Identification of Mustard Aldehyde Dehydrogenase (ALDH) Gene Family and Expression Analysis Under Salt and Drought Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Their Treatment
2.2. Identification of the BjALDH Gene Family Members and Chromosome Localization
2.3. Phylogenetic and Evolutionary Analysis of the ALDH Proteins in Mustard
2.4. Gene Structure and Protein Conserved Domain Analysis of BjALDH
2.5. Analyzed by qRT-PCR Analysis of the ALDH Expression Pattern
3. Results
3.1. Identification and Chromosome Mapping of Gene Families
3.2. Physicochemical Properties and Subcellular Localization Prediction of the Proteins
3.3. Phylogenetic Analysis of the Proteins
3.4. Analysis of Gene Structure, Conserved Motifs, and Promoter Cis-Acting Elements for BjALDH
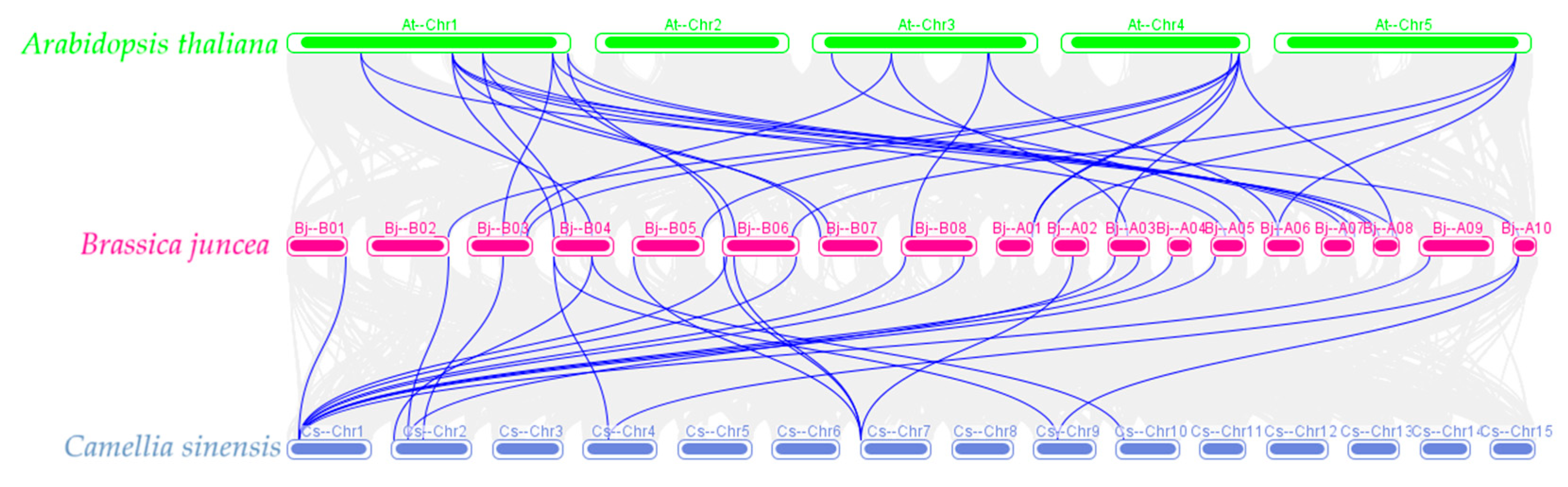
3.5. Collinear Analysis of the ALDH Gene Family
3.6. Expression Pattern of BjALDH Under Drought and High Salt Conditions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ALDH | Aldehyde Dehydrogenase |
| qRT-PCR | quantitative Real-Time Polymerase Chain Reaction |
| NaCl | Sodium Chloride |
| PBS | Phosphate Buffered Saline |
| DNA | Deoxyribonucleic Acid |
| RNA | Ribonucleic Acid |
| Gm. | Glycine max |
| Ca. | Capsicum annuum |
| At. | Arabidopsis thaliana |
| St. | Solanum tuberosum |
| Bj. | Brassica juncea |
| Cs. | Camellia sinensis |
| SS | salt stress |
| DS | drought stress |
| CK | control |
| mM | millimoles |
| Os. | Oryza sativa |
| FLC | Flowering Locus C |
| ABA | Abscisic acid |
References
- Kang, L.; Qian, L.W.; Zheng, M.; Chen, L.Y.; Chen, H.; Yang, L.; You, L.; Yang, B.; Yan, M.L.; Gu, Y.G.; et al. Genomic insights into the origin, domestication and diversification of Brassica juncea. Nat. Genet. 2021, 53, 1392–1402. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, D.; Wang, X.; Ji, C.; Cheng, F.; Liu, B.; Hu, Z.; Chen, S.; Pental, D.; Ju, Y.; et al. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 2016, 48, 1225–1232. [Google Scholar] [CrossRef] [PubMed]
- Jabeen, A.; Narejo, G.A.; Mirbahar, A.A.; Yasin, S.; Afzal, T.; Sadia, H. Effect of foliar-applied Si in alleviating cadmium toxicity to different Raya (Brassica Junceae, L.) genotypes. Silicon 2024, 16, 3951–3970. [Google Scholar] [CrossRef]
- Singh, K.H.; Singh, G.; Singh, L.; Aftab, N.; Thakur, A.K. Delineation of inbred lines of Indian mustard into diverse gene pools based on agro-morphological traits. Czech J. Genet. Plant Breed. 2023, 59, 109–116. [Google Scholar] [CrossRef]
- Wang, P.; Zhao, Z.; Zhang, Z.; Cai, Z.; Liao, J.; Tan, Q.; Xiang, M.; Chang, L.; Xu, D.; Tian, Q.; et al. Genome-wide identification and analysis of NPR family genes in Brassica juncea var. tumida. Gene 2020, 769, 145210. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant. Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Farooq, U.; Rehman, A.; Ashraf, M.A.; Rasheed, R.; Shahid, M.; Ali, S.; Sarker, P.K. Taurine priming improves redox balance, osmotic adjustment, and nutrient acquisition to lessen phytotoxic effects of neutral and alkaline salts on pea (Pisum sativum L.). Plant Signal Behav. 2025, 20, 2480224. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hazell, P.; Wood, S. Drivers of change in global agriculture. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008, 363, 495–515. [Google Scholar] [CrossRef]
- Wang, J.; Gao, X.; Wang, X.; Song, W.; Wang, Q.; Wang, X.; Li, S.; Fu, B. Exogenous melatonin ameliorates drought stress in Agropyron mongolicum by regulating flavonoid biosynthesis and carbohydrate metabolism. Front. Plant Sci. 2022, 13, 1051165. [Google Scholar] [CrossRef]
- Liu, J.; Wu, Y.; Dong, G.; Zhu, G.; Zhou, G. Progress of Research on the Physiology and Molecular Regulation of Sorghum Growth under Salt Stress by Gibberellin. Int. J. Mol. Sci. 2023, 24, 6777. [Google Scholar] [CrossRef]
- Jiang, Z.; Li, Y.P.; Gai, P.Z.; Gao, J.; Xu, L. Exogenously applied ABA alleviates dysplasia of maize (Zea mays L.) ear under drought stress by altering photosynthesis and sucrose transport. Plant Signal Behav. 2025, 20, 2462497. [Google Scholar] [PubMed]
- Tola, A.J.; Jaballi, A.; Germain, H.; Missihoun, T.D. Recent development on plant aldehyde dehydrogenase enzymes and their functions in plant development and stress signaling. Genes 2020, 12, 51. [Google Scholar] [CrossRef]
- Islam, S.; Mohtasim, M.; Islam, T.; Ghosh, A. Aldehyde dehydrogenase superfamily in sorghum: Genome-wide identification, evolution, and transcript profiling during development stages and stress conditions. BMC Plant Biol. 2022, 22, 316. [Google Scholar] [CrossRef]
- Guo, X.; Wang, Y.; Lu, H.; Cai, X.; Wang, X.; Zhou, Z.; Wang, C.; Wang, Y.; Zhang, Z.; Wang, K.; et al. Genome-wide characterization and expression analysis of the aldehyde dehydrogenase (ALDH) gene superfamily under abiotic stresses cotton. Gene 2017, 628, 230–245. [Google Scholar] [CrossRef]
- Kotchoni, S.O.; Jimenez-Lopez, J.C.; Kayodé, A.P.P.; Gachomo, E.W.; Baba-Moussa, L. The soybean aldehyde dehydrogenase (ALDH) protein superfamily. Gene 2012, 495, 128–133. [Google Scholar] [CrossRef]
- Yang, D.; Chen, H.; Zhang, Y.; Wang, Y.; Zhai, Y.; Xu, G.; Ding, Q.; Wang, M.; Zhang, Q.-A.; Lu, X.; et al. Genome-Wide Identification and Expression Analysis of the Melon Aldehyde Dehydrogenase (ALDH) Gene Family in Response to Abiotic and Biotic Stresses. Plants 2024, 13, 2939. [Google Scholar] [CrossRef] [PubMed]
- Bhuya, A.R.; Shuvo, M.R.K.; Nahid, A.A.; Ghosh, A. Genome-wide identification, classification, and expression profiling of the aldehyde dehydrogenase gene family in pepper. Plant Physiol. Biochem. 2024, 219, 109–413. [Google Scholar] [CrossRef] [PubMed]
- Kirch, H.H.; Schlingensiepen, S.; Kotchoni, S.; Sunkar, R.; Bartels, D. Detailed expression analysis of selected genes of the aldehyde dehydrogenase (ALDH) gene superfamily in Arabidopsis thaliana. Plant Mol. Biol. 2005, 57, 315–332. [Google Scholar] [CrossRef]
- Islam, S.; Hasan, S.; Hasan, N.; Prodhan, S.H.; Islam, T.; Ghosh, A. Genome-wide identification, evolution, and transcript profiling of Aldehyde dehydrogenase superfamily in potato during development stages and stress conditions. Sci. Rep. 2021, 11, 18284. [Google Scholar] [CrossRef]
- Zhang, Y.; Mao, L.; Wang, H.; Brocker, C.; Yin, X.; Vasiliou, V.; Fei, Z.; Wang, X. Genome-wide identification and analysis of grape aldehyde dehydrogenase (ALDH) gene superfamily. PLoS ONE 2012, 7, e32153. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2017, 5, e11335. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11:Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Nystrom, S.; McKay, D. Memes: A motif analysis environment in R using tools from the MEME Suite. PLoS Comput. Biol. 2021, 17, e1008991. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Chen, L.; Tan, X.; Ming, R.; Huang, D.; Tan, Y.; Li, L.; Huang, R.; Yao, S. Genome-Wide Identification of the bHLH Gene Family in Callerya speciosa Reveals Its Potential Role in the Regulation of Isoflavonoid Biosynthesis. Int. J. Mol. Sci. 2024, 25, 11900. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Ming, C.; Zhao-Shi, X.; Lian-Cheng, L.; Xue-Ping, C.; You-Zhi, M. Characteristics and Expression Patterns of the Aldehyde Dehydrogenase (ALDH) Gene Superfamily of Foxtail Millet (Setaria italica L.). PLoS ONE 2014, 9, e101136. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Liu, L.; Huang, H.; Shang, C.; Pan, H.; Fan, H.; Han, X.; Qiu, W.; Lu, Z.; Qiao, G.; et al. Genome-wide characterization and gene expression analyses of ALDH gene family in response to drought stress in moso bamboo (Phyllostachys edulis). Plant Physiol. Biochem. 2023, 202, 107954. [Google Scholar] [CrossRef] [PubMed]
- Stiti, N.; Giarola, V.; Bartels, D. From algae to vascular plants: The multistep evolutionary trajectory of the ALDH superfamily towards functional promiscuity and the emergence of structural characteristics, Environmental and Experimental. Botany 2021, 185, 104376. [Google Scholar] [CrossRef]
- Chen, J.; Wan, H.; Zhao, H.; Dai, X.; Wu, W.; Liu, J.; Xu, J.; Yang, R.; Xu, B.; Zeng, C.; et al. Identification and expression analysis of the Xyloglucan transglycosylase/hydrolase (XTH) gene family under abiotic stress in oilseed (Brassica napus L.). BMC Plant Biol. 2024, 24, 400. [Google Scholar] [CrossRef]
- Islam, M.S.; Ghosh, A. Evolution, family expansion, and functional diversification of plant aldehyde dehydrogenases. Gene 2022, 829, 146522. [Google Scholar] [CrossRef]
- Jimenez-Lopez, J.C.; Lopez-Valverde, F.J.; Robles-Bolivar, P.; Lima-Cabello, E.; Gachomo, E.W.; Kotchoni, S.O. Genome-Wide Identification and Functional Classification of Tomato (Solanum lycopersicum) Aldehyde Dehydrogenase (ALDH) Gene Superfamily. PLoS ONE 2016, 11, e0164798. [Google Scholar] [CrossRef]
- Guo, J.; Sun, W.; Liu, H.; Chi, J.; Odiba, A.S.; Li, G.; Jin, L.; Xin, C. Aldehyde dehydrogenase plays crucial roles in response to lower temperature stress in Solanum tuberosum and Nicotiana benthamiana. Plant Sci. 2020, 297, 110525. [Google Scholar] [CrossRef]
- Gao, C.; Han, B. Evolutionary and expression study of the aldehyde dehydrogenase (ALDH) gene superfamily in rice (Oryza sativa). Gene 2009, 431, 86–94. [Google Scholar] [CrossRef]
- Li, X.; Guo, R.; Li, J.; Singer, S.D.; Zhang, Y.; Yin, X.; Zheng, Y.; Fan, C.; Wang, X. Genome-wide identification and analysis of the aldehyde dehydrogenase (ALDH) gene superfamily in apple (Malus × domestica Borkh.). Plant Physiol. Biochem. 2013, 71, 268–282. [Google Scholar] [CrossRef]
- Liu, Y.; Li, J.; Shi, J.; Pan, Y.; Yang, S.; Xue, Y. Combined metabolome and transcriptome analysis reveals the key pathways involved in the responses of soybean plants to high Se stress. Ecotoxicol. Environ. Saf. 2024, 287, 117262. [Google Scholar] [CrossRef] [PubMed]





| Gene Name | Forward Primer (5′ 3′) | Reverse Primer (5′ 3′) |
|---|---|---|
| BjALDH3 | CTCGGAAGGTTGGCCCTG | TGCAAATTCAGCCGCAGC |
| BjALDH5 | GGTCGGCCACATCATCCC | CTGCTGGCTTGACCACCA |
| BjALDH7 | TGTGAGGACGCCGATGTC | CACCCGCACAGTTTTGCC |
| BjALDH8 | GCAAGCTGCGGGTGAAGT | GGACCATAAGGCCAGCGG |
| BjALDH11 | TGGGGTGGAGTCAAACGC | GCATCCCCATGGGTCGTT |
| BjALDH15 | GCCGTGAAGGAGACTGTGG | CTCCGAGCTGTGCCTTCC |
| BjALDH21 | CAACTGCTGGAGCTGCCA | GATTGCTTGCTGCTGCGG |
| BjALDH22 | GCCATGGAGGAGACTGTGG | TCCGAGCTGTGCCTTCCT |
| BjALDH25 | GCACTGGTCTGCGGAAAC | TGGCACCGGGTAAACTGT |
| GAPDHF | TCAGTTGTTGACCTCACGGTT | CTGTCACCAACGAAGTCAGT |
| Gene ID | Gene Name | Number of Amino Acids | Formula Weight/Da Molecular Weight | Isoelectric Point pI | Instability Index Instability Index | Average Hydrophobicity Gravy |
|---|---|---|---|---|---|---|
| BjuA01g40020S | BjALDH1 | 585 | 64,272.48 | 8.35 | 40.06 | 0.082 |
| BjuA01g41590S | BjALDH2 | 481 | 53,511.23 | 7.56 | 36.24 | −0.023 |
| BjuA02g19160S | BjALDH3 | 205 | 22,174.78 | 8.27 | 31.46 | 0.219 |
| BjuA03g04550S | BjALDH4 | 484 | 53,896.72 | 8.29 | 35.02 | −0.035 |
| BjuA03g21040S | BjALDH5 | 501 | 54,329.36 | 5.99 | 31.97 | −0.085 |
| BjuA05g18570S | BjALDH6 | 573 | 63,434.63 | 8.47 | 40.46 | 0.068 |
| BjuA05g35930S | BjALDH7 | 666 | 73,865.62 | 8.45 | 42.26 | 0.098 |
| BjuA06g17280S | BjALDH8 | 557 | 61,799.07 | 6.45 | 36.19 | −0.137 |
| BjuA06g19000S | BjALDH9 | 503 | 54,788.03 | 5.83 | 28.18 | −0.049 |
| BjuA06g19030S | BjALDH10 | 537 | 58,771.01 | 8.63 | 33.06 | −0.147 |
| BjuA07g24680S | BjALDH11 | 909 | 99,378.23 | 5.44 | 36.32 | 0.009 |
| BjuA07g37710S | BjALDH12 | 556 | 59,787.01 | 7.07 | 43.29 | 0.088 |
| BjuA08g01080S | BjALDH13 | 509 | 54,209.34 | 5.44 | 34.6 | 0.052 |
| BjuA08g05370S | BjALDH14 | 482 | 53,035.76 | 8.11 | 38.35 | 0.062 |
| BjuA08g18560S | BjALDH15 | 492 | 54,640.65 | 8.33 | 34.98 | −0.013 |
| BjuA08g23210S | BjALDH16 | 537 | 58,617.06 | 7.19 | 27.45 | −0.089 |
| BjuA10g06430S | BjALDH17 | 254 | 28,363.27 | 9.08 | 44.8 | 0.017 |
| BjuB02g32810S | BjALDH18 | 58 | 5911.94 | 8.21 | 26.45 | 0.66 |
| BjuB02g74420S | BjALDH19 | 484 | 53,958.68 | 8.05 | 36.4 | −0.062 |
| BjuB03g14960S | BjALDH20 | 126 | 13,802.94 | 4.27 | 34.53 | 0.329 |
| BjuB03g38000S | BjALDH21 | 501 | 54,415.44 | 5.93 | 35.62 | −0.077 |
| BjuB03g47710S | BjALDH22 | 488 | 54,232.29 | 8.47 | 33.9 | 0.011 |
| BjuB04g00710S | BjALDH23 | 475 | 52,727.58 | 9.34 | 37.01 | −0.005 |
| BjuB04g01700S | BjALDH24 | 205 | 22,113.56 | 5.73 | 29.67 | 0.074 |
| BjuB04g08660S | BjALDH25 | 124 | 13,520.87 | 4.99 | 34.83 | 0.513 |
| BjuB04g21940S | BjALDH26 | 523 | 56,972.18 | 7.67 | 26.38 | −0.054 |
| BjuB05g01010S | BjALDH27 | 133 | 14,099.38 | 8.46 | 42.56 | 0.237 |
| BjuB05g60440S | BjALDH28 | 481 | 53,471.12 | 7.07 | 37.24 | −0.029 |
| BjuB06g01260S | BjALDH29 | 528 | 56,475.96 | 7.54 | 41.71 | 0.005 |
| BjuB06g14620S | BjALDH30 | 86 | 9294.79 | 9.46 | 28.44 | −0.206 |
| BjuB06g16690S | BjALDH31 | 906 | 98,933.92 | 5.59 | 35.44 | 0.03 |
| BjuB06g26910S | BjALDH32 | 482 | 53,166.87 | 8.13 | 40.76 | 0.068 |
| BjuB06g30770S | BjALDH33 | 253 | 26,986.02 | 6.44 | 22.42 | 0.068 |
| BjuB06g31160S | BjALDH34 | 96 | 10,132.99 | 9.69 | 36.5 | 0.493 |
| BjuB06g47790S | BjALDH35 | 520 | 57,688.28 | 6.9 | 35.8 | −0.171 |
| BjuB07g01690S | BjALDH36 | 508 | 54,135.26 | 5.44 | 35 | 0.05 |
| BjuB07g08290S | BjALDH37 | 555 | 60,815.45 | 8.01 | 42.87 | 0.018 |
| BjuB07g14010S | BjALDH38 | 242 | 26,062.94 | 6.44 | 25.97 | 0.002 |
| BjuB08g11910S | BjALDH39 | 538 | 58,501.85 | 8.06 | 32.23 | −0.073 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Y.; Wang, S.; He, L.; Chen, R.; Zhang, W.; He, H.; Hu, H.; Liu, X.; Wan, H.; Wu, C. Identification of Mustard Aldehyde Dehydrogenase (ALDH) Gene Family and Expression Analysis Under Salt and Drought Stress. Genes 2025, 16, 559. https://doi.org/10.3390/genes16050559
Zheng Y, Wang S, He L, Chen R, Zhang W, He H, Hu H, Liu X, Wan H, Wu C. Identification of Mustard Aldehyde Dehydrogenase (ALDH) Gene Family and Expression Analysis Under Salt and Drought Stress. Genes. 2025; 16(5):559. https://doi.org/10.3390/genes16050559
Chicago/Turabian StyleZheng, Yuling, Shanshan Wang, Ling He, Rui Chen, Wei Zhang, Huachuan He, Hanbing Hu, Xiaoyun Liu, Heping Wan, and Chunhong Wu. 2025. "Identification of Mustard Aldehyde Dehydrogenase (ALDH) Gene Family and Expression Analysis Under Salt and Drought Stress" Genes 16, no. 5: 559. https://doi.org/10.3390/genes16050559
APA StyleZheng, Y., Wang, S., He, L., Chen, R., Zhang, W., He, H., Hu, H., Liu, X., Wan, H., & Wu, C. (2025). Identification of Mustard Aldehyde Dehydrogenase (ALDH) Gene Family and Expression Analysis Under Salt and Drought Stress. Genes, 16(5), 559. https://doi.org/10.3390/genes16050559






