Genome-Wide Characterization and Analysis of the FH Gene Family in Medicago truncatula Under Abiotic Stresses
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Physicochemical Properties Analysis of MtFH Gene Family
2.2. Phylogenetic Tree of MtFH Gene Family and Analysis of Gene Structure, Domain, and Conserved Motifs
2.3. Chromosome Localization, Collinearity, and Ka/Ks Analysis of MtFH Gene Family
2.4. Analysis of Cis-Acting Elements of MtFH Gene Family
2.5. MtFH Gene miRNA Prediction and GO Enrichment Analysis
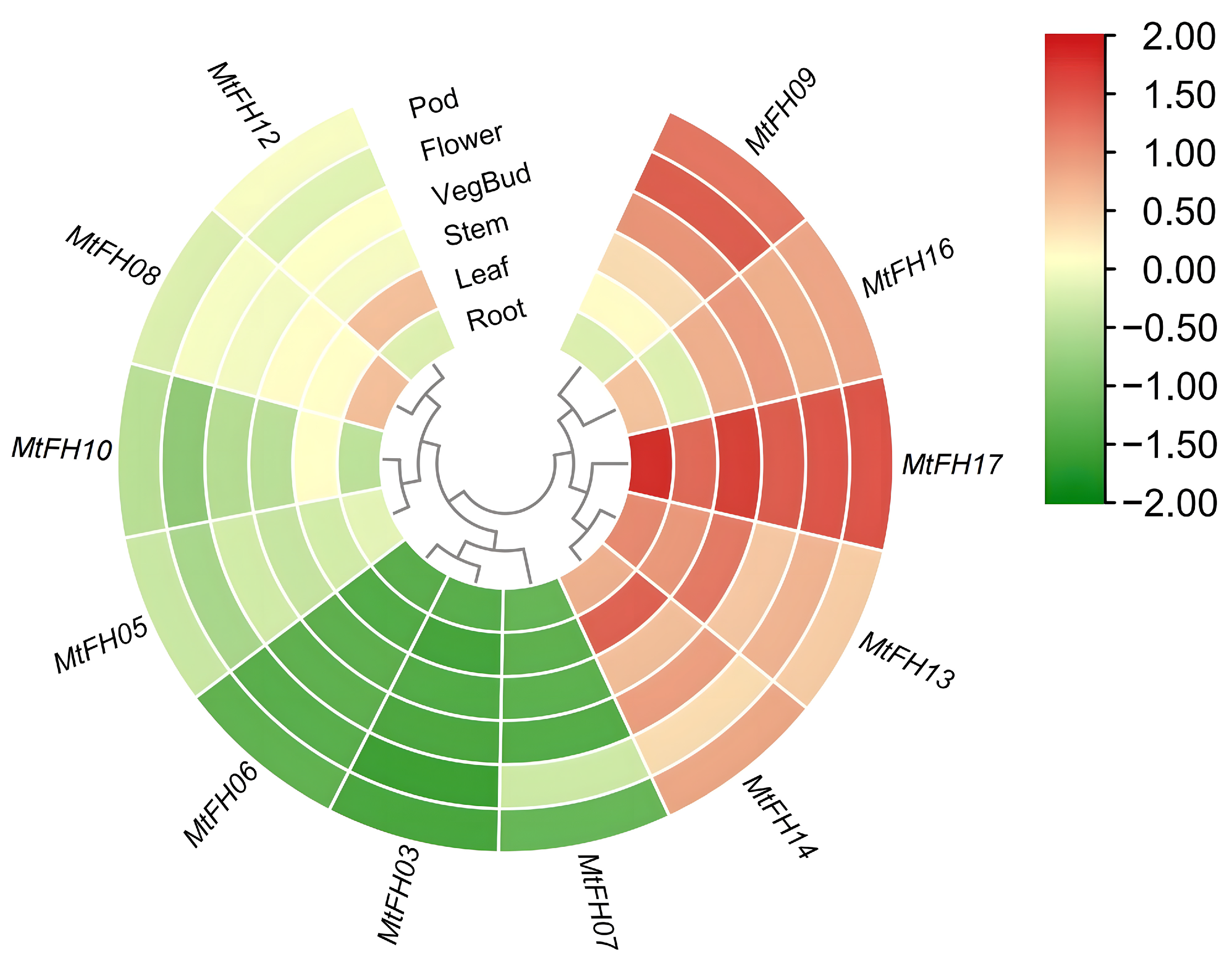
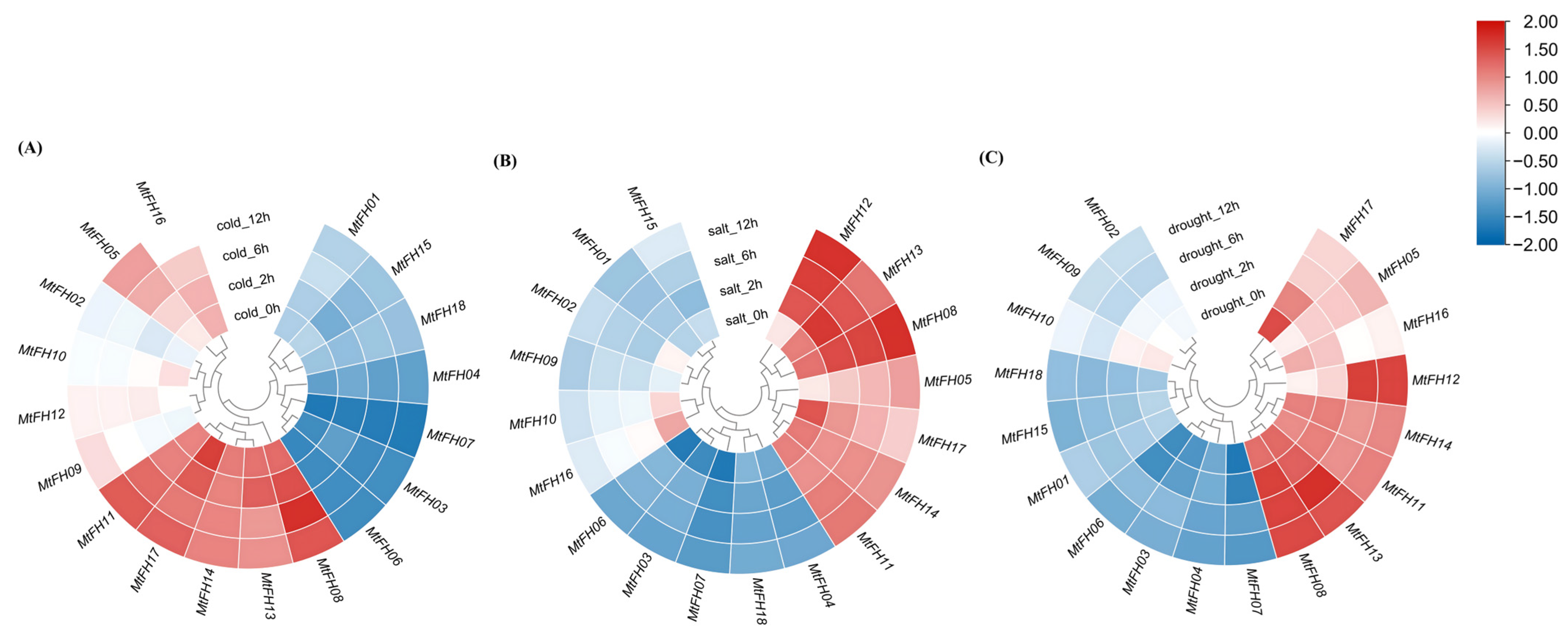
2.6. Tissue-Specific and Stress-Related Expression Profiles of MtFH Gene Family
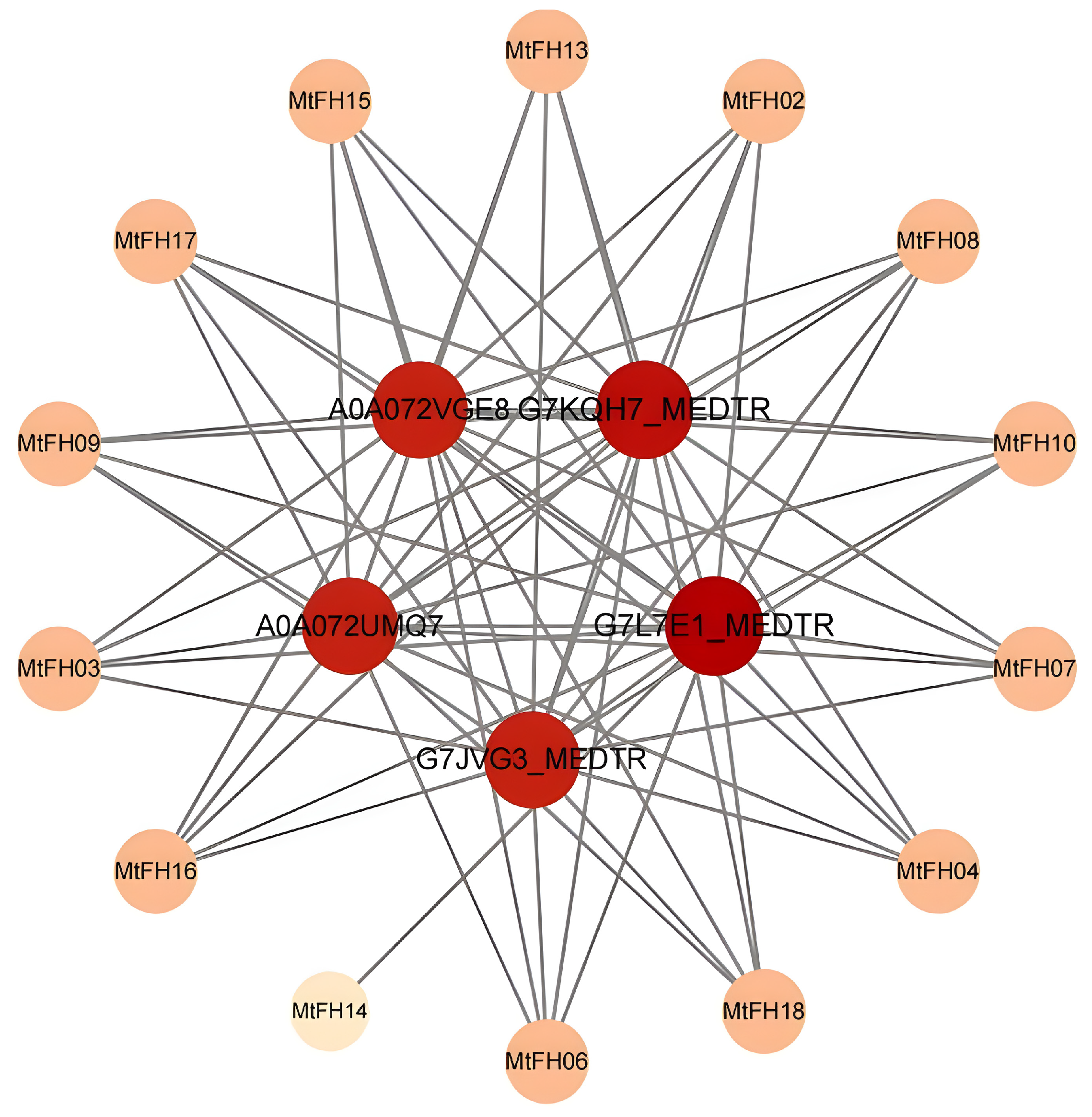
2.7. PPI Interaction Network
3. Results
3.1. Identification and Information of FH Gene Family in M. truncatula
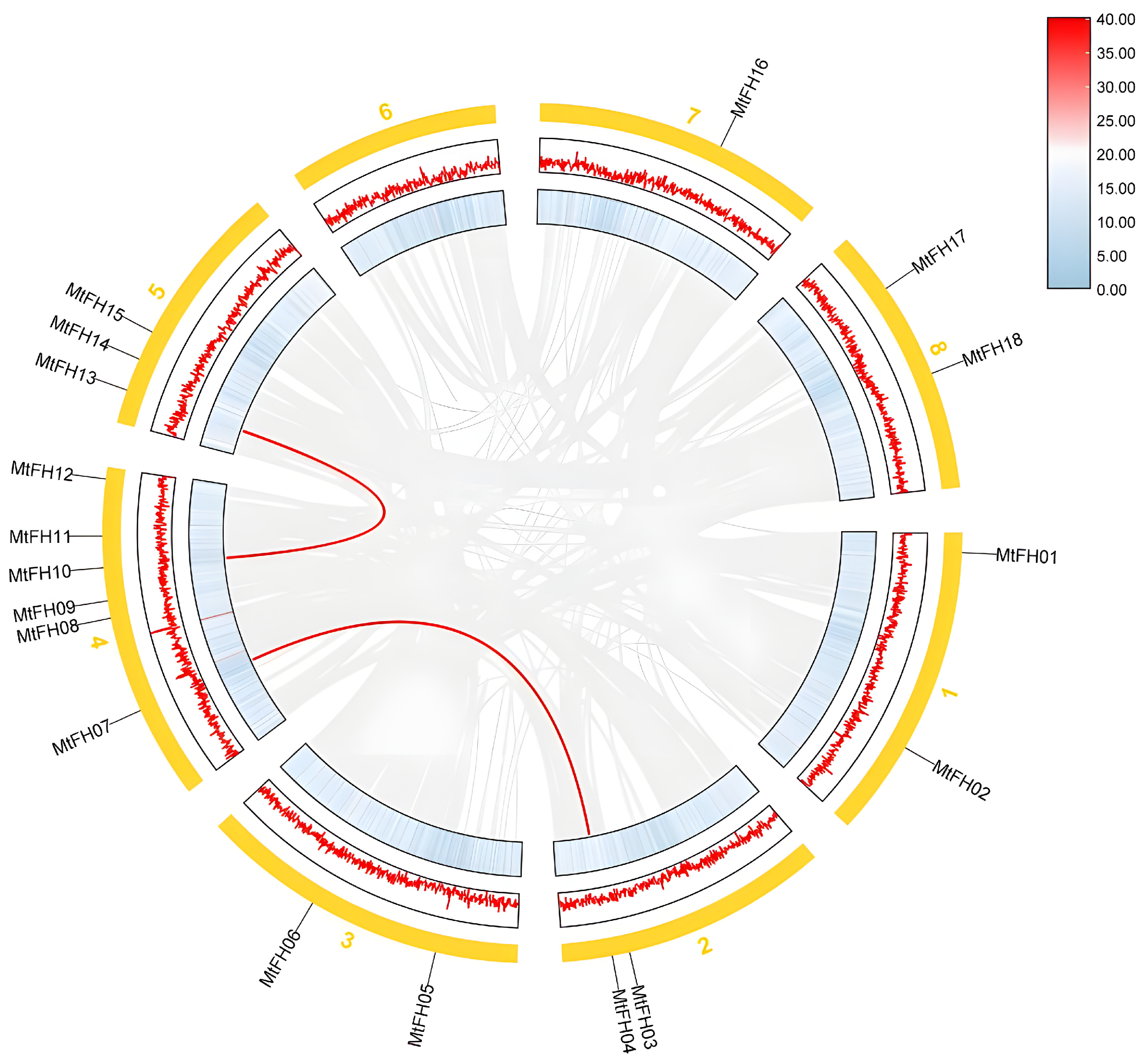
3.2. Chromosomal Distribution of FH Gene Family in M. truncatula
3.3. Phylogenetic Analysis of FH Family in M. truncatula
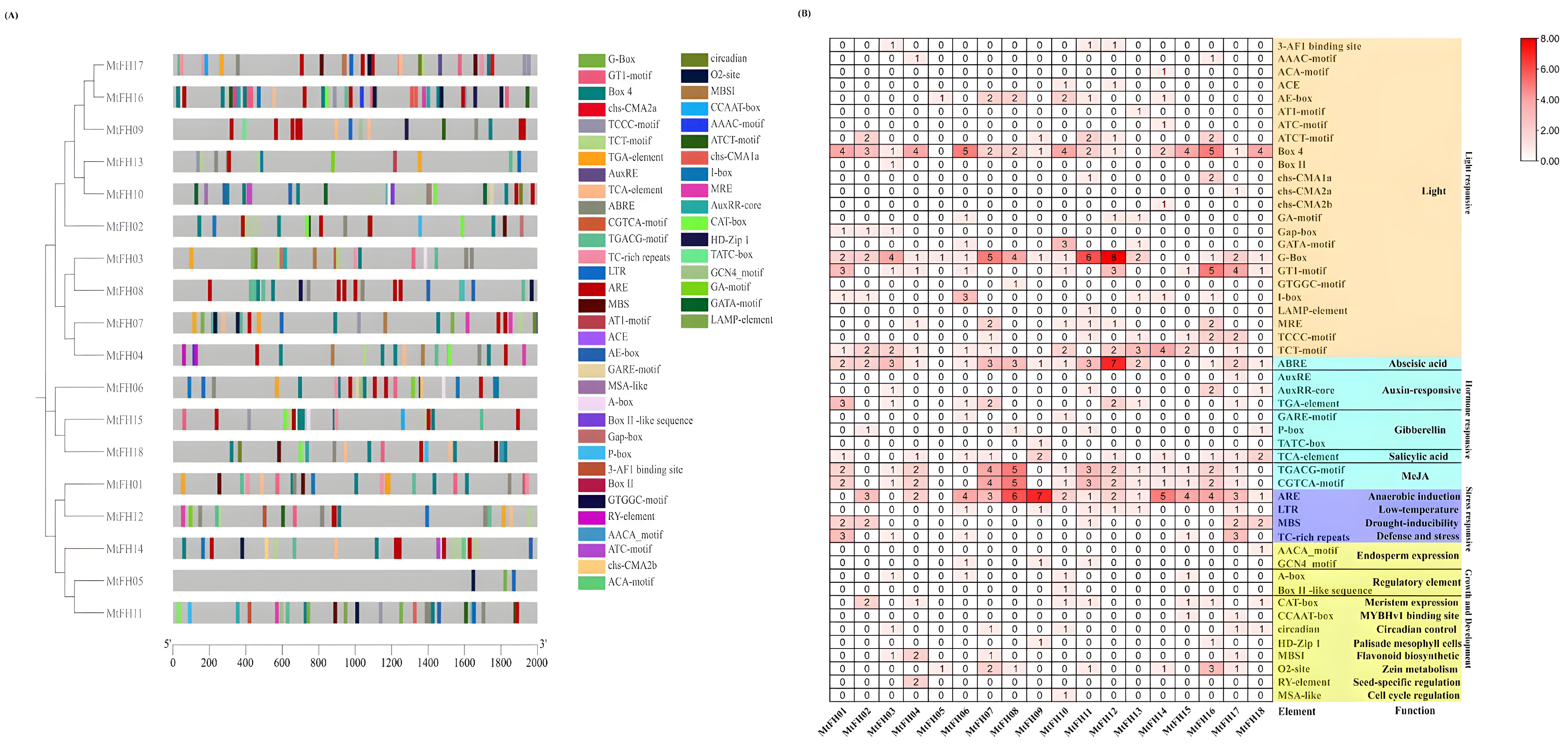
3.4. Evolutionary Relationship, Motif, Conserved Domain, and Gene Structure Analysis of FH Gene Family in M. truncatula
3.5. Intermediate Collinearity Analysis of M. truncatula
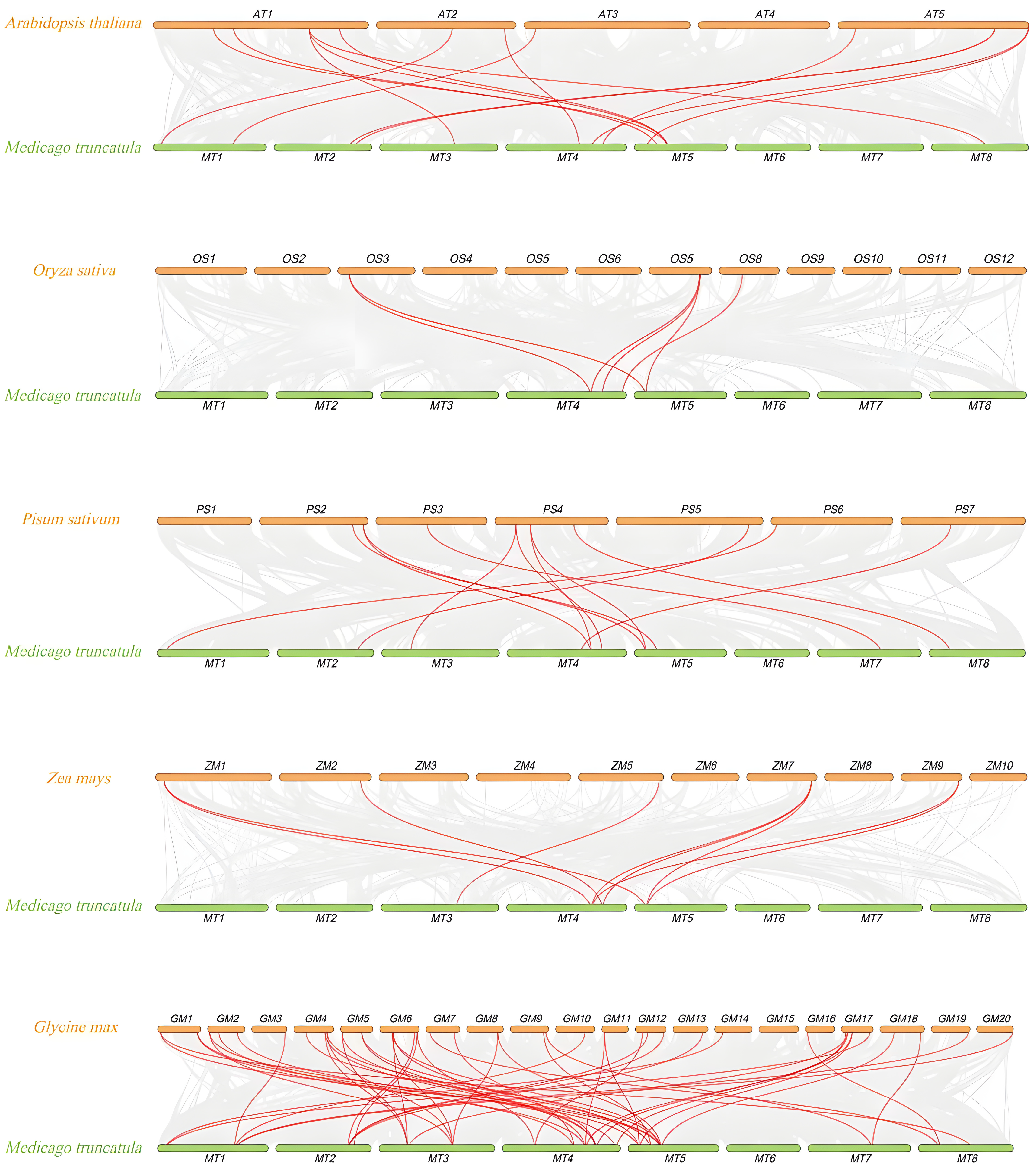
3.6. Collinearity Analysis Between M. truncatula and Other Plants
3.7. Analysis of the MtFH Gene’s Cis-Acting Elements
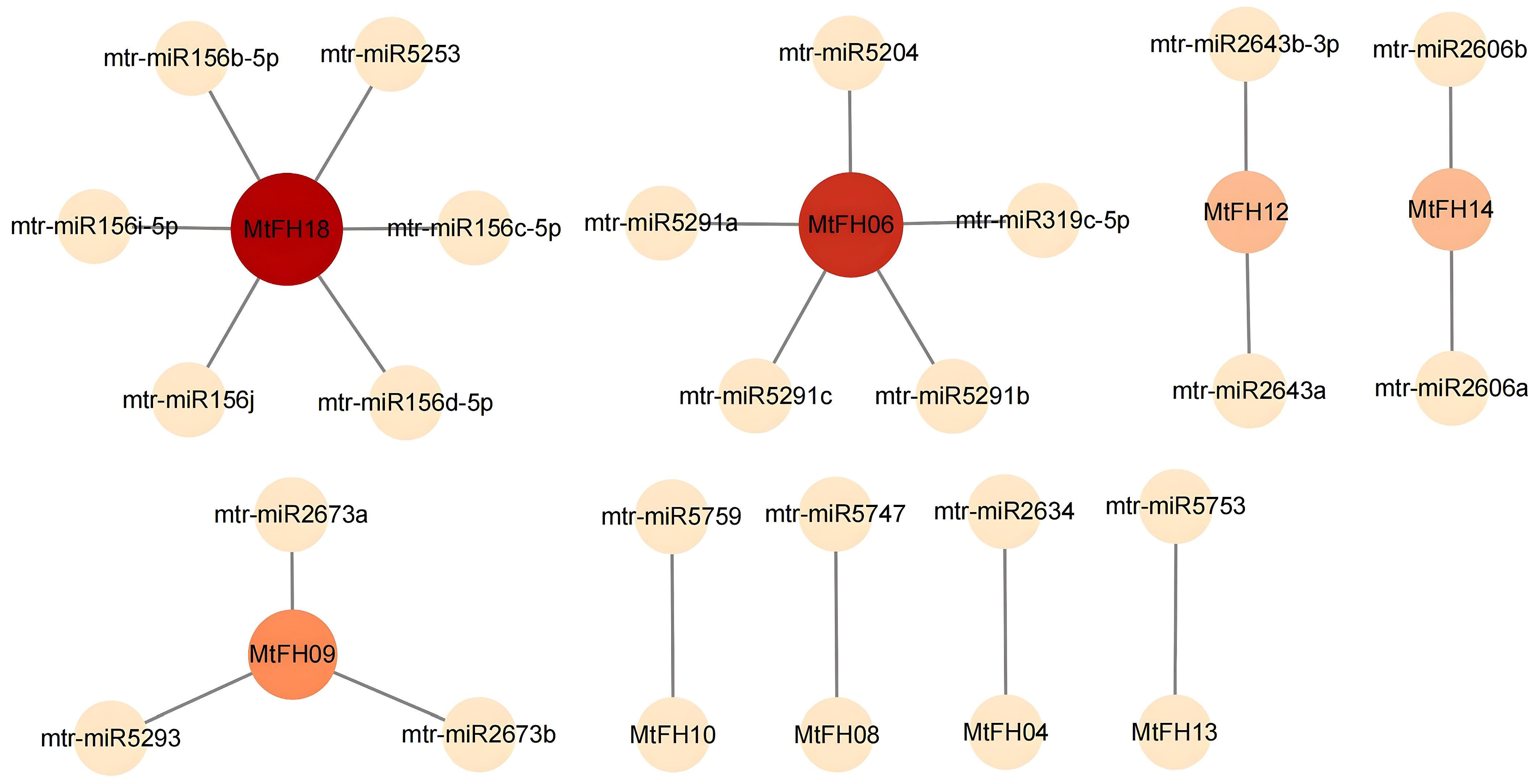
3.8. Prediction of miRNAs for MtFH Gene
3.9. GO Enrichment Analysis
3.10. Analysis of Expression Patterns of MtFH Gene in Different Tissues of M. truncatula
3.11. Expression Pattern of MtFH Gene Under Abiotic Stress
3.12. Analysis of Protein–Protein Interaction Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Evangelista, M.; Zigmond, S.; Boone, C. Formins: Signaling effectors for assembly and polarization of actin filaments. J. Cell Sci. 2003, 116, 2603–2611. [Google Scholar] [CrossRef] [PubMed]
- Volkmann, N.; Page, C.; Li, R.; Hanein, D. Three-dimensional reconstructions of actin filaments capped by Arp2/3 complex. Eur. J. Cell Biol. 2014, 93, 179–183. [Google Scholar] [CrossRef]
- Zimmermann, D.; Kovar, D.R. Feeling the force: Formin’s role in mechanotransduction. Curr. Opin. Cell Biol. 2019, 56, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Akin, O.; Mullins, R.D. Capping Protein Increases the Rate of Actin-Based Motility by Promoting Filament Nucleation by the Arp2/3 Complex. Cell 2008, 133, 841–851. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Cao, L.; Mladenov, M.; Jegou, A.; Way, M.; Moores, C.A. Cortactin stabilizes actin branches by bridging activated Arp2/3 to its nucleated actin filament. Nat. Struct. Mol. Biol. 2024, 31, 801–809. [Google Scholar] [CrossRef]
- Baum, B.; Kunda, P. Actin Nucleation: Spire—Actin Nucleator in a Class of Its Own. Curr. Biol. 2005, 15, R305–R308. [Google Scholar] [CrossRef]
- Sagot, I.; Rodal, A.A.; Moseley, J.; Goode, B.L.; Pellman, D. An actin nucleation mechanism mediated by Bni1 and Profilin. Nat. Cell Biol. 2002, 4, 626–631. [Google Scholar] [CrossRef]
- Rodnick-Smith, M.; Luan, Q.; Liu, S.-L.; Nolen, B.J. Role and structural mechanism of WASP-triggered conformational changes in branched actin filament nucleation by Arp2/3 complex. Proc. Natl. Acad. Sci. USA 2016, 113, E3834–E3843. [Google Scholar] [CrossRef]
- Quinlan, M.E.; Heuser, J.E.; Kerkhoff, E.; Dyche Mullins, R. Drosophila Spire is an actin nucleation factor. Nature 2005, 433, 382–388. [Google Scholar] [CrossRef]
- Valencia, D.A.; Quinlan, M.E. Formins. Curr. Biol. 2021, 31, R517–R522. [Google Scholar] [CrossRef]
- Moseley, J.B.; Sagot, I.; Manning, A.L.; Xu, Y.; Eck, M.J.; Pellman, D.; Goode, B.L. A Conserved Mechanism for Bni1- and mDia1-induced Actin Assembly and Dual Regulation of Bni1 by Bud6 and Profilin. Mol. Biol. Cell 2004, 15, 896–907. [Google Scholar] [CrossRef]
- Evangelista, M.; Pruyne, D.; Amberg, D.C.; Boone, C.; Bretscher, A. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 2002, 4, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Pruyne, D.; Evangelista, M.; Yang, C.; Bi, E.; Zigmond, S.; Bretscher, A.; Boone, C. Role of Formins in Actin Assembly: Nucleation and Barbed-End Association. Science 2002, 297, 612–615. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Moseley, J.B.; Sagot, I.; Poy, F.; Pellman, D.; Goode, B.L.; Eck, M.J. Crystal Structures of a Formin Homology-2 Domain Reveal a Tethered Dimer Architecture. Cell 2004, 116, 711–723. [Google Scholar] [CrossRef]
- Kleinebrecht, J.; Selow, J.; Winkler, W. The mouse mutant limb-deformity (ld). Anat Anz 1982, 152, 313–324. [Google Scholar]
- Tanaka, K. Formin Family Proteins in Cytoskeletal Control. Biochem. Biophys. Res. Commun. 2000, 267, 479–481. [Google Scholar] [CrossRef] [PubMed]
- Evangelista, M.; Blundell, K.; Longtine, M.S.; Chow, C.J.; Adames, N.; Pringle, J.R.; Peter, M.; Boone, C. Bni1p, a Yeast Formin Linking Cdc42p and the Actin Cytoskeleton During Polarized Morphogenesis. Science 1997, 276, 118–122. [Google Scholar] [CrossRef]
- Duan, W.-j.; Liu, Z.-h.; Bai, J.-f.; Yuan, S.-h.; Li, Y.-m.; Lu, F.-k.; Zhang, T.-b.; Sun, J.-h.; Zhang, F.-t.; Zhao, C.-p.; et al. Comprehensive analysis of formin gene family highlights candidate genes related to pollen cytoskeleton and male fertility in wheat (Triticum aestivum L.). BMC Genom. 2021, 22, 570. [Google Scholar] [CrossRef]
- Courtemanche, N. Pathways for Actin Polymerization Mediated by Formins. Biophys. J. 2020, 118, 32a. [Google Scholar] [CrossRef]
- Cvrčková, F.; Novotný, M.; Pícková, D.; Žárský, V. Formin homology 2 domains occur in multiple contexts in angiosperms. BMC Genom. 2004, 5, 44. [Google Scholar] [CrossRef]
- Blanchoin, L.; Staiger, C.J. Plant formins: Diverse isoforms and unique molecular mechanism. Biochim. Biophys. Acta (BBA)—Mol. Cell Res. 2010, 1803, 201–206. [Google Scholar] [CrossRef]
- Paul, A.; Pollard, T. The Role of the FH1 Domain and Profilin in Formin-Mediated Actin-Filament Elongation and Nucleation. Curr. Biol. 2008, 18, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Faix, J.; Grosse, R. Staying in Shape with Formins. Dev. Cell 2006, 10, 693–706. [Google Scholar] [CrossRef]
- Zweifel, M.E.; Courtemanche, N. Profilin’s Affinity for Formin Regulates the Availability of Filament Ends for Actin Monomer Binding. J. Mol. Biol. 2020, 432, 166688. [Google Scholar] [CrossRef] [PubMed]
- Kovar, D.R. Molecular details of formin-mediated actin assembly. Curr. Opin. Cell Biol. 2006, 18, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Vavylonis, D.; Kovar, D.R.; O’Shaughnessy, B.; Pollard, T.D. Model of Formin-Associated Actin Filament Elongation. Mol. Cell 2006, 21, 455–466. [Google Scholar] [CrossRef]
- Petersen, J.; Nielsen, O.; Egel, R.; Hagan, I.M. FH3, A Domain Found in Formins, Targets the Fission Yeast Formin Fus1 to the Projection Tip During Conjugation. J. Cell Biol. 1998, 141, 1217–1228. [Google Scholar] [CrossRef]
- Kato, T.; Watanabe, N.; Morishima, Y.; Fujita, A.; Ishizaki, T.; Narumiya, S. Localization of a mammalian homolog of diaphanous, mDia1, to the mitotic spindle in HeLa cells. J. Cell Sci. 2001, 114, 775–784. [Google Scholar] [CrossRef]
- Deeks, M.J.; Hussey, P.J.; Davies, B. Formins: Intermediates in signal-transduction cascades that affect cytoskeletal reorganization. Trends Plant Sci. 2002, 7, 492–498. [Google Scholar] [CrossRef]
- Grunt, M.; Žárský, V.; Cvrčková, F. Roots of angiosperm formins: The evolutionary history of plant FH2 domain-containing proteins. BMC Evol. Biol. 2008, 8, 115. [Google Scholar] [CrossRef]
- van Gisbergen, P.A.C.; Bezanilla, M. Plant formins: Membrane anchors for actin polymerization. Trends Cell Biol. 2013, 23, 227–233. [Google Scholar] [CrossRef]
- Huang, J.; Kim, C.M.; Xuan, Y.-h.; Liu, J.; Kim, T.H.; Kim, B.-K.; Han, C.-d. Formin homology 1 (OsFH1) regulates root-hair elongation in rice (Oryza sativa). Planta 2013, 237, 1227–1239. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, Y.; Tan, H.; Wang, Y.; Li, G.; Liang, W.; Yuan, Z.; Hu, J.; Ren, H.; Zhang, D. RICE MORPHOLOGY DETERMINANT Encodes the Type II Formin FH5 and Regulates Rice Morphogenesis. Plant Cell 2011, 23, 681–700. [Google Scholar] [CrossRef]
- Lan, Y.; Liu, X.; Fu, Y.; Huang, S. Arabidopsis class I formins control membrane-originated actin polymerization at pollen tube tips. PLoS Genet. 2018, 14, e1007789. [Google Scholar] [CrossRef] [PubMed]
- Yi, K.; Guo, C.; Chen, D.; Zhao, B.; Yang, B.; Ren, H. Cloning and Functional Characterization of a Formin-Like Protein (AtFH8) from Arabidopsis. Plant Physiol. 2005, 138, 1071–1082. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, Z.; Shan, M.; Amjad, Z.; Xue, J.; Zhang, Z.; Wang, J.; Guo, Y. Genome-Wide Studies of FH Family Members in Soybean (Glycine max) and Their Responses under Abiotic Stresses. Plants 2024, 13, 276. [Google Scholar] [CrossRef]
- Khatun, M.S.; Islam, M.S.U.; Shing, P.; Zohra, F.T.; Rashid, S.B.; Rahman, S.M.; Sarkar, M.A.R. Genome-wide identification and characterization of FORMIN gene family in potato (Solanum tuberosum L.) and their expression profiles in response to drought stress condition. PLoS ONE 2024, 19, e0309353. [Google Scholar] [CrossRef]
- Smýkal, P.; von Wettberg, E.J.B.; McPhee, K. Legume Genetics and Biology: From Mendel’s Pea to Legume Genomics. Int. J. Mol. Sci. 2020, 21, 3336. [Google Scholar] [CrossRef]
- Tang, H.; Krishnakumar, V.; Bidwell, S.; Rosen, B.; Chan, A.; Zhou, S.; Gentzbittel, L.; Childs, K.L.; Yandell, M.; Gundlach, H.; et al. An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genom. 2014, 15, 312. [Google Scholar] [CrossRef]
- Raza, A.; Razzaq, A.; Mehmood, S.S.; Zou, X.; Zhang, X.; Lv, Y.; Xu, J. Impact of Climate Change on Crops Adaptation and Strategies to Tackle Its Outcome: A Review. Plants 2019, 8, 34. [Google Scholar] [CrossRef]
- Naramoto, S.; Hata, Y.; Fujita, T.; Kyozuka, J. The bryophytes Physcomitrium patens and Marchantia polymorpha as model systems for studying evolutionary cell and developmental biology in plants. Plant Cell 2021, 34, 228–246. [Google Scholar] [CrossRef] [PubMed]
- Young, N.D.; Udvardi, M. Translating Medicago truncatula genomics to crop legumes. Curr. Opin. Plant Biol. 2009, 12, 193–201. [Google Scholar] [CrossRef]
- Zhang, X.; Sun, Y.; Qiu, X.; Lu, H.; Hwang, I.; Wang, T. Tolerant mechanism of model legume plant Medicago truncatula to drought, salt, and cold stresses. Front. Plant Sci. 2022, 13, 847166. [Google Scholar] [CrossRef]
- Yan, L.; Su, B.-X.; Li, J.-J.; Li, Y.-Y.; Chen, S.-Y.; Feng, C.-Y.; Tian, Y.; Ai, Y.; Zhang, Q.-H. Genome-Wide Identification of Nucleotide-Binding Site–Leucine-Rich Repeat Gene Family in Cymbidium ensifolium and Expression Profiles in Response to Fusarium Wilt Infection. Horticulturae 2024, 10, 634. [Google Scholar] [CrossRef]
- Tian, J.; Chang, K.; Lei, Y.; Li, S.; Wang, J.; Huang, C.; Zhong, F. Genome-Wide Identification of Proline Transporter Gene Family in Non-Heading Chinese Cabbage and Functional Analysis of BchProT1 under Heat Stress. Int. J. Mol. Sci. 2024, 25, 99. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Wu, Y.; Zhao, X.; Miao, J.-L.; Deng, S.; Li, S.; Gao, R.; Liu, Z.-J.; Zhai, J. Genome-Wide Identification and Expression Analysis of WNK Kinase Gene Family in Acorus. Int. J. Mol. Sci. 2023, 24, 17594. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhang, B.; Li, T.; Yao, X.; Feng, T.; Ai, H.; Huang, X. Identification and Characterization of the BZR Transcription Factor Genes Family in Potato (Solanum tuberosum L.) and Their Expression Profiles in Response to Abiotic Stresses. Plants 2024, 13, 407. [Google Scholar] [CrossRef]
- Zheng, Q.; Huang, Y.; He, X.; Zhang, M.-M.; Liu, Z.-J. Genome-Wide Identification and Expression Pattern Analysis of GATA Gene Family in Orchidaceae. Genes 2024, 15, 915. [Google Scholar] [CrossRef]
- Yang, F.; Sun, X.; Wu, G.; He, X.; Liu, W.; Wang, Y.; Sun, Q.; Zhao, Y.; Xu, D.; Dai, X.; et al. Genome-Wide Identification and Expression Profiling of the ABF Transcription Factor Family in Wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2024, 25, 3783. [Google Scholar] [CrossRef]
- Breitsprecher, D.; Goode, B.L. Formins at a glance. J. Cell Sci. 2013, 126, 1–7. [Google Scholar] [CrossRef]
- Goode, B.L.; Eck, M.J. Mechanism and Function of Formins in the Control of Actin Assembly. Annu. Rev. Biochem. 2007, 76, 593–627. [Google Scholar] [CrossRef] [PubMed]
- Wasteneys, G.O.; Galway, M.E. Remodeling the Cytoskeleton for Growth and Form: An Overview with Some New Views. Annu. Rev. Plant Biol. 2003, 54, 691–722. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Li, X.; Yang, J.; Zhang, Q.; Zhang, L.; Cheng, F.; Xu, W. Phosphorus-Use-Efficiency Gene Identification in Fabaceae and RSL2 Expansion in Lupinus albus Is Associated with Low-Phosphorus Adaptation. Genes 2024, 15, 1049. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Fan, C.; Sun, J.; Chang, Y.; Lu, J.; Sun, J.; Wang, C.; Liu, J. Genome-Wide Identification, Evolution, and Expression Analysis of the TCP Gene Family in Rose (Rosa chinensis Jacq.). Horticulturae 2022, 8, 961. [Google Scholar] [CrossRef]
- Fiallos-Salguero, M.S.; Li, J.; Li, Y.; Xu, J.; Fang, P.; Wang, Y.; Zhang, L.; Tao, A. Identification of AREB/ABF Gene Family Involved in the Response of ABA under Salt and Drought Stresses in Jute (Corchorus olitorius L.). Plants 2023, 12, 1161. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.A.; Hsu, P.-K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic Acid and Abiotic Stress Tolerance in Crop Plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef]
- Yang, Y.-Z.; Tan, B.-C. A Distal ABA Responsive Element in AtNCED3 Promoter Is Required for Positive Feedback Regulation of ABA Biosynthesis in Arabidopsis. PLoS ONE 2014, 9, e87283. [Google Scholar] [CrossRef]
- Ahmadi, F.I.; Karimi, K.; Struik, P.C. Effect of exogenous application of methyl jasmonate on physiological and biochemical characteristics of Brassica napus L. cv. Talaye under salinity stress. S. Afr. J. Bot. 2018, 115, 5–11. [Google Scholar] [CrossRef]
- Tavallali, V.; Karimi, S. Methyl jasmonate enhances salt tolerance of almond rootstocks by regulating endogenous phytohormones, antioxidant activity and gas-exchange. J. Plant Physiol. 2019, 234–235, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Riffo-Campos, Á.L.; Riquelme, I.; Brebi-Mieville, P. Tools for Sequence-Based miRNA Target Prediction: What to Choose? Int. J. Mol. Sci. 2016, 17, 1987. [Google Scholar] [CrossRef]
- Zhang, Y.; Peng, Y.; Zhang, H.; Gao, Q.; Song, F.; Cui, X.; Mo, F. Genome-Wide Identification of APX Gene Family in Citrus maxima and Expression Analysis at Different Postharvest Preservation Times. Genes 2024, 15, 911. [Google Scholar] [CrossRef] [PubMed]
- Arshad, M.; Feyissa, B.A.; Amyot, L.; Aung, B.; Hannoufa, A. MicroRNA156 improves drought stress tolerance in alfalfa (Medicago sativa) by silencing SPL13. Plant Sci. 2017, 258, 122–136. [Google Scholar] [CrossRef]
- Guo, M.; Li, H.; Zhu, L.; Wu, Z.; Li, J.; Li, Z. Genome-wide identification of microRNAs associated with osmotic stress and elucidation of the role of miR319 in Medicago ruthenica seedlings. Plant Physiol. Biochem. 2021, 168, 53–61. [Google Scholar] [CrossRef]
- Cao, C.; Long, R.; Zhang, T.; Kang, J.; Wang, Z.; Wang, P.; Sun, H.; Yu, J.; Yang, Q. Genome-Wide Identification of microRNAs in Response to Salt/Alkali Stress in Medicago truncatula through High-Throughput Sequencing. Int. J. Mol. Sci. 2018, 19, 4076. [Google Scholar] [CrossRef]
- Shing, P.; Islam, M.S.U.; Khatun, M.S.; Zohra, F.T.; Hasan, N.; Rahman, S.M.; Sarkar, M.A.R. Genome-wide identification, characterization and expression profiles of FORMIN gene family in cotton (Gossypium raimondii L.). BMC Genom. Data 2024, 25, 105. [Google Scholar] [CrossRef]
- Paul, S.K.; Islam, M.S.U.; Akter, N.; Zohra, F.T.; Rashid, S.B.; Ahmed, M.S.; Rahman, S.M.; Sarkar, M.A.R. Genome-wide identification and characterization of FORMIN gene family in cotton (Gossypium hirsutum L.) and their expression profiles in response to multiple abiotic stress treatments. PLoS ONE 2025, 20, e0319176. [Google Scholar] [CrossRef]

 represents FH members of Glycine max,
represents FH members of Glycine max,  represents FH members of Oryza sativa,
represents FH members of Oryza sativa,  represents FH members of Arabidopsis thaliana.
represents FH members of Arabidopsis thaliana.
 represents FH members of Glycine max,
represents FH members of Glycine max,  represents FH members of Oryza sativa,
represents FH members of Oryza sativa,  represents FH members of Arabidopsis thaliana.
represents FH members of Arabidopsis thaliana.


| Symbol | Gene ID | Chr | Start | End | aa | MW | pI | Instability Index | Aliphatic Index | GRAVY | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MtFH01 | Medtr1g013800.1 | Chr1 | 3233402 | 3244659 | 1778 | 194,744.01 | 5.84 | 60.86 | 79.26 | −0.447 | Nucleus |
| MtFH02 | Medtr1g083260.1 | Chr1 | 37051517 | 37055541 | 909 | 101,594.46 | 7.39 | 60.08 | 77.48 | −0.547 | Nucleus |
| MtFH03 | Medtr2g082190.1 | Chr2 | 34624278 | 34629796 | 860 | 95,004.64 | 8.58 | 45.73 | 77.21 | −0.555 | Chloroplast |
| MtFH04 | Medtr2g089040.1 | Chr2 | 37560923 | 37565770 | 847 | 95,007.30 | 8.34 | 47.33 | 79.22 | −0.571 | Chloroplast |
| MtFH05 | Medtr3g037080.1 | Chr3 | 13564187 | 13578034 | 1928 | 206,923.34 | 6.18 | 80.43 | 62.04 | −0.618 | Nucleus |
| MtFH06 | Medtr3g078623.1 | Chr3 | 35438534 | 35441319 | 689 | 77,181.65 | 8.42 | 48.24 | 79.83 | −0.431 | Nucleus |
| MtFH07 | Medtr4g045670.1 | Chr4 | 15509608 | 15513682 | 860 | 94,395.75 | 6.15 | 55.77 | 76.64 | −0.529 | Nucleus |
| MtFH08 | Medtr4g081410.1 | Chr4 | 31560742 | 31567076 | 984 | 106,691.00 | 8.52 | 55.67 | 77.73 | −0.429 | Nucleus |
| MtFH09 | Medtr4g087890.1 | Chr4 | 34465056 | 34468637 | 889 | 98,723.37 | 6.07 | 62.15 | 73.69 | −0.523 | Nucleus |
| MtFH10 | Medtr4g095780.1 | Chr4 | 39940525 | 39944800 | 857 | 94,815.78 | 8.88 | 48.34 | 77.58 | −0.494 | Nucleus |
| MtFH11 | Medtr4g109040.1 | Chr4 | 45203395 | 45215879 | 1576 | 171,050.56 | 6.40 | 69.23 | 65.86 | −0.578 | Nucleus |
| MtFH12 | Medtr4g131020.1 | Chr4 | 54634518 | 54646617 | 1198 | 133,396.00 | 8.38 | 58.39 | 77.82 | −0.496 | Nucleus |
| MtFH13 | Medtr5g015690.1 | Chr5 | 5489429 | 5494498 | 908 | 99,681.28 | 9.14 | 52.06 | 73.25 | −0.599 | Nucleus |
| MtFH14 | Medtr5g026645.1 | Chr5 | 10985497 | 10993656 | 1211 | 131,552.96 | 7.54 | 55.84 | 74.35 | −0.422 | Nucleus |
| MtFH15 | Medtr5g036540.1 | Chr5 | 15943741 | 15946226 | 797 | 87,539.55 | 9.21 | 57.17 | 80.36 | −0.495 | Nucleus |
| MtFH16 | Medtr7g080920.1 | Chr7 | 30828192 | 30832234 | 1012 | 112,425.62 | 6.89 | 66.84 | 72.23 | −0.571 | Nucleus |
| MtFH17 | Medtr8g027995.1 | Chr8 | 10323250 | 10328257 | 1071 | 117,088.69 | 8.62 | 69.42 | 73.44 | −0.459 | Nucleus |
| MtFH18 | Medtr8g062830.1 | Chr8 | 26282122 | 26285210 | 740 | 82,239.76 | 9.02 | 49.26 | 85.91 | −0.492 | Vacuole |
| Gene 1 | Gene 2 | Ka | Ks | Ka/Ks | Purifying Selection | Duplicate Type |
|---|---|---|---|---|---|---|
| MtFH04 | MtFH07 | 0.281891053 | 0.99328562 | 0.283796571 | Yes | segmental |
| MtFH10 | MtFH13 | 0.227763917 | 1.194490726 | 0.190678682 | Yes | segmental |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Zhou, C. Genome-Wide Characterization and Analysis of the FH Gene Family in Medicago truncatula Under Abiotic Stresses. Genes 2025, 16, 555. https://doi.org/10.3390/genes16050555
Wang J, Zhou C. Genome-Wide Characterization and Analysis of the FH Gene Family in Medicago truncatula Under Abiotic Stresses. Genes. 2025; 16(5):555. https://doi.org/10.3390/genes16050555
Chicago/Turabian StyleWang, Jiatong, and Chunyang Zhou. 2025. "Genome-Wide Characterization and Analysis of the FH Gene Family in Medicago truncatula Under Abiotic Stresses" Genes 16, no. 5: 555. https://doi.org/10.3390/genes16050555
APA StyleWang, J., & Zhou, C. (2025). Genome-Wide Characterization and Analysis of the FH Gene Family in Medicago truncatula Under Abiotic Stresses. Genes, 16(5), 555. https://doi.org/10.3390/genes16050555





