Body Mass Index as an Example of a Negative Confounder: Evidence and Solutions
Abstract
1. Introduction
2. Methods
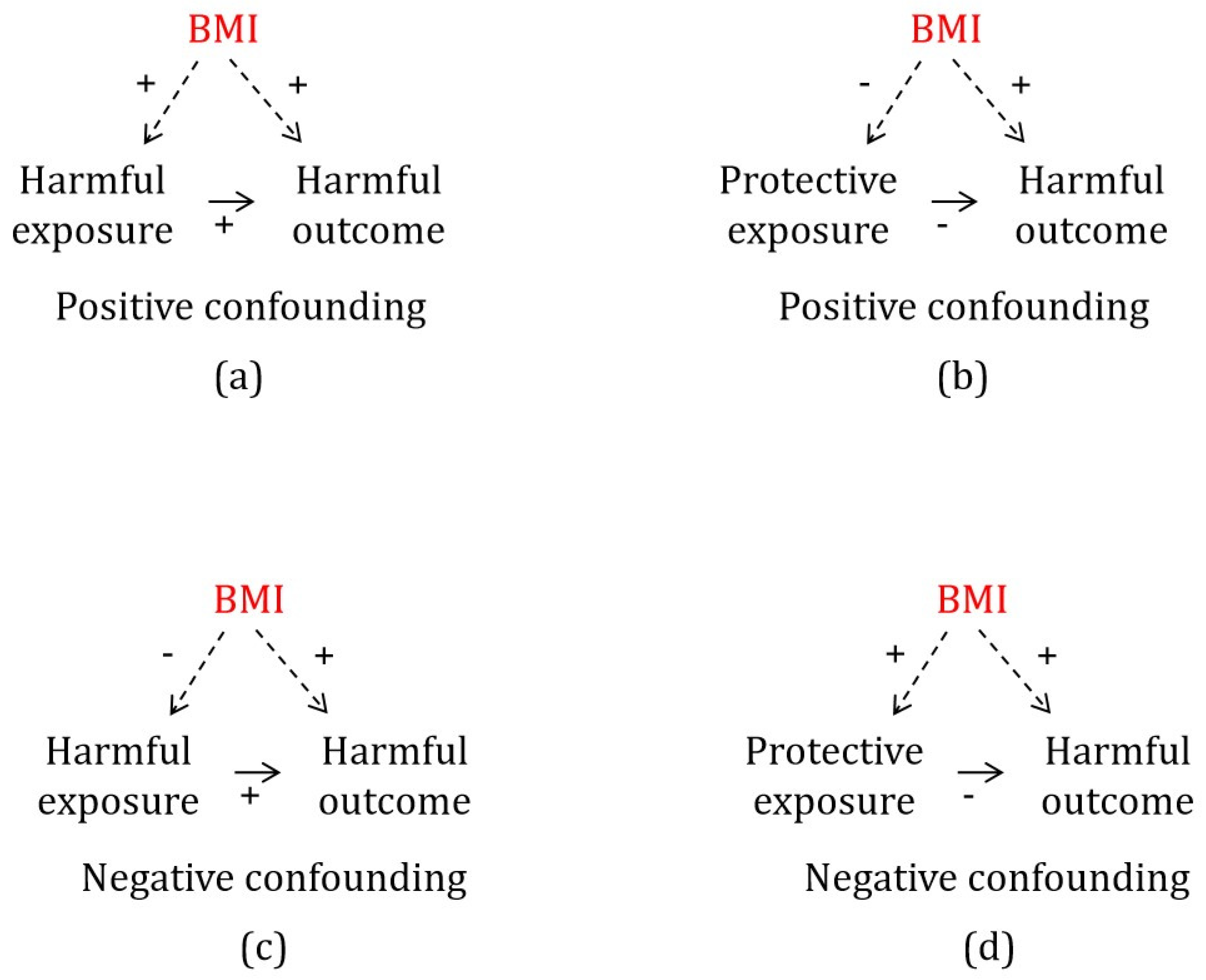
2.1. Negative Confounding by BMI
2.2. Study Design
2.3. Assumptions of MR
2.4. Data Sources
2.4.1. Body Mass Index
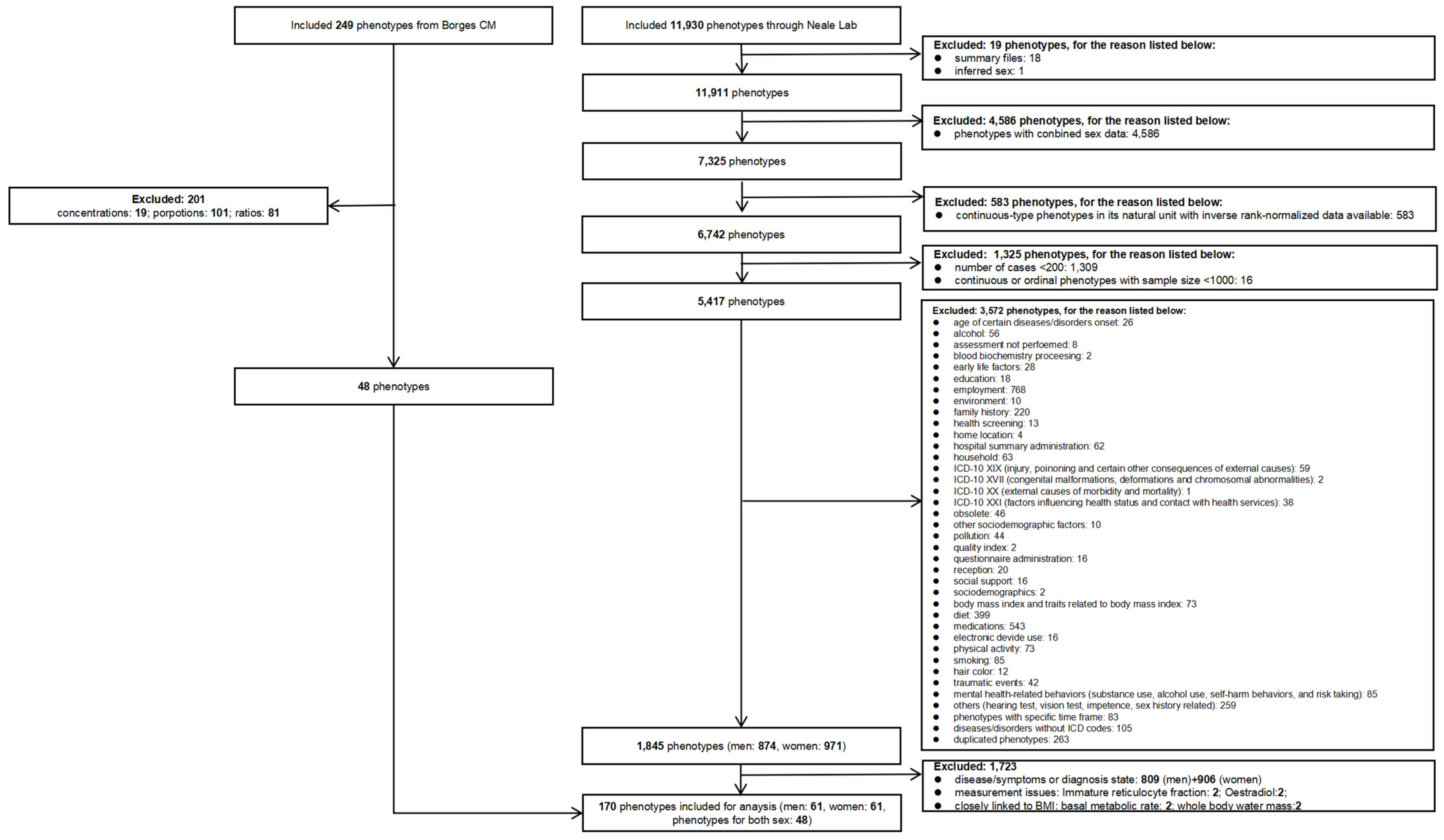
2.4.2. Inclusion and Exclusion Criteria for Exposures
2.4.3. Categorization of Exposures
2.4.4. Selection of Genetic Instruments for BMI
2.4.5. Statistical Analysis
3. Results
3.1. Exposure Selection
3.2. Negative Confounding by BMI for a Harmful Outcome
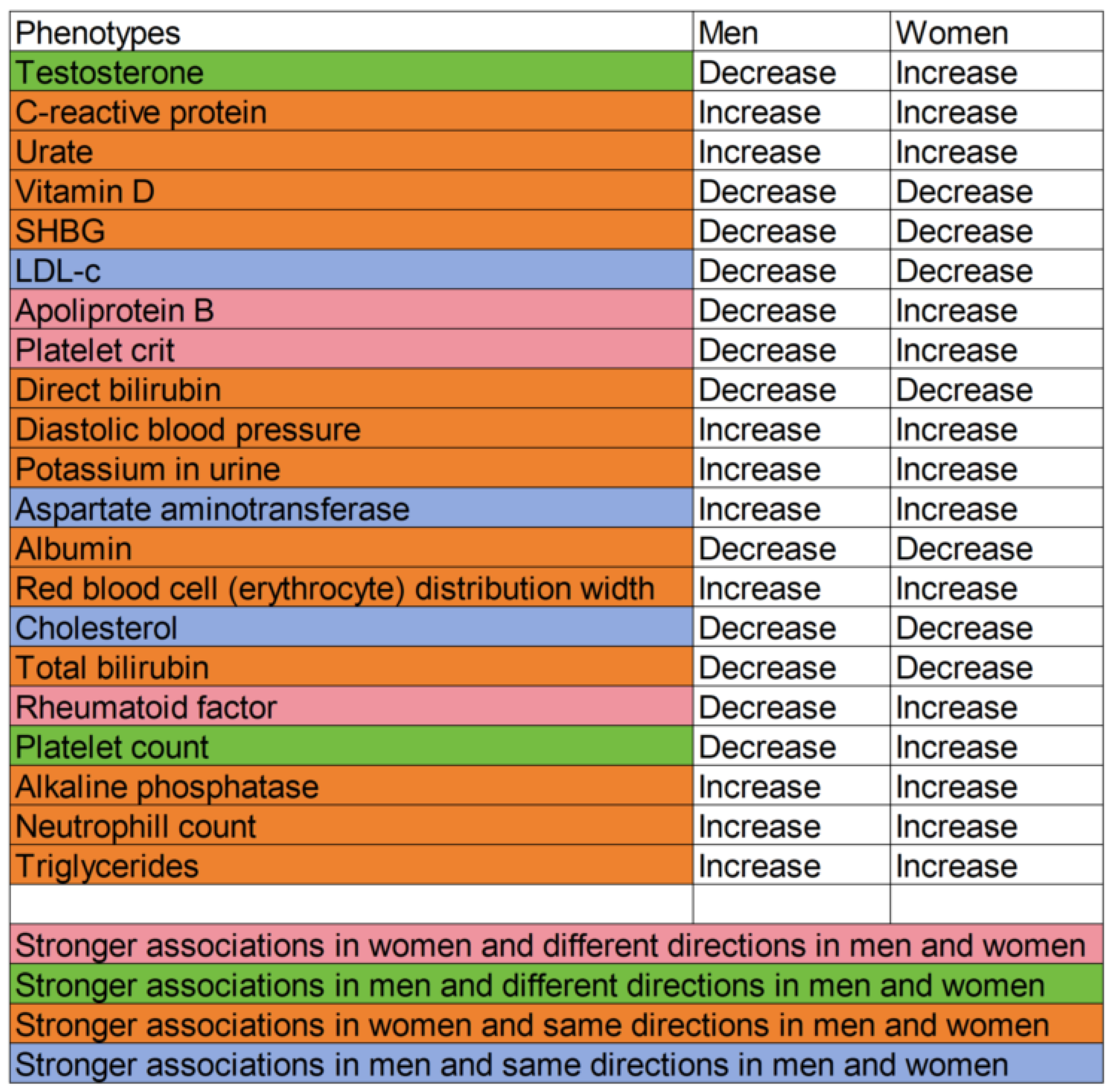
3.2.1. Potentially Harmful Exposures Reduced by BMI
3.2.2. Potentially Beneficial Exposures Increased by BMI
3.3. Positive Confounding by BMI for a Harmful Outcome
3.3.1. Potentially Harmful Exposures Increased by BMI
3.3.2. Potentially Beneficial Exposures Reduced by BMI
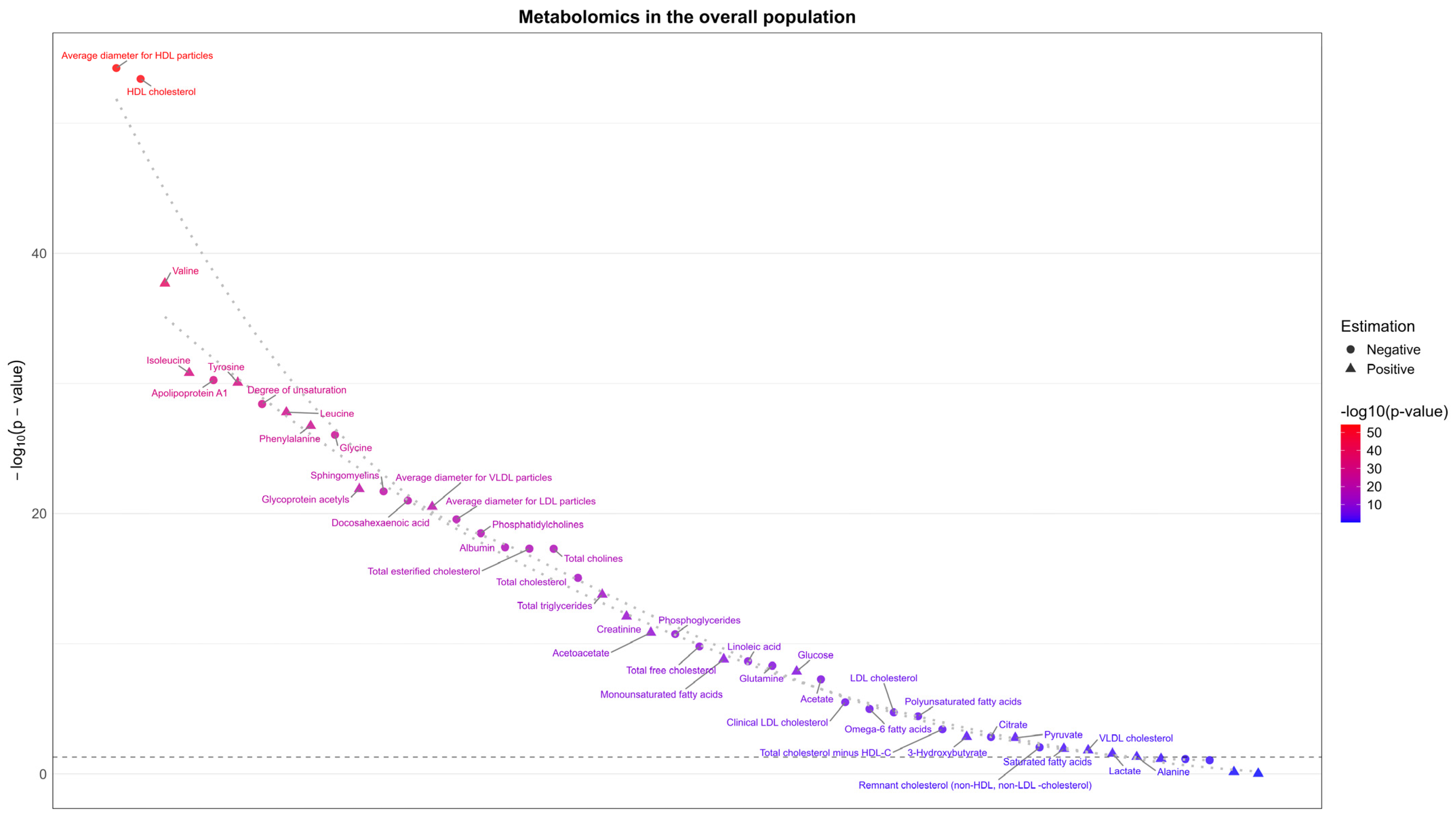
3.4. Sensitivity Analysis
3.5. Replication
4. Discussion
4.1. Comparison with Previous Studies
4.2. Implications for Observational Studies
4.3. Public Health Implications
4.4. Strengths and Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Miettinen, O. Confounding and effect-modification. Am. J. Epidemiol. 1974, 100, 350–353. [Google Scholar] [CrossRef] [PubMed]
- Rothman, K.J.; Greenland, S.; Lash, T.L. Modern Epidemiology; Wolters Kluwer Health/Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2008; Volume 3. [Google Scholar]
- Greenland, S.; Neutra, R. Control of confounding in the assessment of medical technology. Int. J. Epidemiol. 1980, 9, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Krieger, N. Theories for social epidemiology in the 21st century: An ecosocial perspective. Int. J. Epidemiol. 2001, 30, 668–677. [Google Scholar] [CrossRef] [PubMed]
- Mlinarić, A.; Horvat, M.; Šupak Smolčić, V. Dealing with the positive publication bias: Why you should really publish your negative results. Biochem. Medica 2017, 27, 447–452. [Google Scholar] [CrossRef]
- Gelman, A.; Loken, E. The statistical crisis in science. Am. Sci. 2014, 102, 460–465. [Google Scholar] [CrossRef]
- Choi, A.L.; Cordier, S.; Weihe, P.; Grandjean, P. Negative confounding in the evaluation of toxicity: The case of methylmercury in fish and seafood. Crit. Rev. Toxicol. 2008, 38, 877–893. [Google Scholar] [CrossRef]
- Darrous, L.; Hemani, G.; Davey Smith, G.; Kutalik, Z. PheWAS-based clustering of Mendelian Randomisation instruments reveals distinct mechanism-specific causal effects between obesity and educational attainment. Nat. Commun. 2024, 15, 1420. [Google Scholar] [CrossRef]
- Holmes, M.V.; Lange, L.A.; Palmer, T.; Lanktree, M.B.; North, K.E.; Almoguera, B.; Buxbaum, S.; Chandrupatla, H.R.; Elbers, C.C.; Guo, Y. Causal effects of body mass index on cardiometabolic traits and events: A Mendelian randomization analysis. Am. J. Hum. Genet. 2014, 94, 198–208. [Google Scholar] [CrossRef]
- MacDonald, C.-J.; Laouali, N.; Madika, A.-L.; Mancini, F.R.; Boutron-Ruault, M.-C. Dietary inflammatory index, risk of incident hypertension, and effect modification from BMI. Nutr. J. 2020, 19, 1–8. [Google Scholar] [CrossRef]
- Szamreta, E.A.; Qin, B.; Ohman-Strickland, P.A.; Devine, K.A.; Stapleton, J.L.; Ferrante, J.M.; Bandera, E.V. Associations of anthropometric, behavioral, and social factors on level of body esteem in peripubertal girls. J. Dev. Behav. Pediatr. 2017, 38, 58–64. [Google Scholar] [CrossRef]
- Bredella, M.A. Sex differences in body composition. In Sex and Gender Factors Affecting Metabolic Homeostasis, Diabetes and Obesity; Springer: Berlin/Heidelberg, Germany, 2017; pp. 9–27. [Google Scholar]
- Costanzo, M.; Caterino, M.; Sotgiu, G.; Ruoppolo, M.; Franconi, F.; Campesi, I. Sex differences in the human metabolome. Biol. Sex Differ. 2022, 13, 30. [Google Scholar] [CrossRef] [PubMed]
- Power, M.L.; Schulkin, J. Sex differences in fat storage, fat metabolism, and the health risks from obesity: Possible evolutionary origins. Br. J. Nutr. 2008, 99, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Richardson, T.G.; Sanderson, E.; Palmer, T.M.; Ala-Korpela, M.; Ference, B.A.; Davey Smith, G.; Holmes, M.V. Evaluating the relationship between circulating lipoprotein lipids and apolipoproteins with risk of coronary heart disease: A multivariable Mendelian randomisation analysis. PLoS Med. 2020, 17, e1003062. [Google Scholar] [CrossRef]
- Jensen, N.J.; Wodschow, H.Z.; Nilsson, M.; Rungby, J. Effects of Ketone Bodies on Brain Metabolism and Function in Neurodegenerative Diseases. Int. J. Mol. Sci. 2020, 21, 8767. [Google Scholar] [CrossRef]
- Sae-Jie, W.; Supasai, S.; Kivimaki, M.; Price, J.F.; Wong, A.; Kumari, M.; Engmann, J.; Shah, T.; Schmidt, A.F.; Gaunt, T.R.; et al. Triangulating evidence from observational and Mendelian randomization studies of ketone bodies for cognitive performance. BMC Med. 2023, 21, 340. [Google Scholar] [CrossRef]
- Davey Smith, G.; Ebrahim, S. ‘Mendelian randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003, 32, 1–22. [Google Scholar] [CrossRef]
- Burgess, S.; Scott, R.A.; Timpson, N.J.; Davey Smith, G.; Thompson, S.G.; Consortium, E.-I. Using published data in Mendelian randomization: A blueprint for efficient identification of causal risk factors. Eur. J. Epidemiol. 2015, 30, 543–552. [Google Scholar] [CrossRef]
- Smith, G.D.; Lawlor, D.A.; Harbord, R.; Timpson, N.; Day, I.; Ebrahim, S. Clustered environments and randomized genes: A fundamental distinction between conventional and genetic epidemiology. PLoS Med. 2007, 4, e352. [Google Scholar] [CrossRef]
- Didelez, V.; Sheehan, N. Mendelian randomization as an instrumental variable approach to causal inference. Stat. Methods Med. Res. 2007, 16, 309–330. [Google Scholar] [CrossRef]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’Connell, J.; et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef]
- Sudlow, C.; Gallacher, J.; Allen, N.; Beral, V.; Burton, P.; Danesh, J.; Downey, P.; Elliott, P.; Green, J.; Landray, M.; et al. UK biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015, 12, e1001779. [Google Scholar] [CrossRef] [PubMed]
- Privé, F.; Luu, K.; Blum, M.G.; McGrath, J.J.; Vilhjálmsson, B.J. Efficient toolkit implementing best practices for principal component analysis of population genetic data. Bioinformatics 2020, 36, 4449–4457. [Google Scholar] [CrossRef] [PubMed]
- Minelli, C.; Del Greco M, F.; van der Plaat, D.A.; Bowden, J.; Sheehan, N.A.; Thompson, J. The use of two-sample methods for Mendelian randomization analyses on single large datasets. Int. J. Epidemiol. 2021, 50, 1651–1659. [Google Scholar] [CrossRef]
- Locke, A.E.; Kahali, B.; Berndt, S.I.; Justice, A.E.; Pers, T.H.; Day, F.R.; Powell, C.; Vedantam, S.; Buchkovich, M.L.; Yang, J.; et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015, 518, 197–206. [Google Scholar] [CrossRef]
- Yengo, L.; Sidorenko, J.; Kemper, K.E.; Zheng, Z.; Wood, A.R.; Weedon, M.N.; Frayling, T.M.; Hirschhorn, J.; Yang, J.; Visscher, P.M. Meta-analysis of genome-wide association studies for height and body mass index in ∼700,000 individuals of European ancestry. Hum. Mol. Genet. 2018, 27, 3641–3649. [Google Scholar] [CrossRef]
- Verma, A.; Bradford, Y.; Dudek, S.; Lucas, A.M.; Verma, S.S.; Pendergrass, S.A.; Ritchie, M.D. A simulation study investigating power estimates in phenome-wide association studies. BMC Bioinform. 2018, 19, 120. [Google Scholar] [CrossRef]
- Aschard, H.; Vilhjálmsson, B.J.; Joshi, A.D.; Price, A.L.; Kraft, P. Adjusting for heritable covariates can bias effect estimates in genome-wide association studies. Am. J. Hum. Genet. 2015, 96, 329–339. [Google Scholar] [CrossRef]
- Newman, J.D.; Handelsman, D.J. Challenges to the measurement of oestradiol: Comments on an endocrine society position statement. Clin. Biochem. Rev. 2014, 35, 75. [Google Scholar]
- Piva, E.; Brugnara, C.; Spolaore, F.; Plebani, M. Clinical utility of reticulocyte parameters. Clin. Lab. Med. 2015, 35, 133–163. [Google Scholar] [CrossRef]
- Bowden, J.; Del Greco M, F.; Minelli, C.; Davey Smith, G.; Sheehan, N.A.; Thompson, J.R. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: The role of the I 2 statistic. Int. J. Epidemiol. 2016, 45, 1961–1974. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 2017, 32, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Bowden, J.; Del Greco, M.F.; Minelli, C.; Davey Smith, G.; Sheehan, N.; Thompson, J. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat. Med. 2017, 36, 1783–1802. [Google Scholar] [CrossRef] [PubMed]
- Bowden, J.; Davey Smith, G.; Haycock, P.C.; Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef]
- Burgess, S.; Bowden, J.; Fall, T.; Ingelsson, E.; Thompson, S.G. Sensitivity analyses for robust causal inference from Mendelian randomization analyses with multiple genetic variants. Epidemiology 2017, 28, 30–42. [Google Scholar] [CrossRef]
- Bowden, J.; Davey Smith, G.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef]
- Altman, D.G.; Bland, J.M. Interaction revisited: The difference between two estimates. BMJ 2003, 326, 219. [Google Scholar] [CrossRef]
- Borges, M.C.; Haycock, P.C.; Zheng, J.; Hemani, G.; Holmes, M.V.; Davey Smith, G.; Hingorani, A.D.; Lawlor, D.A. Role of circulating polyunsaturated fatty acids on cardiovascular diseases risk: Analysis using Mendelian randomization and fatty acid genetic association data from over 114,000 UK Biobank participants. BMC Med. 2022, 20, 210. [Google Scholar] [CrossRef]
- Ference, B.A.; Yoo, W.; Alesh, I.; Mahajan, N.; Mirowska, K.K.; Mewada, A.; Kahn, J.; Afonso, L.; Williams, K.A.; Flack, J.M. Effect of long-term exposure to lower low-density lipoprotein cholesterol beginning early in life on the risk of coronary heart disease: A Mendelian randomization analysis. J. Am. Coll. Cardiol. 2012, 60, 2631–2639. [Google Scholar] [CrossRef]
- Navarese, E.P.; Vine, D.; Proctor, S.; Grzelakowska, K.; Berti, S.; Kubica, J.; Raggi, P. Independent causal effect of remnant cholesterol on atherosclerotic cardiovascular outcomes: A Mendelian randomization study. Arterioscler. Thromb. Vasc. Biol. 2023, 43, e373–e380. [Google Scholar] [CrossRef]
- Luo, S.; Yeung, S.L.A.; Zhao, J.V.; Burgess, S.; Schooling, C.M. Association of genetically predicted testosterone with thromboembolism, heart failure, and myocardial infarction: Mendelian randomisation study in UK Biobank. BMJ 2019, 364, l476. [Google Scholar] [CrossRef]
- Larsson, S.C.; Michaëlsson, K.; Burgess, S. IGF-1 and cardiometabolic diseases: A Mendelian randomisation study. Diabetologia 2020, 63, 1775–1782. [Google Scholar] [CrossRef] [PubMed]
- Thibord, F.; Klarin, D.; Brody, J.A.; Chen, M.-H.; Levin, M.G.; Chasman, D.I.; Goode, E.L.; Hveem, K.; Teder-Laving, M.; Martinez-Perez, A. Cross-ancestry investigation of venous thromboembolism genomic predictors. Circulation 2022, 146, 1225–1242. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Sun, Z.; Mei, Z.; Zeng, H.; Zhao, M.; Hu, J.; Xia, M.; Huang, T.; Wang, C.; Gao, X. The causal associations of circulating amino acids with blood pressure: A Mendelian randomization study. BMC Med. 2022, 20, 414. [Google Scholar] [CrossRef]
- Zhao, J.V.; Schooling, C.M. Effect of linoleic acid on ischemic heart disease and its risk factors: A Mendelian randomization study. BMC Med. 2019, 17, 61. [Google Scholar] [CrossRef]
- Louck, L.E.; Cara, K.C.; Klatt, K.; Wallace, T.C.; Chung, M. The relationship of circulating choline and choline-related metabolite levels with health outcomes: A scoping review of genome-wide association studies and Mendelian randomization studies. Adv. Nutr. 2024, 15, 100164. [Google Scholar] [CrossRef]
- Zanetti, D.; Bergman, H.; Burgess, S.; Assimes, T.L.; Bhalla, V.; Ingelsson, E. Urinary albumin, sodium, and potassium and cardiovascular outcomes in the UK Biobank: Observational and Mendelian randomization analyses. Hypertension 2020, 75, 714–722. [Google Scholar] [CrossRef]
- Holmes, M.V.; Asselbergs, F.W.; Palmer, T.M.; Drenos, F.; Lanktree, M.B.; Nelson, C.P.; Dale, C.E.; Padmanabhan, S.; Finan, C.; Swerdlow, D.I. Mendelian randomization of blood lipids for coronary heart disease. Eur. Heart J. 2015, 36, 539–550. [Google Scholar] [CrossRef]
- Joshi, A.D.; McCormick, N.; Yokose, C.; Yu, B.; Tin, A.; Terkeltaub, R.; Merriman, T.R.; Eliassen, A.H.; Curhan, G.C.; Raffield, L.M. Prediagnostic Glycoprotein Acetyl Levels and Incident and Recurrent Flare Risk Accounting for Serum Urate Levels: A Population-Based, Prospective Study and Mendelian Randomization Analysis. Arthritis Rheumatol. 2023, 75, 1648–1657. [Google Scholar] [CrossRef]
- Casanova, F.; Wood, A.R.; Yaghootkar, H.; Beaumont, R.N.; Jones, S.E.; Gooding, K.M.; Aizawa, K.; Strain, W.D.; Hattersley, A.T.; Khan, F. A mendelian randomization study provides evidence that adiposity and dyslipidemia lead to lower urinary albumin-to-creatinine ratio, a marker of microvascular function. Diabetes 2020, 69, 1072–1082. [Google Scholar] [CrossRef]
- Zeng, Y.; Cao, S.; Tang, J.; Lin, G. Effects of saturated and monounsaturated fatty acids on cognitive impairment: Evidence from Mendelian randomization study. Eur. J. Clin. Nutr. 2024, 17, 585–590. [Google Scholar] [CrossRef]
- Kiltschewskij, D.J.; Reay, W.R.; Cairns, M.J. Evidence of genetic overlap and causal relationships between blood-based biochemical traits and human cortical anatomy. Transl. Psychiatry 2022, 12, 373. [Google Scholar] [CrossRef] [PubMed]
- Tan, E.M.; Smolen, J.S. Historical observations contributing insights on etiopathogenesis of rheumatoid arthritis and role of rheumatoid factor. J. Exp. Med. 2016, 213, 1937–1950. [Google Scholar] [CrossRef] [PubMed]
- Haworth, S.; Mitchell, R.; Corbin, L.; Wade, K.; Dudding, T.; Budu-Aggrey, A. Apparent latent structure within the UK Biobank sample has implications for epidemiological analysis. Nat. Commun. 2019, 10, 333. [Google Scholar] [CrossRef] [PubMed]
- Janiaud, P.; Agarwal, A.; Tzoulaki, I.; Theodoratou, E.; Tsilidis, K.K.; Evangelou, E.; Ioannidis, J.P. Validity of observational evidence on putative risk and protective factors: Appraisal of 3744 meta-analyses on 57 topics. BMC Med. 2021, 19, 157. [Google Scholar] [CrossRef]
- Schooling, C.M.; Lopez, P.M.; Yang, Z.; Zhao, J.; Yeung, A.; Lun, S.; Huang, J.V. Use of multivariable Mendelian randomization to address biases due to competing risk before recruitment. Front. Genet. 2021, 11, 610852. [Google Scholar] [CrossRef]

| Trait | Data Source | Ancestry | Sex | Sample Size | Adjusted Covariates | R2 | F Statistics in Univariable MR |
|---|---|---|---|---|---|---|---|
| Body mass index | UK Biobank (Neale lab) | European ancestry | Men | 167,020 | age, age2, and the first 20 principal components | 4.1% | 50.7 |
| Body mass index | UK Biobank (Neale lab) | European ancestry | Women | 194,174 | age, age2, and the first 20 principal components | 3.8% | 49.8 |
| Body mass index | GIANT [26] | European ancestry | Men | 152,893 | age, age2, and study-specific covariates | 1.5% | 62.8 |
| Body mass index | GIANT [26] | European ancestry | Women | 171,977 | age, age2, and study-specific covariates | 1.9% | 65.7 |
| Body mass index | GIANT [27] (includes approximately 64% from the UK Biobank) | European ancestry | Men and women | 681,275 | age, sex, and study-specific covariates | 5.3% | 72.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiesisibieke, Z.L.; Schooling, C.M. Body Mass Index as an Example of a Negative Confounder: Evidence and Solutions. Genes 2025, 16, 564. https://doi.org/10.3390/genes16050564
Jiesisibieke ZL, Schooling CM. Body Mass Index as an Example of a Negative Confounder: Evidence and Solutions. Genes. 2025; 16(5):564. https://doi.org/10.3390/genes16050564
Chicago/Turabian StyleJiesisibieke, Zhu Liduzi, and C. Mary Schooling. 2025. "Body Mass Index as an Example of a Negative Confounder: Evidence and Solutions" Genes 16, no. 5: 564. https://doi.org/10.3390/genes16050564
APA StyleJiesisibieke, Z. L., & Schooling, C. M. (2025). Body Mass Index as an Example of a Negative Confounder: Evidence and Solutions. Genes, 16(5), 564. https://doi.org/10.3390/genes16050564






