Romanov, M.N. A pair of gametologous genes provides further insights into avian comparative cytogenomics. Biologia 2023, 78, 2737–2746. [Google Scholar] [CrossRef]
- Tuttle, E.; Korody, M.; Lear, T.; Gonser, R.; Houck, M.; Ryder, O.; Romanov, M.; Balakrishnan, C.; Bergland, A.; Warren, W. Whole Genome Sequence of the Behaviorally Polymorphic White-throated Sparrow. 1: Mapping Genes for Sociogenomics. In Proceedings of the Evolution 2014 Conference, Raleigh, NC, USA, 20–24 June 2014; Society for the Study of Evolution (SSE): St. Louis, MO, USA; Society of Systematic Biologists (SSB): Windsor, WI, USA; American Society of Naturalists (ASN): Pacific Grove, CA, USA, 2014; p. 183. Available online: https://kar.kent.ac.uk/46698/ (accessed on 21 April 2025).
- Griffin, D.K.; Kretschmer, R.; Srikulnath, K.; Singchat, W.; O’Connor, R.E.; Romanov, M.N. Insights into avian molecular cytogenetics—With reptilian comparisons. Mol. Cytogenet. 2024, 17, 24. [Google Scholar] [CrossRef] [PubMed]
- Sazanov, A.A.; Sazanova, A.L.; Romanov, M.N.; Stekol’nikova, V.A.; Malewski, T.; Korczak, M.; Jaszczak, K.; Smirnov, A.F. Molecular Organization of Chicken Genome. In Proceedings of the Conference “Genomic and Microarray Analysis in Biology and Medicine”, Sucha Beskidzka, Poland, 25–28 June 2005; p. 26. Available online: https://kar.kent.ac.uk/46616/ (accessed on 21 April 2025).
- Jahuey-Martínez, F.J.; Martínez-Quintana, J.A.; Rodríguez-Almeida, F.A.; Parra-Bracamonte, G.M. Exploration and enrichment analysis of the QTLome for important traits in livestock species. Genes 2024, 15, 1513. [Google Scholar] [CrossRef]
- Salvi, S.; Tuberosa, R. The crop QTLome comes of age. Curr. Opin. Biotechnol. 2015, 32, 179–185. [Google Scholar] [CrossRef]
- Ji, H.; Xu, Y.; Teng, G. Predicting egg production rate and egg weight of broiler breeders based on machine learning and Shapley additive explanations. Poult. Sci. 2025, 104, 104458. [Google Scholar] [CrossRef]
- Wolc, A.; Jankowski, T.; Arango, J.; Settar, P.; Fulton, J.E.; O’Sullivan, N.P.; Dekkers, J.C.M. Investigating the genetic determination of clutch traits in laying hens. Poult. Sci. 2019, 98, 39–45. [Google Scholar] [CrossRef]
- Miao, X.; Huang, Z.; Liu, J.; Zhang, L.; Feng, Y.; Zhang, Y.; Li, D.; Ning, Z. Genomically selected genes associated with a high rate of egg production in Puan Panjiang black-bone chickens. Animals 2025, 15, 363. [Google Scholar] [CrossRef]
- Lei, Q.; Zhang, S.; Wang, J.; Qi, C.; Liu, J.; Cao, D.; Li, F.; Han, H.; Liu, W.; Li, D.; et al. Genome-wide association studies of egg production traits by whole genome sequencing of Laiwu Black chicken. Poult. Sci. 2024, 103, 103705. [Google Scholar] [CrossRef]
- Wang, D.; Tan, L.; Zhi, Y.; Bu, L.; Wang, Y.; Wang, Z.; Guo, Y.; Tian, W.; Xu, C.; Li, D.; et al. Genome-wide variation study and inter-tissue communication analysis unveil regulatory mechanisms of egg-laying performance in chickens. Nat. Commun. 2024, 15, 7069. [Google Scholar] [CrossRef]
- Du, Y.; Liu, L.; He, Y.; Dou, T.; Jia, J.; Ge, C. Endocrine and genetic factors affecting egg laying performance in chickens: A review. Br. Poult. Sci. 2020, 61, 538–549. [Google Scholar] [CrossRef]
- Zhao, X.; Nie, C.; Zhang, J.; Li, X.; Zhu, T.; Guan, Z.; Chen, Y.; Wang, L.; Lv, X.Z.; Yang, W.; et al. Identification of candidate genomic regions for chicken egg number traits based on genome-wide association study. BMC Genom. 2021, 22, 610. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Wu, Y.; Shen, J.; Pan, A.; Zhang, H.; Sun, J.; Liang, Z.; Huang, T.; Du, J.; Pi, J. Genome-wide association study of egg production traits in Shuanglian chickens using whole genome sequencing. Genes 2023, 14, 2129. [Google Scholar] [CrossRef] [PubMed]
- Yi, G.; Shen, M.; Yuan, J.; Sun, C.; Duan, Z.; Qu, L.; Dou, T.; Ma, M.; Lu, J.; Guo, J.; et al. Genome-wide association study dissects genetic architecture underlying longitudinal egg weights in chickens. BMC Genom. 2015, 16, 746. [Google Scholar] [CrossRef]
- Ma, X.; Ying, F.; Li, Z.; Bai, L.; Wang, M.; Zhu, D.; Liu, D.; Wen, J.; Zhao, G.; Liu, R. New insights into the genetic loci related to egg weight and age at first egg traits in broiler breeder. Poult. Sci. 2024, 103, 103613. [Google Scholar] [CrossRef]
- Yang, S.; Ning, C.; Yang, C.; Li, W.; Zhang, Q.; Wang, D.; Tang, H. Identify candidate genes associated with the weight and egg quality traits in Wenshui green shell-laying chickens by the copy number variation-based genome-wide association study. Vet. Sci. 2024, 11, 76. [Google Scholar] [CrossRef]
- Munoz-Sanjuan, I.; Smallwood, P.M.; Nathans, J. Isoform diversity among fibroblast growth factor homologous factors is generated by alternative promoter usage and differential splicing. J. Biol. Chem. 2000, 275, 2589–2597. [Google Scholar] [CrossRef] [PubMed]
- Berradi, H.; Taouis, M.; Cassy, S.; Rideau, N. Glucokinase in chicken (Gallus gallus). Partial cDNA cloning, immunodetection and activity determination. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2005, 141, 129–139. [Google Scholar] [CrossRef]
- Li, G.; Yang, X.; Li, J.; Zhang, B. Genome-wide analysis of lncRNA and mRNA expression in the uterus of laying hens during aging. Genes 2023, 14, 639. [Google Scholar] [CrossRef]
- Chen, X.; Li, X.; Zhong, C.; Jiang, X.; Wu, G.; Li, G.; Yan, Y.; Yang, N.; Sun, C. Genetic patterns and genome-wide association analysis of eggshell quality traits of egg-type chicken across an extended laying period. Poult. Sci. 2024, 103, 103458. [Google Scholar] [CrossRef]
- Rideau, N.; Berradi, H.; Skiba-Cassy, S.; Panserat, S.; Cailleau-Audouin, E.; Dupont, J. Induction of glucokinase in chicken liver by dietary carbohydrates. Gen. Comp. Endocrinol. 2008, 158, 173–177. [Google Scholar] [CrossRef]
- Rideau, N.; Derouet, M.; Grimsby, J.; Simon, J. Glucokinase activation induces potent hypoglycemia without recruiting insulin and inhibits food intake in chicken. Gen. Comp. Endocrinol. 2010, 169, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Berradi, H.; Bernadet, M.D.; Guy, G.; Rideau, N. Expression of the glucokinase gene in mule duck liver and glucokinase activities in chicken and mule duck livers. Poult. Sci. 2007, 86, 2216–2220. [Google Scholar] [CrossRef] [PubMed]
- Kanakachari, M.; Ashwini, R.; Chatterjee, R.N.; Bhattacharya, T.K. Embryonic transcriptome unravels mechanisms and pathways underlying embryonic development with respect to muscle growth, egg production, and plumage formation in native and broiler chickens. Front. Genet. 2022, 13, 990849. [Google Scholar] [CrossRef]
- Christensen, V.L.; Grimes, J.L.; Donaldson, W.E.; Lerner, S. Correlation of body weight with hatchling blood glucose concentration and its relationship to embryonic survival. Poult. Sci. 2000, 79, 1817–1822. [Google Scholar] [CrossRef]
- Shanawany, M.M. Inter-relationship between egg weight, parental age and embryonic development. Br. Poult. Sci. 1984, 25, 449–455. [Google Scholar] [CrossRef]
- Fathi, M.; Abou-Emera, O.; Al-Homidan, I.; Galal, A.; Rayan, G. Effect of genotype and egg weight on hatchability properties and embryonic mortality pattern of native chicken populations. Poult. Sci. 2022, 101, 102129. [Google Scholar] [CrossRef]
- Duman, M.; Şekeroğlu, A. Effect of egg weights on hatching results, broiler performance and some stress parameters. Rev. Bras. Cienc. Avic. 2017, 19, 255–262. [Google Scholar] [CrossRef]
- Siopes, T.D.; Millam, J.R.; Steinman, M.Q. Initiating egg production in turkey breeder hens: Thyroid hormone involvement. Poult. Sci. 2010, 89, 2265–2272. [Google Scholar] [CrossRef]
- Liu, H.; Wang, L.; Guo, Z.; Xu, Q.; Fan, W.; Xu, Y.; Hu, J.; Zhang, Y.; Tang, J.; Xie, M.; et al. Genome-wide association and selective sweep analyses reveal genetic loci for FCR of egg production traits in ducks. Genet. Sel. Evol. 2021, 53, 98. [Google Scholar] [CrossRef]
- Harms, R.H.; Waldroup, P.W. Length of laying cycle as influenced by dietary protein level. Poult. Sci. 1963, 42, 1195–1197. [Google Scholar] [CrossRef]
- Arulnathan, V.; Turner, I.; Bamber, N.; Ferdous, J.; Grassauer, F.; Doyon, M.; Pelletier, N. A systematic review of potential productivity, egg quality, and animal welfare implications of extended lay cycles in commercial laying hens in Canada. Poult. Sci. 2024, 103, 103475. [Google Scholar] [CrossRef] [PubMed]
- Bain, M.M.; Nys, Y.; Dunn, I.C. Increasing persistency in lay and stabilising egg quality in longer laying cycles. What are the challenges? Br. Poult. Sci. 2016, 57, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Fan, X.; Tian, K.; Yan, S.; Xu, C.; Tian, Y.; Xiao, C.; Jia, X.; Shi, J.; Bai, Y.; et al. Dynamic expression profile of follicles at different stages in high- and low-production laying hens. Genes 2023, 15, 40. [Google Scholar] [CrossRef]
- Bouvarel, I.; Nys, Y.; Lescoat, P. 12-Hen nutrition for sustained egg quality. In Egg Chemistry, Production and Consumption, Woodhead Publishing Series in Food Science, Technology and Nutrition, Improving the Safety and Quality of Eggs and Egg Products; Nys, Y., Bain, M., Van Immerseel, F., Eds.; Woodhead Publishing Ltd.: Sawston, UK, 2011; pp. 261–299. ISBN 9781845697549. [Google Scholar] [CrossRef]
- Kazemi, V.; Zarghi, H.; Golian, A. The effect of dietary energy and nutrients density on performance, egg components, egg quality, and profits of Hy-Line W-36 during the peak stage of first laying cycle. Ital. J. Anim. Sci. 2022, 21, 1034–1046. [Google Scholar] [CrossRef]
- Jahan, A.A.; Dao, T.H.; Morgan, N.K.; Crowley, T.M.; Moss, A.F. Effects of AM/PM diets on laying performance, egg quality, and nutrient utilisation in free-range laying hens. Appl. Sci. 2024, 14, 2163. [Google Scholar] [CrossRef]
- Volyanskaya, A.R.; Akberdin, I.R.; Kulyashov, M.A.; Yevshin, I.S.; Romanov, M.N.; Shagimardanova, E.I.; Gusev, O.A.; Kolpakov, F.A. A bird’s-eye overview of molecular mechanisms regulating feed intake in chickens—With mammalian comparisons. Anim. Nutr. 2024, 17, 61–74. [Google Scholar] [CrossRef]
- Proszkowiec-Weglarz, M.; Dupont, J.; Rideau, N.; Gespach, C.; Simon, J.; Porter, T.E. Insulin immuno-neutralization decreases food intake in chickens without altering hypothalamic transcripts involved in food intake and metabolism. Poult. Sci. 2017, 96, 4409–4418. [Google Scholar] [CrossRef]
- Bédécarrats, G.Y.; Hanlon, C. Chapter 7-Effect of lighting and photoperiod on chicken egg production and quality. In Egg Innovations and Strategies for Improvements; Hester, P.Y., Ed.; Academic Press: Cambridge, MA, USA; Elsevier: London, UK, 2017; pp. 65–75. ISBN 9780128008799. [Google Scholar] [CrossRef]
- Roly, Z.Y.; Godini, R.; Estermann, M.A.; Major, A.T.; Pocock, R.; Smith, C.A. Transcriptional landscape of the embryonic chicken Müllerian duct. BMC Genom. 2020, 21, 688. [Google Scholar] [CrossRef]
- Qin, N.; Fan, X.C.; Zhang, Y.Y.; Xu, X.X.; Tyasi, T.L.; Jing, Y.; Mu, F.; Wei, M.L.; Xu, R.F. New insights into implication of the SLIT/ROBO pathway in the prehierarchical follicle development of hen ovary. Poult. Sci. 2015, 94, 2235–2246. [Google Scholar] [CrossRef]
- Gao, J.; Xu, W.; Zeng, T.; Tian, Y.; Wu, C.; Liu, S.; Zhao, Y.; Zhou, S.; Lin, X.; Cao, H.; et al. Genome-wide association study of egg-laying traits and egg quality in LingKun chickens. Front. Vet. Sci. 2022, 9, 877739. [Google Scholar] [CrossRef]
- Liu, Z.; Zheng, Q.; Zhang, X.; Lu, L. Microarray analysis of genes involved with shell strength in layer shell gland at the early stage of active calcification. Asian-Australas. J. Anim. Sci. 2013, 26, 609–624. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.-T.; Lin, C.-Y.; Liou, J.-S.; Fan, Y.-H.; Chiou, S.-H.; Huang, C.-W.; Wu, C.-P.; Lin, E.-C.; Chen, C.-F.; Lee, Y.-P.; et al. Differentially expressed transcripts in shell glands from low and high egg production strains of chickens using cDNA microarrays. Anim. Reprod. Sci. 2007, 101, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.; Sun, C.; Shen, M.; Wang, K.; Yang, N.; Zheng, J.; Xu, G. Genetic architecture dissection by genome-wide association analysis reveals avian eggshell ultrastructure traits. Sci. Rep. 2016, 6, 28836. [Google Scholar] [CrossRef]
- Sanchez-Ajofrin, I.; Iniesta-Cuerda, M.; Martin-Maestro, A.; Peris-Frau, P.; Garde, J.J.; Soler, A.J. The effect of maternal hyperthermia on oocyte quality in sheep. Reprod. Domest. Anim. 2018, 53 (Suppl. S2), 192–193. [Google Scholar]
- Jehl, F.; Désert, C.; Klopp, C.; Brenet, M.; Rau, A.; Leroux, S.; Boutin, M.; Lagoutte, L.; Muret, K.; Blum, Y.; et al. Chicken adaptive response to low energy diet: Main role of the hypothalamic lipid metabolism revealed by a phenotypic and multi-tissue transcriptomic approach. BMC Genom. 2019, 20, 1033. [Google Scholar] [CrossRef]
- Yuan, J.; Wang, K.; Yi, G.; Ma, M.; Dou, T.; Sun, C.; Qu, L.-J.; Shen, M.; Qu, L.; Yang, N. Genome-wide association studies for feed intake and efficiency in two laying periods of chickens. Genet. Sel. Evol. 2015, 47, 82. [Google Scholar] [CrossRef]
- Chen, A.; Zhao, X.; Zhao, X.; Wang, G.; Zhang, X.; Ren, X.; Zhang, Y.; Cheng, X.; Yu, X.; Wang, H.; et al. Genetic foundation of male spur length and its correlation with female egg production in chickens. Animals 2024, 14, 1780. [Google Scholar] [CrossRef]
- Dou, D.; Shen, L.; Zhou, J.; Cao, Z.; Luan, P.; Li, Y.; Xiao, F.; Guo, H.; Li, H.; Zhang, H. Genome-wide association studies for growth traits in broilers. BMC Genom. Data 2022, 23, 1. [Google Scholar] [CrossRef]
- Moroudi, R.S.; Mahyari, S.A.; Torshizi, R.V.; Lanjanian, H.; Masoudi-Nejad, A. Identification of new genes and quantitative trait locis associated with growth curve parameters in F2 chicken population using genome-wide association study. Anim. Genet. 2021, 52, 171–184. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Y.; Li, Y.; Xiao, F.; Guo, H.; Gao, H.; Wang, N.; Zhang, H.; Li, H. Genome-wide association study and selective sweep analysis reveal the genetic architecture of body weights in a chicken F2 resource population. Front. Vet. Sci. 2022, 9, 875454. [Google Scholar] [CrossRef]
- Abdalla, E.A.E.; Makanjuola, B.O.; Wood, B.J.; Baes, C.F. Genome-wide association study reveals candidate genes relevant to body weight in female turkeys (Meleagris gallopavo). PLoS ONE 2022, 17, e0264838. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Liu, H.; Xi, Y.; Lu, Y.; Han, X.; He, X.; Qi, J.; Zhu, Y.; He, H.; Wang, J.; et al. Genome-wide association study for bone quality of ducks during the laying period. Poult. Sci. 2024, 103, 103575. [Google Scholar] [CrossRef]
- Burge, S.; Kelly, E.; Lonsdale, D.; Mutowo-Muellenet, P.; McAnulla, C.; Mitchell, A.; Sangrador-Vegas, A.; Yong, S.Y.; Mulder, N.; Hunter, S. Manual GO annotation of predictive protein signatures: The InterPro approach to GO curation. Database 2012, 2012, bar068. [Google Scholar] [CrossRef]
- Ye, H.; Ji, C.; Liu, X.; Bello, S.F.; Guo, L.; Fang, X.; Lin, D.; Mo, Y.; Lei, Z.; Cai, B.; et al. Improvement of the accuracy of breeding value prediction for egg production traits in Muscovy duck using low-coverage whole-genome sequence data. Poult. Sci. 2025, 104, 104812. [Google Scholar] [CrossRef]
- Guo, Y.; Chen, Y.; Guo, H.; Wang, B.; Xiong, Y.; Ding, J.; Li, J. Genome-wide association study revealed candidate genes associated with egg-laying time traits in layer chicken. Poult. Sci. 2025, 104, 105255. [Google Scholar] [CrossRef]
- Romanov, M.N.; Sazanov, A.A.; Moiseyeva, I.G.; Smirnov, A.F. Poultry. In Genome Mapping and Genomics in Animals, Vol. 3: Genome Mapping and Genomics in Domestic Animals; Cockett, N.E., Kole, C., Eds.; Springer: Berlin/Heidelberg, Germany; New York, NY, USA, 2009; pp. 75–141. ISBN 978-3-540-73834-3/978-3-540-73835-0. [Google Scholar] [CrossRef]
- Yang, W.; Yu, S.; Song, D.; Lin, W.; Xu, H.; Lang, X.; Zhang, C.; Guo, L.; Chen, X. A genome-wide association study identified candidate genes associated with egg quality traits in Muscovy duck. BMC Genom. 2025, 26, 422. [Google Scholar] [CrossRef]

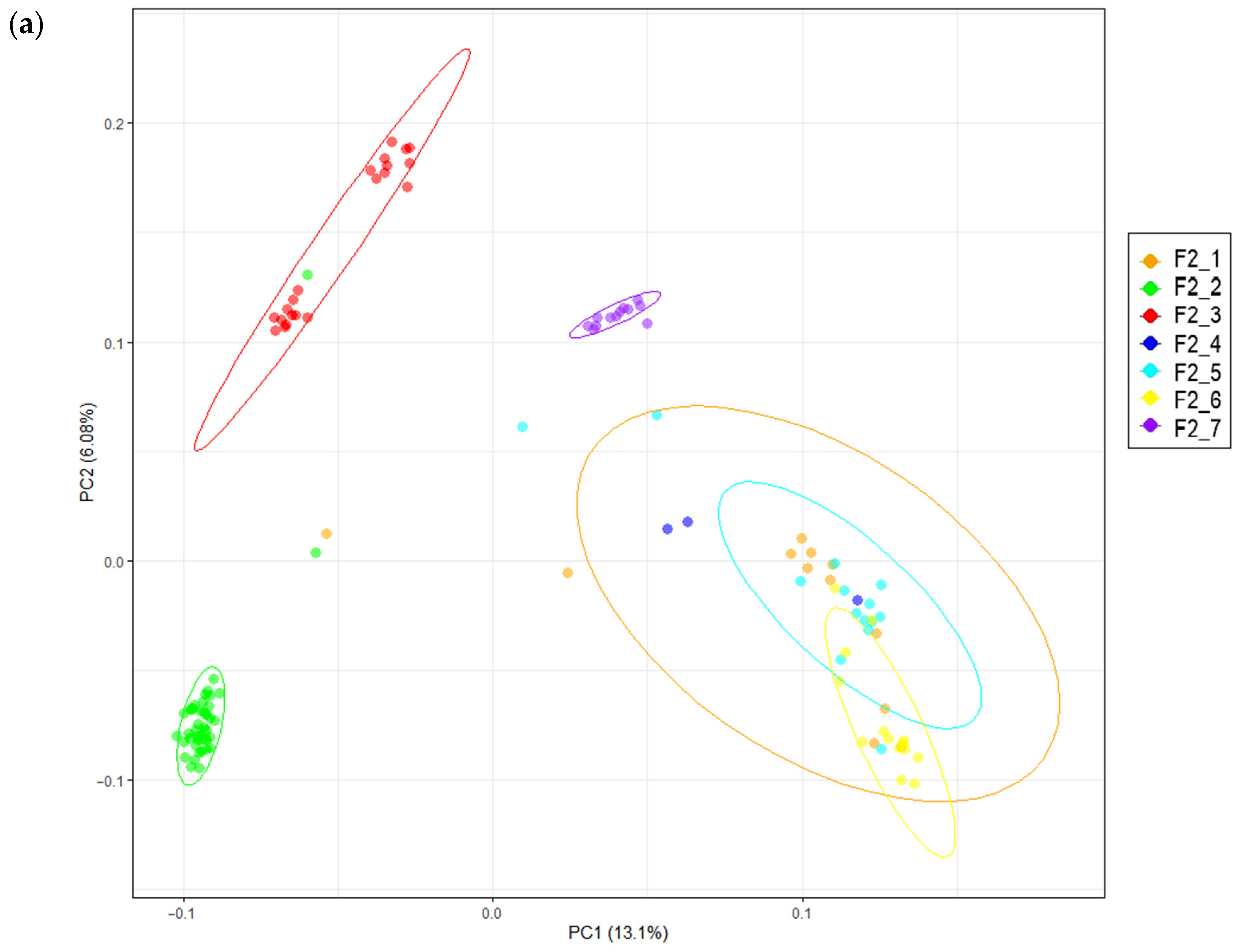
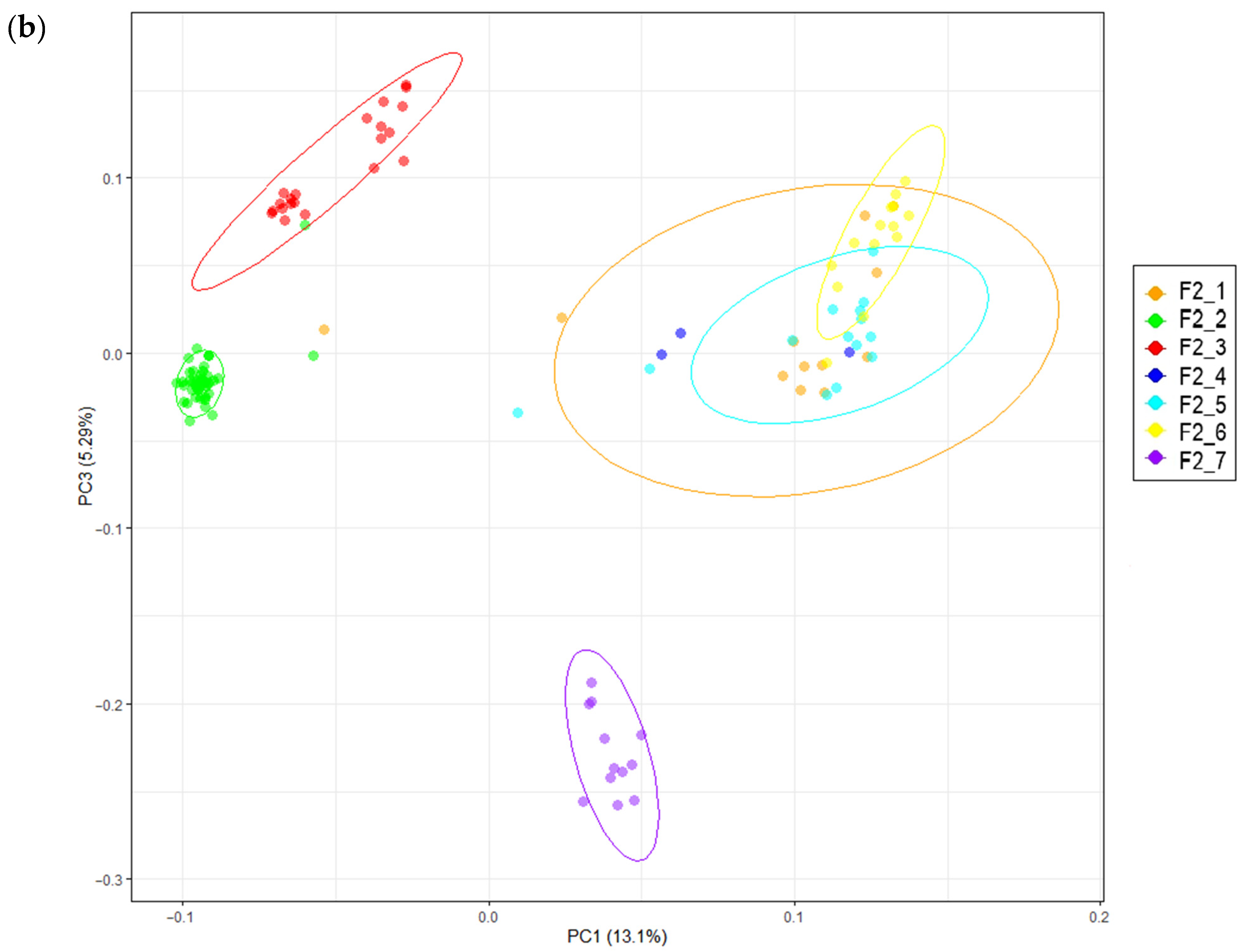
| Trait | F2 Population (n = 142) | Russian White (n = 20) | Cornish White (n = 15) | |||
|---|---|---|---|---|---|---|
| Mean ± SD | CV, % | Mean ± SD | CV, % | Mean ± SD | CV, % | |
| Age at first egg, days | 131.6 ± 18.4 | 13.9 | 144.5 ± 6.3 | 4.4 | 180.6 ± 17.8 | 9.9 |
| Duration of egg laying, days | 232.4 ± 18.4 | 7.9 | 220.5 ± 6.3 | 2.9 | 184.4 ± 17.8 | 9.6 |
| Egg number for 238 days | 126.2 ± 31.6 | 25.1 | 150.3 ± 10.8 | 7.2 | 82.0 ± 11.1 | 13.5 |
| Egg laying cycle, days | 2.4 ± 0.8 | 33.4 | N/A | N/A | N/A | N/A |
| Egg laying interval, days | 2.4 ± 1.0 | 41.4 | N/A | N/A | N/A | N/A |
| Egg weight at age of 18–28 weeks, g | 43.0 ± 3.5 | 8.2 | 44.1 ± 3.2 | 7.2 | N/A | N/A |
| Egg weight at age of 29–41 weeks, g | 50.8 ± 4.7 | 9.3 | 50.6 ± 4.3 | 8.7 | 52.2 ± 4.2 | 8.0 |
| Egg weight at age of 42–52 weeks, g | 57.8 ± 3.9 | 6.7 | 55.1 ± 2.4 | 4.3 | 63.1 ± 4.6 | 7.4 |
| Trait | No. of SNPs | Chromosomes 1 |
|---|---|---|
| Duration of egg laying | 4 | GGA1, GGA12, GGA14 |
| Egg laying cycle | 3 | GGA2, GGA25 |
| Egg laying interval | 38 | GGA1–GGA6, GGA9–GGA11, GGA13–GGA15, GGA19, GGA22, GGA28 |
| Egg weight at age of 18–28 weeks | 1 | GGA8 |
| Egg weight at age of 42–52 weeks | 5 | GGA1, GGA10, GGA22, GGA27 |
| Trait | GGA 1 | SNP | Location, bp | Gene | p-Value |
|---|---|---|---|---|---|
| Egg weight at 42–52 weeks of age | 1 | Gga_rs13950763 | 144,541,672 | FGF14 | 7.893 × 10−7 |
| Gga_rs13950783 | 144,581,256 | FGF14 | 7.893 × 10−7 | ||
| 22 | Gga_rs16733701 | 5,254,215 | GCK | 8.868 × 10−7 | |
| Duration of egg laying | 12 | Gga_rs14048080 | 18,439,246 | CNTN4 | 2.497 × 10−7 |
| Egg laying cycle | 2 | Gga_rs13772998 | 136,095,111 | SAMD12 | 1.565 × 10−7 |
| Egg laying interval | 1 | GGaluGA016975 | 49,631,028 | PHF5A | 8.431 × 10−10 |
| GGaluGA021889 | 62,111,416 | AKR1B1 | 3.519 × 10−8 | ||
| Gga_rs14835481 | 62,385,811 | CALD1 | 4.072 × 10−15 | ||
| Gga_rs13973123 | 171,655,178 | ATP7B | 7.489 × 10−8 | ||
| 2 | Gga_rs13670867 | 41,560,503 | PIK3R4 | 1.337 × 10−8 | |
| Gga_rs14258322 | 145,755,426 | PTK2 | 2.003 × 10−7 | ||
| 3 | Gga_rs14329753 | 26,472,162 | PRKCE | 7.511 × 10−7 | |
| 4 | Gga_rs15598417 | 61,793,533 | FAT1 | 2.166 × 10−9 | |
| Gga_rs15600128 | 62,825,026 | PCM1 | 4.072 × 10−15 | ||
| GGaluGA266321 | 76,726,036 | CC2D2A | 5.887 × 10−15 | ||
| 6 | Gga_rs14564900 | 5,790,608 | BMS1 | 2.811 × 10−9 | |
| 10 | GGaluGA068824 | 10,301,717 | SEMA6D | 3.172 × 10−7 | |
| 11 | GGaluGA078973 | 16,137,404 | CDH13 | 4.874 × 10−7 | |
| 13 | GGaluGA092132 | 5,413,088 | SLIT3 | 3.573 × 10−7 | |
| Gga_rs14050895 | 8,419,842 | ATP10B | 4.550 × 10−7 | ||
| 15 | Gga_rs15773720 | 6,817,919 | ISCU | 2.166 × 10−9 | |
| 19 | GGaluGA126763 | 5,248,070 | LRRC75A | 3.247 × 10−7 | |
| 22 | Gga_rs14684608 | 2,639,829 | LETM2 | 9.628 × 10−7 | |
| 28 | Gga_rs16210664 | 2,694,485 | ANKRD24 | 1.184 × 10−7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Volkova, N.A.; Romanov, M.N.; Dzhagaev, A.Y.; Larionova, P.V.; Volkova, L.A.; Abdelmanova, A.S.; Vetokh, A.N.; Griffin, D.K.; Zinovieva, N.A. Genome-Wide Association Studies and Candidate Genes for Egg Production Traits in Layers from an F2 Crossbred Population Produced Using Two Divergently Selected Chicken Breeds, Russian White and Cornish White. Genes 2025, 16, 583. https://doi.org/10.3390/genes16050583
Volkova NA, Romanov MN, Dzhagaev AY, Larionova PV, Volkova LA, Abdelmanova AS, Vetokh AN, Griffin DK, Zinovieva NA. Genome-Wide Association Studies and Candidate Genes for Egg Production Traits in Layers from an F2 Crossbred Population Produced Using Two Divergently Selected Chicken Breeds, Russian White and Cornish White. Genes. 2025; 16(5):583. https://doi.org/10.3390/genes16050583
Chicago/Turabian StyleVolkova, Natalia A., Michael N. Romanov, Alan Yu. Dzhagaev, Polina V. Larionova, Ludmila A. Volkova, Alexandra S. Abdelmanova, Anastasia N. Vetokh, Darren K. Griffin, and Natalia A. Zinovieva. 2025. "Genome-Wide Association Studies and Candidate Genes for Egg Production Traits in Layers from an F2 Crossbred Population Produced Using Two Divergently Selected Chicken Breeds, Russian White and Cornish White" Genes 16, no. 5: 583. https://doi.org/10.3390/genes16050583
APA StyleVolkova, N. A., Romanov, M. N., Dzhagaev, A. Y., Larionova, P. V., Volkova, L. A., Abdelmanova, A. S., Vetokh, A. N., Griffin, D. K., & Zinovieva, N. A. (2025). Genome-Wide Association Studies and Candidate Genes for Egg Production Traits in Layers from an F2 Crossbred Population Produced Using Two Divergently Selected Chicken Breeds, Russian White and Cornish White. Genes, 16(5), 583. https://doi.org/10.3390/genes16050583










