Comparative Mitochondrial Genomic and Phylogenetic Study of Eight Species of the Family Lonchodidae (Phasmatodea: Euphasmatodea)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Taxon Sampling and Sequencing
2.2. Genome Annotation and Sequence Analysis
2.3. Phylogenetic Analysis
3. Results and Discussion
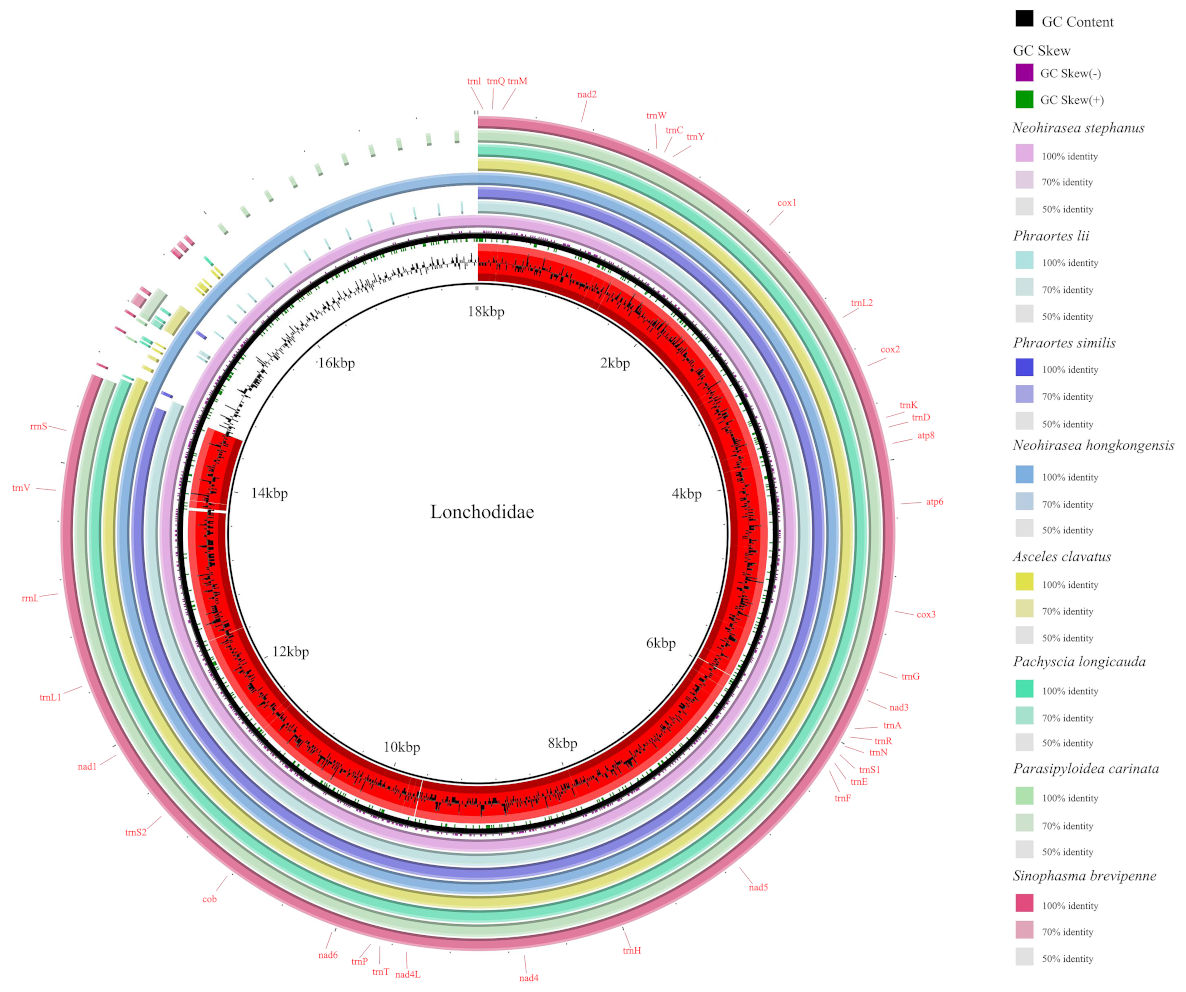
3.1. Mitochondrial Genomic Characterization of Eight Species
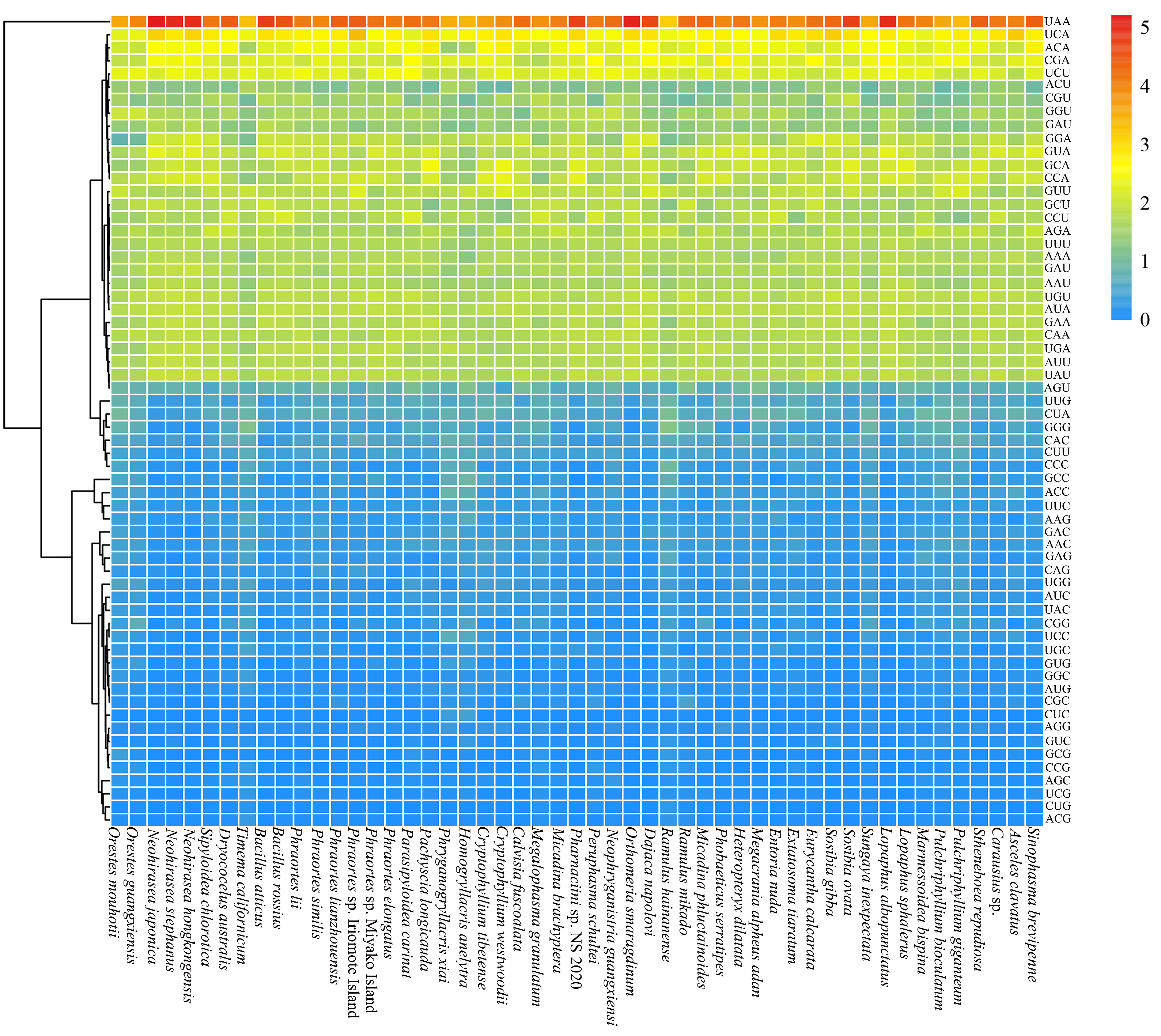
3.2. Codon Usage
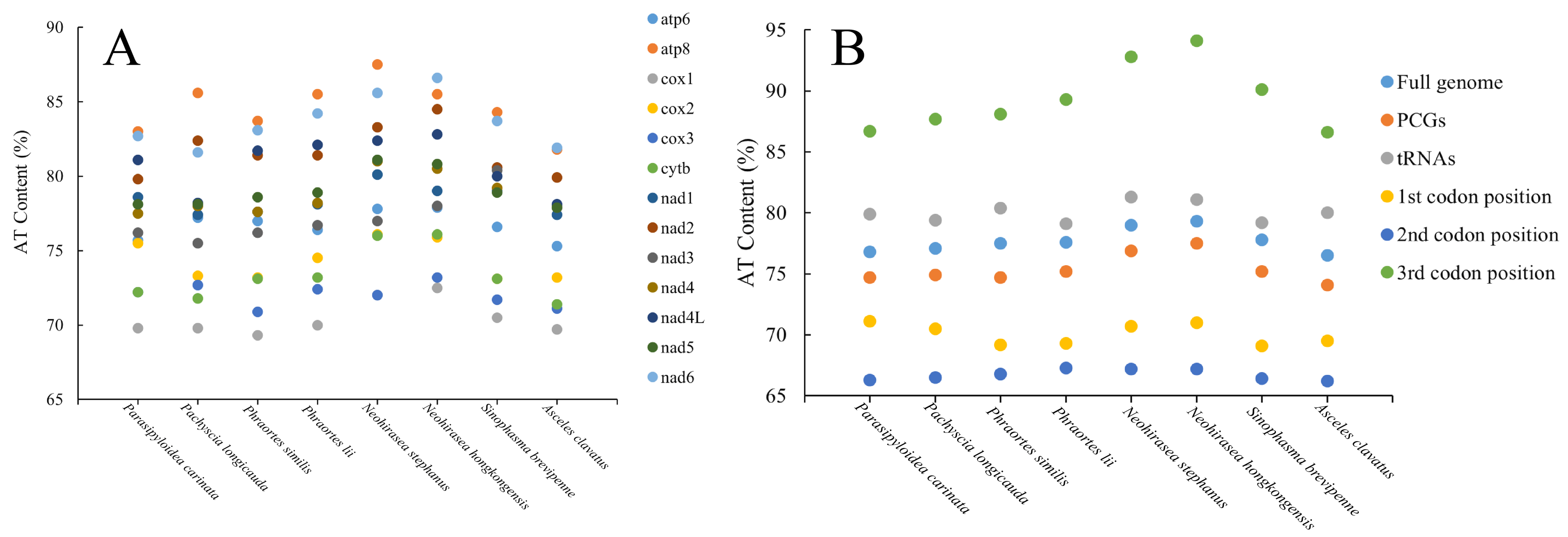
3.3. AT Bias
3.4. Genetic Rearrangement
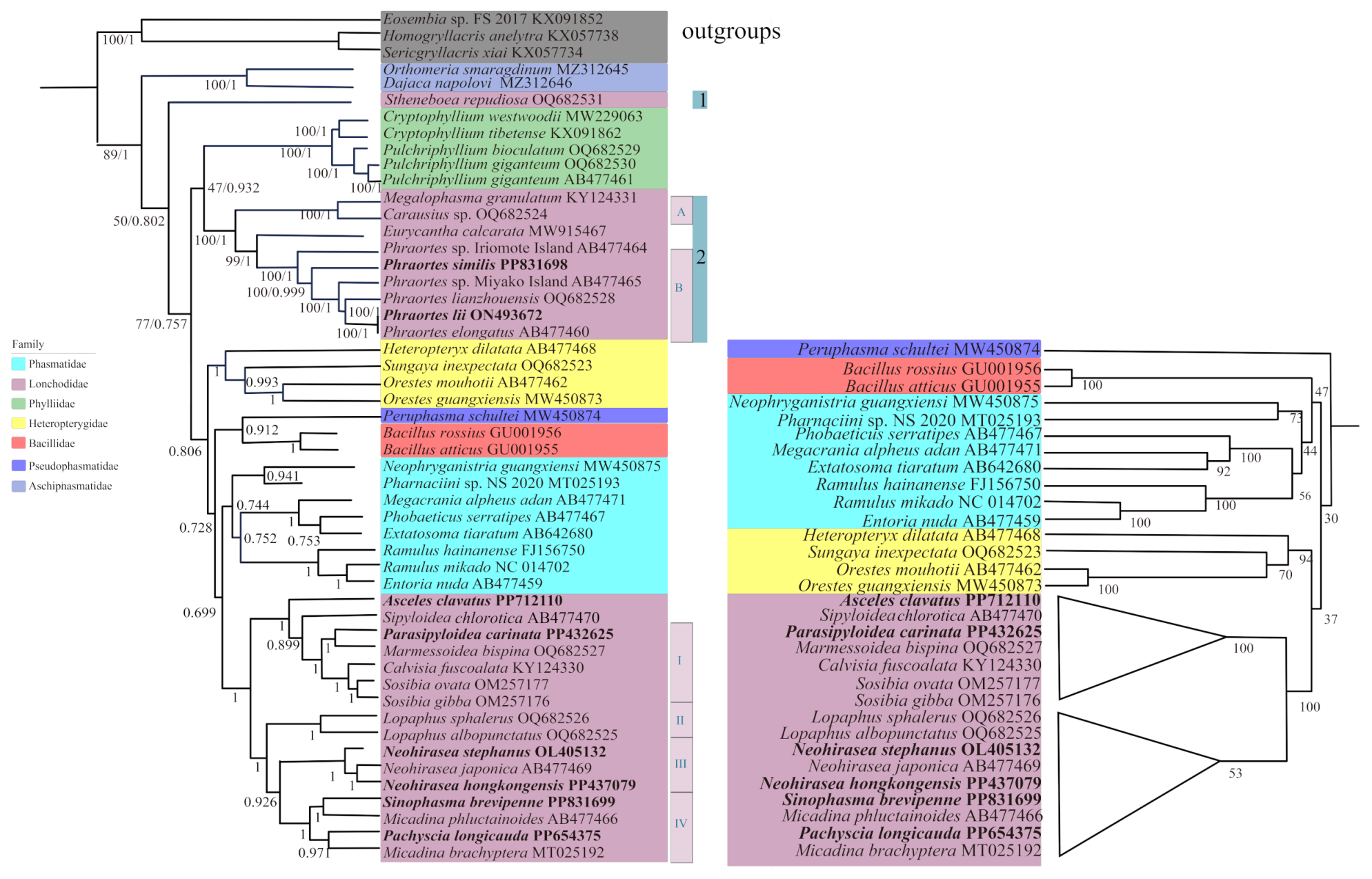
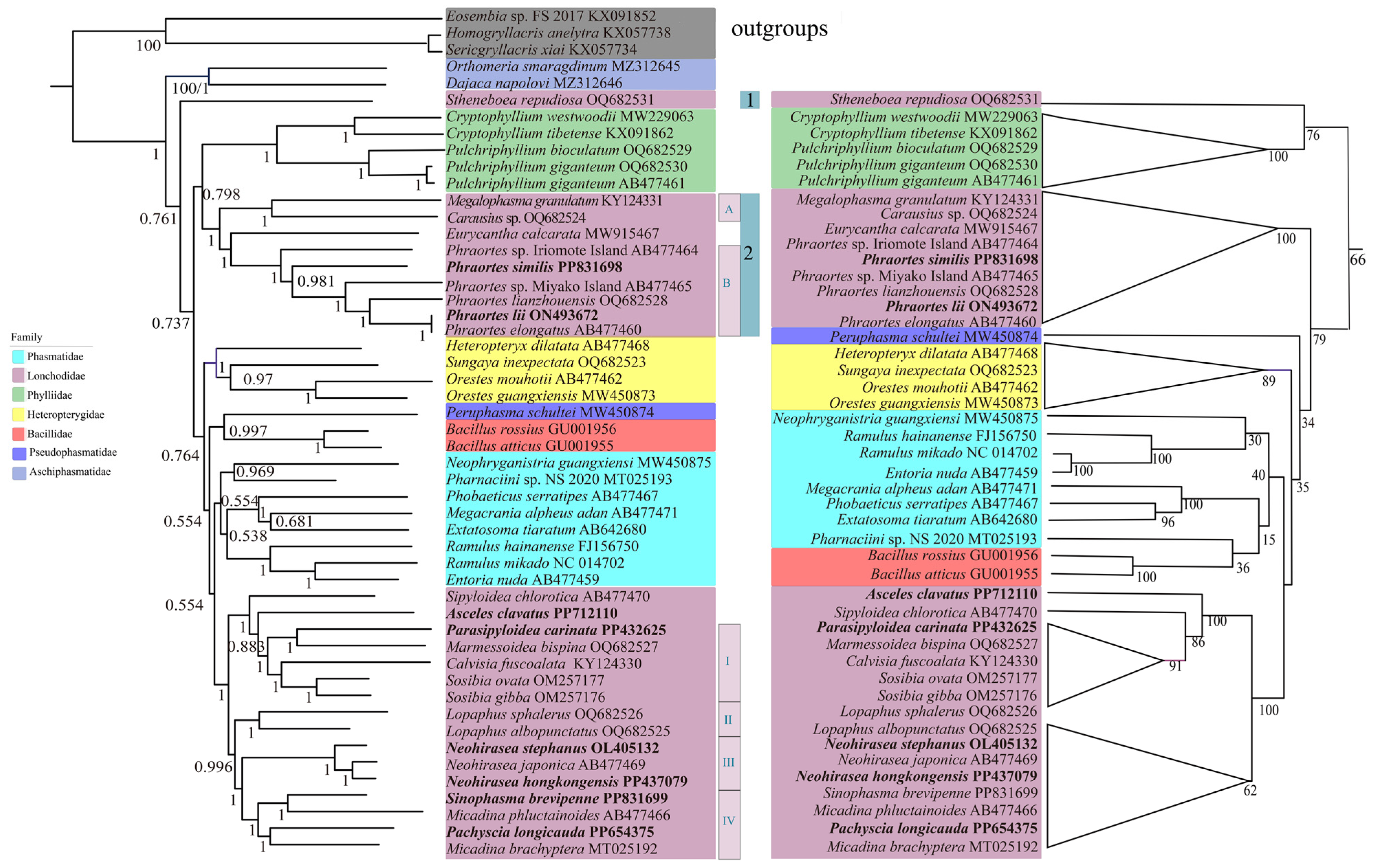
3.5. Phylogenetic Relationships
3.6. Relationships Between Gene Rearrangement and Phylogeny
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- O’Hanlon, J.C.; Jones, B.R.; Bulbert, M.W. The dynamic eggs of the Phasmatodea and their apparent convergence with plants. Sci. Nat. 2020, 107, 34. [Google Scholar] [CrossRef] [PubMed]
- Song, N.; Li, X.H.; Na, R.S. Mitochondrial genomes of stick insects (Phasmatodea) and phylogenetic considerations. PLoS ONE 2020, 15, e0240186. [Google Scholar] [CrossRef] [PubMed]
- Whiting, M.F.; Bradler, S.; Maxwell, T. Loss and recovery of wings in stick insects. Nature 2003, 421, 264–267. [Google Scholar] [CrossRef]
- Bradler, S.; Robertson, J.A.; Whiting, M.F. A molecular phylogeny of Phasmatodea with emphasis on Necrosciinae, the most species-rich subfamily of stick insects. Syst. Entomol. 2014, 39, 205–222. [Google Scholar] [CrossRef]
- Brock, P.D.; Büscher, T.H.; Baker, E. Phasmida Species File Online. Available online: https://phasmida.speciesfile.org/otus/852544/overview (accessed on 26 February 2025).
- Hennemann, F.H.; Conle, O.V. Studies on Neotropical Phasmatodea XXVI: Taxonomic review of Cladomorformia tax. n., a lineage of Diapheromerinae stick insects, with the descriptions of seven new genera and 41 new species (Phasmatodea: Occidophasmata: Diapheromerinae). Zootaxa 2024, 5444, 1–454. [Google Scholar] [CrossRef]
- Chen, Y.; Yuan, Y.; Yang, W.; Storey, K.B.; Zhang, J.; Yu, D. Insight into the Phylogenetic Relationships of Phasmatodea and Selection Pressure Analysis of Phraortes liaoningensis Chen & He, 1991 (Phasmatodea: Lonchodidae) Using Mitogenomes. Insects 2024, 15, 858. [Google Scholar] [CrossRef]
- Forni, G.; Plazzi, F.; Cussigh, A.; Conle, O.; Hennemann, F.; Luchetti, A.; Mantovani, B. Phylomitogenomics provides new perspectives on the Euphasmatodea radiation (Insecta: Phasmatodea). Mol. Phylogenet. Evol. 2020, 155, 106983. [Google Scholar] [CrossRef]
- Bradler, S.; Buckley, T.R. Biodiversity of Phasmatodea. Insect Biodivers. Sci. Soc. 2018, 2, 281–313. [Google Scholar] [CrossRef]
- Buckley, T.R.; Attanayake, D.; Bradler, S. Extreme convergence in stick insect evolution: Phylogenetic placement of the Lord Howe Island tree lobster. Proc. Biol. Sci. 2009, 276, 1055–1062. [Google Scholar] [CrossRef]
- Salinas-Giegé, T.; Giegé, R.; Giegé, P. tRNA Biology in Mitochondria. Int. J. Mol. Sci. 2015, 16, 4518–4559. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed]
- Macino, G.; Scazzocchio, C.; Waring, R.B.; Berks, M.M.; Wayne, D.R. Conservation and rearrangement of mitochondrial structural gene sequences. Nature 1980, 288, 404–406. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, M.M.; Miya, M.U.; Nishida, M. Use of a PCR-based approach for sequencing whole mitochondrial genomes of insects: Two examples (cockroach and dragonfly) based on the method developed for decapod crustaceans. Insect Mol. Biol. 2004, 13, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Pang, S.; Zhang, Q.; Liang, L.; Qin, Y.; Li, S.; Bian, X. Comparative Mitogenomics and Phylogenetic Implications for Nine Species of the Subfamily Meconematinae (Orthoptera: Tettigoniidae). Insects 2024, 15, 413. [Google Scholar] [CrossRef]
- Ghanavi, H.R.; Twort, V.; Hartman, T.J.; Zahiri, R.; Wahlberg, N. The (non) accuracy of mitochondrial genomes for family-level phylogenetics in Erebidae (Lepidoptera). Zool. Scr. 2022, 51, 695–707. [Google Scholar] [CrossRef]
- De Mandal, S.; Chhakchhuak, L.; Gurusubramanian, G.; Kumar, N.S. Mitochondrial markers for identification and phylogenetic studies in insects–A Review. DNA Barcodes 2014, 2, 1–9. [Google Scholar] [CrossRef]
- Dong, Z.; Wang, Y.; Li, C.; Li, L.; Men, X. Mitochondrial DNA as a molecular marker in insect ecology: Current status and future prospects. Ann. Entomol. Soc. 2021, 114, 470–476. [Google Scholar] [CrossRef]
- Cameron, S.L.; Whiting, M.F. The complete mitochondrial genome of the tobacco hornworm, Manduca sexta, (Insecta: Lepidoptera: Sphingidae), and an examination of mitochondrial gene variability within butterflies and moths. Gene 2008, 408, 112–123. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef]
- Lu, H.; He, B.; Hao, Y.; Zhou, Z.; Su, C.; Huang, D. Comparative Mitogenomic Analysis of Two Cuckoo Bees (Apoidea: Anthophila: Megachilidae) with Phylogenetic Implications. Insects 2021, 12, 29. [Google Scholar] [CrossRef]
- Shao, R.; Campbell, N.J.; Schmidt, E.R.; Barker, S.C. Increased rate of gene rearrangement in the mitochondrial genomes of three orders of hemipteroid insects. Mol. Biol. Evol. 2001, 18, 1828–1832. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, K.; Chakraborty, R.; Cameron, S.L.; Sweet, A.D.; Chandra, K.; Kumar, V. Rearrangement and evolution of mitochondrial genomes in Thysanoptera (Insecta). Sci. Rep. 2020, 10, 695. [Google Scholar] [CrossRef]
- Li, R.; Lei, Z.; Li, W.; Zhang, W.; Zhou, C. Comparative Mitogenomic Analysis of Heptageniid Mayflies (Insecta: Ephemeroptera): Conserved Intergenic Spacer and tRNA Gene Duplication. Insects 2021, 12, 170. [Google Scholar] [CrossRef]
- Jiang, P.; Li, H.; Song, F.; Cai, Y.; Wang, J.; Liu, J.; Cai, W. Duplication and Remolding of tRNA Genes in the Mitochondrial Genome of Reduvius tenebrosus (Hemiptera: Reduviidae). Int. J. Mol. Sci. 2016, 17, 951. [Google Scholar] [CrossRef]
- Sun, Z.; Liu, Y.; Wilson, J.J.; Chen, Z.; Song, F.; Cai, W.; Li, H. Mitochondrial genome of Phalantus geniculatus (Hemiptera: Reduviidae): TrnT duplication and phylogenetic implications. Int. J. Biol. Macromol. 2019, 129, 110–115. [Google Scholar] [CrossRef]
- Liu, Q.; He, J.; Song, F.; Tian, L.; Cai, W.; Li, H. Positive Correlation of the Gene Rearrangements and Evolutionary Rates in the Mitochondrial Genomes of Thrips (Insecta: Thysanoptera). Insects 2022, 13, 585. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhang, L.; Li, K.; Hong, Y.; Storey, K.B.; Zhang, J.; Yu, D. Nine Mitochondrial Genomes of Phasmatodea with Two Novel Mitochondrial Gene Rearrangements and Phylogeny. Insects 2023, 14, 485. [Google Scholar] [CrossRef]
- Zhou, Z.; Guan, B.; Chai, J.; Che, X. Next-generation sequencing data used to determine the mitochondrial genomes and a preliminary phylogeny of Verophasmatodea insects. J. Asia-Pacif. Entomol. 2017, 20, 713–719. [Google Scholar] [CrossRef]
- Li, Y.; Wang, S.; Zhou, J.; Li, T.; Jiang, K.; Zhang, Y.; Zheng, C.; Liang, J.; Bu, W. The phylogenic position of Aschiphasmatidae in Euphasmatodea based on mitochondrial genomic evidence. Gene 2022, 808, 145974. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information (NCBI). Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information. 1988. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 23 December 2024).
- Robertson, J.A.; Bradler, S.; Whiting, M.F. Evolution of oviposition techniques in stick and leaf insects (Phasmatodea). Front. Ecol. Evol. 2018, 6, 216. [Google Scholar] [CrossRef]
- Xu, K.K.; Chen, Q.P.; Ayivi, S.P.G. Three complete mitochondrial genomes of Orestes guangxiensis, Peruphasma schultei, and Phryganistria guangxiensis (Insecta: Phasmatodea) and their phylogeny. Insects 2021, 12, 779. [Google Scholar] [CrossRef] [PubMed]
- Kômoto, N.; Yukuhiro, K.; Ueda, K.; Tomita, S. Exploring the molecular phylogeny of phasmids with whole mitochondrial genome sequences. Mol. Phylogenet. Evol. 2011, 58, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Bresseel, J.; Constant, J. Review of the Oriental stick insect genus Trachythorax Redtenbacher, 1908 with two new species from Vietnam and comments on egg parasitism and morphological counteradaptations (Phasmida, Lonchodidae, Necrosciinae). BJE 2021, 120, 1–56. [Google Scholar]
- Li, Y.F.; Wang, S.J.; Chen, J.H.; Zhou, J.Y.; Bu, W.J. Two new stick insect species of Sosibia Stål (Phasmatodea: Lonchodidae: Necrosciinae) from China and the first report on mitochondrial genomes of this genus. Arch. Insect Biochem. Physiol. 2022, 111, e21901. [Google Scholar] [CrossRef]
- Bresseel, J.; Constant, J.; Jiaranaisakul, K.; Huebner, C. A new species of Calvisia (Calvisia) from Thailand and Myanmar and notes on C. (Calvisia) sangarius from Peninsular Malaysia (Phasmida, Lonchodidae, Necrosciinae). BJE 2022, 133, 1–23. [Google Scholar]
- Xie, C.X.; Cai, J.; Qian, Y.H. New species of Andropromachus (Phasmatodea: Lonchodidae: Necrosciinae: Necrosciini) from Yunnan Province, China. Biodivers. Data J. 2022, 10, e78080. [Google Scholar] [CrossRef]
- Gao, H.R.; Huang, J.K.; Wang, C.; Xie, C.X.; Li, Y.H. Revision of the Chinese species of Andropromachus Carl, 1913 (Phasmatodea, Lonchodidae, Necrosciinae). Zootaxa 2022, 5175, 463–477. [Google Scholar] [CrossRef]
- Gao, H.R.; Li, Y.H. First report of the genus Spinomarmessoidea (Phasmatodea, Lonchodidae, Necrosciinae) from China, with the description of a new species. Zootaxa 2023, 5239, 280–288. [Google Scholar] [CrossRef]
- Qian, Y.H.; Xie, C.X.; Wen, J.; Wang, Y. Review of stick insects (Insecta: Phasmatodea) from Yintiaoling Nature Reserve of China, with description of two new species. Zootaxa 2023, 5257, 17–39. [Google Scholar] [CrossRef]
- Bresseel, J.; Constant, J. The new stick insect genus Pterulina gen. nov. a second winged Clitumninae genus from Vietnam with a new combination and a new species (Phasmida, Phasmatidae, Clitumninae, Clitumnini). Belg. J. Entomol. 2020, 96, 1–30. [Google Scholar]
- Hennemann, F.H. Megacraniinae-The Palm Stick Insects: A new subfamily of Old World Phasmatodea and a redefinition of Platycraninae Brunner v. Wattenwyl, 1893 (Phasmatodea: “Anareolatae”). Zootaxa 2020, 4896, 151–179. [Google Scholar] [CrossRef] [PubMed]
- Hennemann, F.H. Stick insects of Sulawesi, Peleng and the Sula Islands, Indonesiaa review including checklists of species and descriptions of new taxa (Insecta: Phasmatodea). Zootaxa 2021, 5073, 1–189. [Google Scholar] [CrossRef] [PubMed]
- Bank, S.; Cumming, R.T.; Li, Y.; Henze, K.; Le Tirant, S.; Bradler, S. A tree of leaves: Phylogeny and historical biogeography of the leaf insects (Phasmatodea: Phylliidae). Commun. Biol. 2021, 4, 932. [Google Scholar] [CrossRef]
- Jones, B.R.; Brock, P.D.; Mantovani, B.; Beasley-Hall, P.; Yeates, D.K.; Lo, N. Integrative taxonomy of the stick insect genus Austrocarausius Brock, 2000 (Phasmatodea: Lonchodidae) reveals cryptic species in remnant Queensland rainforests. Invertebr. Syst. 2022, 36, 849–873. [Google Scholar] [CrossRef]
- Matvienko, M. CLC Genomics Workbench. Plant Anim. Genome. Sr. Field Appl. Sci. CLC Bio Aarhus DE 2015, 1, 1–42. [Google Scholar]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, 4. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Sun, Z.; Wan, D.G.; Murphy, R.W.; Ma, L.; Zhang, X.S.; Huang, D.W. Comparison of base composition and codon usage in insect mitochondrial genomes. Genes. Genom. 2009, 31, 65–71. [Google Scholar] [CrossRef]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- Zhou, Z.J.; Zhao, L.; Liu, N.; Guo, H.F.; Guan, B.; Di, J.X.; Shi, F.M. Towards a higher-level Ensifera phylogeny inferred from mitogenome sequences. Mol. Phylogenet. Evol. 2017, 108, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Kück, P.; Meid, S.A.; Groß, C.; Wägele, J.W.; Misof, B. AliGROOVE–visualization of heterogeneous sequence divergence within multiple sequence alignments and detection of inflated branch support. BMC Bioinf. 2014, 15, 294. [Google Scholar] [CrossRef] [PubMed]
- Jeena, N.S.; Rahuman, S.; Sebastian, W.; Kumar, R.; Sajeela, K.A.; Kizhakudan, J.K.; Menon, K.K.; Roul, S.K.; Gopalakrishnan, A.; Adhakrishnan, E.V. Mitogenomic recognition of incognito lineages in the mud spiny lobster Panulirus polyphagus (Herbst, 1793): A tale of unique genetic structuring and diversification. Int. J. Biol. Macromol. 2024, 277, 134327. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef]
- Xiang, C.Y.; Gao, F.; Jakovlić, I.; Lei, H.P.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.T.; Zhang, D. Using PhyloSuite for molecular phylogeny and tree-based analyses. Imeta 2023, 2, e87. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Hong, M.Y.; Lee, E.M.; Jo, Y.H.; Park, H.C.; Kim, S.R.; Hwang, J.S.; Jin, B.R.; Kang, P.D.; Kim, K.G.; Han, Y.S.; et al. Complete nucleotide sequence and organization of the mitogenome of the silk moth Caligula boisduvalii (Lepidoptera: Saturniidae) and comparison with other lepidopteran insects. Gene 2008, 413, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, H.; Song, F.; Song, W.; Dai, X.; Chang, J.; Cai, W. The mitochondrial genome of Coridius chinensis (Hemiptera: Dinidoridae). Zootaxa 2012, 3537, 29–40. [Google Scholar] [CrossRef]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Zhang, L.; Lu, T.; Ma, J.; Zeng, C.; Yue, B.; Zhang, X. Mitochondrial genomes of blister beetles (Coleoptera, Meloidae) and two large intergenic spacers in Hycleus genera. BMC Genom. 2017, 18, 698. [Google Scholar] [CrossRef]
- Shi, A.; Li, C.; Farhan, M.; Xu, C.; Zhang, Y.; Qian, H.; Zhang, S.; Jing, T. Characterization, Codon Usage Pattern and Phylogenetic Implications of the Waterlily Aphid Rhopalosiphum nymphaeae (Hemiptera: Aphididae) Mitochondrial Genome. Int. J. Mol. Sci. 2024, 25, 11336. [Google Scholar] [CrossRef]
- Tang, J.M.; Li, F.; Cheng, T.Y.; Duan, D.Y.; Liu, G.H. Comparative analyses of the mitochondrial genome of the sheep ked Melophagus ovinus (Diptera: Hippoboscidae) from different geographical origins in China. Parasitol. Res. 2018, 117, 2677–2683. [Google Scholar] [CrossRef]
- Dowton, M.; Castro, L.; Austin, A. Mitochondrial gene rearrangements as phylogenetic characters in the invertebrates: The examination of genome ‘morphology’. Invertebr. Syst. 2002, 16, 345–356. [Google Scholar] [CrossRef]
- Inoue, J.G.; Miya, M.; Tsukamoto, K.; Nishida, M. Complete mitochondrial DNA sequence of Conger myriaster (Teleostei: Anguilliformes): Novel gene order for vertebrate mitochondrial genomes and the phylogenetic implications for anguilliform families. J. Mol. Evol. 2001, 52, 311–320. [Google Scholar] [CrossRef]
- Shi, W.; Gong, L.; Wang, S.Y.; Miao, X.G.; Kong, X.Y. Tandem duplication and random loss for mitogenome rearrangement in Symphurus (Teleost: Pleuronectiformes). BMC Genom. 2015, 16, 355. [Google Scholar] [CrossRef]
- Ma, Z.; Yang, X.; Bercsenyi, M.; Wu, J.; Yu, Y.; Wei, K.; Fan, Q.; Yang, R. Comparative mitogenomics of the genus Odontobutis (Perciformes: Gobioidei: Odontobutidae) revealed conserved gene rearrangement and high sequence variations. Int. J. Mol. Sci. 2015, 16, 25031–25049. [Google Scholar] [CrossRef]
- San, M.D.; Gower, D.J.; Zardoya, R.; Wilkinson, M. A hotspot of gene order rearrangement by tandem duplication and random loss in the vertebrate mitochondrial genome. Mol. Biol. Evol. 2006, 23, 227–234. [Google Scholar] [CrossRef]
- Feng, Z.; Wu, Y.; Yang, C.; Gu, X.; Wilson, J.J.; Li, H.; Cai, W.; Yang, H.; Song, F. Evolution of tRNA gene rearrangement in the mitochondrial genome of ichneumonoid wasps (Hymenoptera: Ichneumonoidea). Int. J. Biol. Macromol. 2020, 164, 540–547. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.Y.; Wang, Y.L.; Liang, L.; Yin, Z.W.; Thayer, M.K.; Newton, A.F.; Zhou, Y.L. Congruence of morphological and molecular phylogenies of the rove beetle subfamily Staphylininae (Coleoptera: Staphylinidae). Sci. Rep. 2019, 9, 15137. [Google Scholar] [CrossRef]
- Glaw, F.; Hawlitschek, O.; Dunz, A.; Goldberg, J.; Bradler, S. When giant stick insects play with colors: Molecular phylogeny of the Achriopterini and description of two new Splendid Species (Phasmatodea: Achrioptera) from Madagascar. Front. Ecol. Evol. 2019, 7, 105. [Google Scholar] [CrossRef]
- Bank, S.; Buckley, T.R.; Büscher, T.H.; Bresseel, J.; Constant, J.; De Haan, M.; Dittmar, D.; Dräger, H.; Kahar, R.S.; Kang, A.; et al. Reconstructing the nonadaptive radiation of an ancient lineage of ground-dwelling stick insects (Phasmatodea: Heteropterygidae). Syst. Entomol. 2021, 46, 487–507. [Google Scholar] [CrossRef]
- Rewitz, K.F.; O’Connor, M.B.; Gilbert, L.I. Molecular evolution of the insect Halloween family of cytochrome P450s: Phylogeny, gene organization and functional conservation. Insect Biochem. Mol. Biol. 2007, 37, 741–753. [Google Scholar] [CrossRef]
- Tamames, J. Evolution of gene order conservation in prokaryotes. Genome Biol. 2001, 2, 1–11. [Google Scholar] [CrossRef]
- Tamames, J.; Ouzounis, C.; Casari, G.; Valencia, A. Conserved clusters of functionally related genes in two bacterial genomes. J. Mol. Evol. 1997, 44, 66–73. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, T.; Zhang, Q.; Pang, S.; Qin, Y.; Zhang, B.; Bian, X. Comparative Mitochondrial Genomic and Phylogenetic Study of Eight Species of the Family Lonchodidae (Phasmatodea: Euphasmatodea). Genes 2025, 16, 565. https://doi.org/10.3390/genes16050565
Luo T, Zhang Q, Pang S, Qin Y, Zhang B, Bian X. Comparative Mitochondrial Genomic and Phylogenetic Study of Eight Species of the Family Lonchodidae (Phasmatodea: Euphasmatodea). Genes. 2025; 16(5):565. https://doi.org/10.3390/genes16050565
Chicago/Turabian StyleLuo, Ting, Qianwen Zhang, Siyu Pang, Yanting Qin, Bin Zhang, and Xun Bian. 2025. "Comparative Mitochondrial Genomic and Phylogenetic Study of Eight Species of the Family Lonchodidae (Phasmatodea: Euphasmatodea)" Genes 16, no. 5: 565. https://doi.org/10.3390/genes16050565
APA StyleLuo, T., Zhang, Q., Pang, S., Qin, Y., Zhang, B., & Bian, X. (2025). Comparative Mitochondrial Genomic and Phylogenetic Study of Eight Species of the Family Lonchodidae (Phasmatodea: Euphasmatodea). Genes, 16(5), 565. https://doi.org/10.3390/genes16050565





