The Complete Mitochondrial Genome of Red Costate Tiger Moth (Aloa lactinea [Cramer, 1777]), and Phylogenetic Analyses of the Subfamily Arctiinae
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. DNA Sequencing and Splicing
2.3. Mitochondrial Genome Annotation
2.4. Phylogenetic Analysis
| Genus | Species | Genbank Sequence Number | |
|---|---|---|---|
| Vamuna | Vamuna virilis | NC_026844.1 | \ |
| Eilema | Eilema ussuricum | MN696172.1 | Unpublished |
| Cyana | Cyana sp. MT-2014 | KM244679.1 | [42] |
| Paraona | Paraona staudingeri | NC_037515.1 | \ |
| Spilosoma | Spilosoma lubricipeda | MK903030.1 | Unpublished |
| Arctia | Arctia plantaginis | NC_057559.1 | [43] |
| Hyphantria | Hyphantria cunea | GU592049.1 | [30] |
| Callimorpha | Callimorpha dominula | NC_027094.1 | [44] |
| Lemyra | Lemyra melli | NC_026692.1 | [33] |
| Nyctemera | Nyctemera arctata albofasciata | KM244681.1 | [42] |
| Nyctemera | Nyctemera adversata | NC_062185.1 | Unpublished |
| Phragmatobia | Phragmatobia fuliginosa | NC_062183.1 | Unpublished |
| Spilarctia | Spilarctia subcarnea | KT258909.1 | Unpublished |
| Spilarctia | Spilarctia casigneta | NC_060594.1 | Unpublished |
| Pareuchaetes | Pareuchaetes insulata | NC_062088.1 | Unpublished |
| Amata | Amata formosae | KC513737.1 | [31] |
| Amerila | Amerila alberti | NC_062176.1 | Unpublished |
| Aloa | Aloa lactinea | \ | This study |
| Miltochrista | Miltochrista miniata | OW121779.1 | [37] |
| Eilema | Eilema sororculum | OU618562.1 | \ |
| Eilema | Eilema depressum | OU612042.1 | \ |
| Cybosia | Cybosia mesomella | OX276419.1 | \ |
| Spilarctia | Spilarctia lutea | OU696502.1 | \ |
3. Results
3.1. Mitochondrial Whole Genome Basic Structure
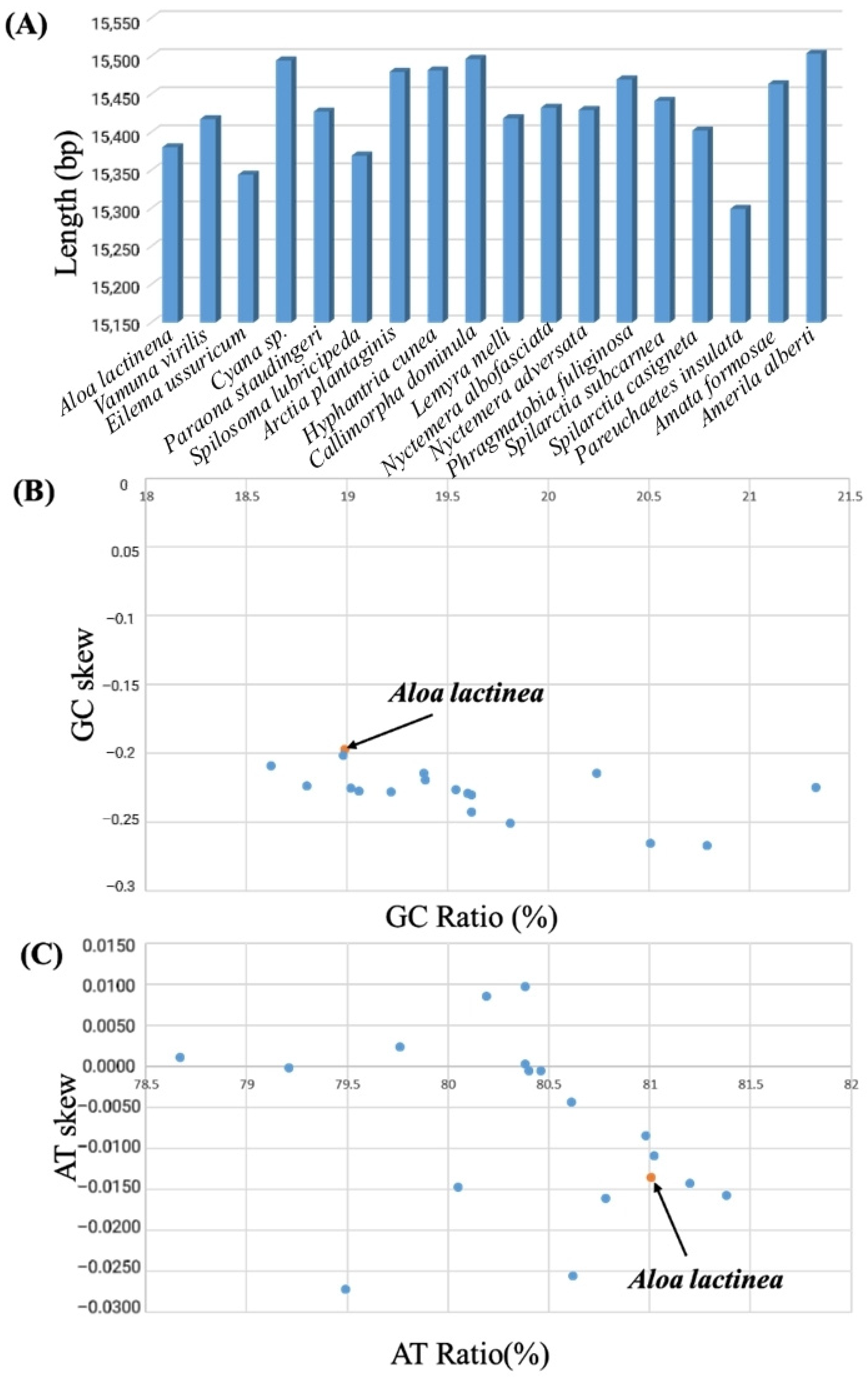
3.2. Nucleotide Composition and Deviation Analysis
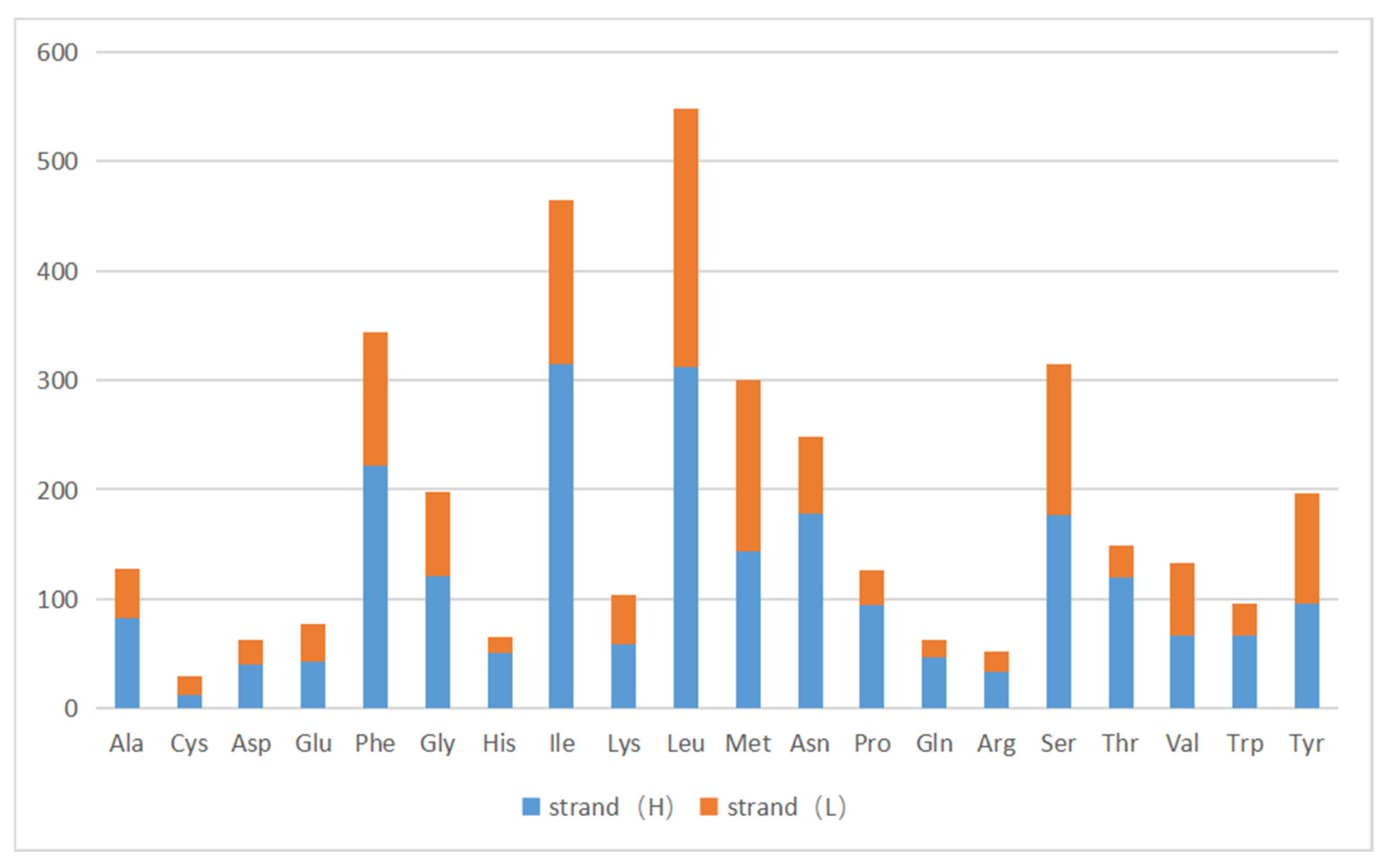
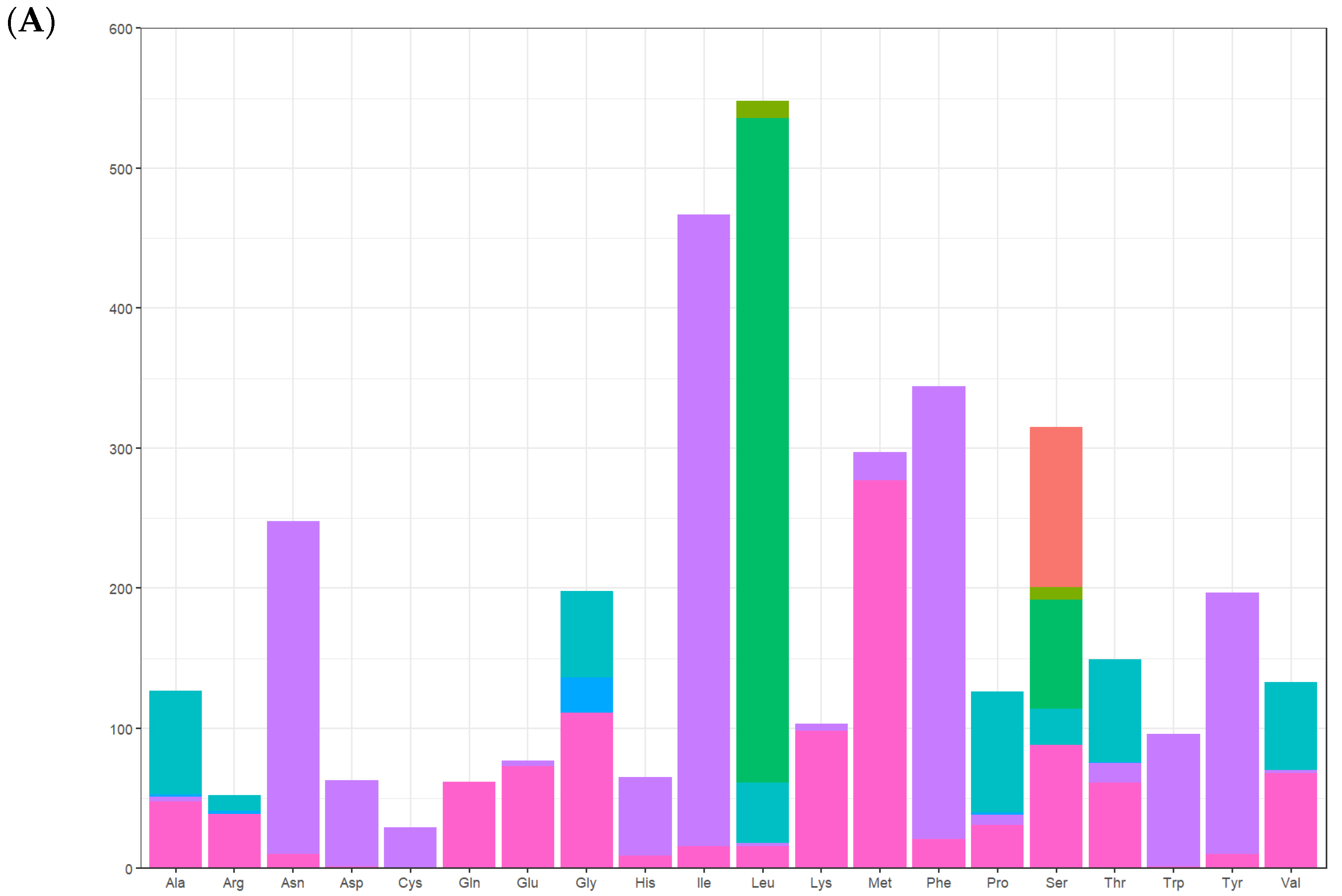
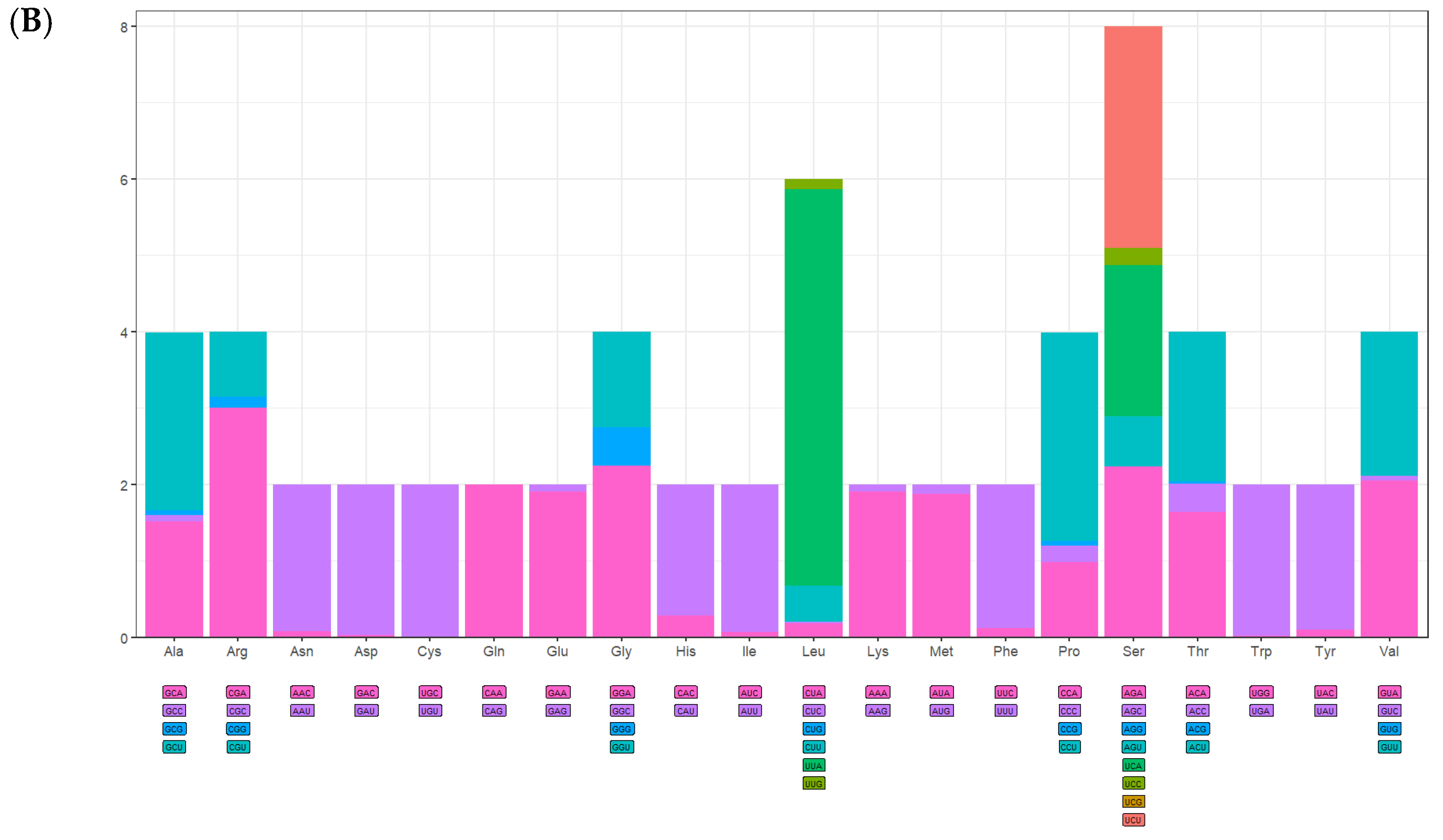
3.3. The Use of Protein-Coding Genes and Codons
3.4. rRNA and tRNA Genes
3.5. Control Area, Non-Coding Area, and Overlapping Area
3.6. Phylogenetic Relationships
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kaleka, A.; Kirti, J.S. Further studies on three indian species of genus aloa walker (Arctiinae: Arctiidae: Lepidoptera). Adv. Biosci. 2000, 19, 47–54. [Google Scholar]
- Fang, C.L. Fauna Sinica; Science Press: Beijing, China, 2000. [Google Scholar]
- Fang, C.L. Zoology of China, Entomoptera 19 Volume Lepidoptera; Science Press: Beijing, China, 2000; pp. 1–295. (In Chinese) [Google Scholar]
- Shu, Y. Taxonomic Study on Arctiinae in Huangshan Scenic Area; Anhui University: Hefei, China, 2023. [Google Scholar]
- Zahiri, R.; Kitching, I.J.; Lafontaine, J.D.; Mutanen, M.; Kaila, L.; Holloway, J.D.; Wahlberg, N. A new molecular phylogeny offers hope for a stable family level classification of the Noctuoidea (Lepidoptera). Zool. Scr. 2011, 40, 158–173. [Google Scholar] [CrossRef]
- Miller, J.S. Cladistics and classification of the Notodontidae (Lepidoptera, Noctuoidea) based on larval and adult morphology. Bull. Am. Mus. Nat. Hist. 1991, 204, 207–221. [Google Scholar]
- Lafontaine, J.D.; Fibiger, M. Revised higher classification of the Noctuoidea (Lepidoptera). Can. Entomol. 2006, 138, 610–635. [Google Scholar] [CrossRef]
- Weller, S.J.; Dacosta, M.; Simmons, R.; Dittmar, K.; Whiting, M. Evolution and taxonomic confusion in Arctiidae. In Tiger Moths and Woolly Bears: Behavior, Ecology, and Evolution of the Arctiidae; Conner, W.E., Ed.; Oxford University Press: New York, NY, USA, 2009; pp. 11–30. [Google Scholar]
- Zahiri, R.; Holloway, J.D.; Kitching, I.J.; Lafontaine, J.D.; Mutanen, M.; Wahlberg, N. Molecular phylogenetics of Erebidae (Lepidoptera, Noctuoidea). Syst. Entomol. 2012, 37, 102–124. [Google Scholar] [CrossRef]
- Song, F.; Li, H.; Jiang, P.; Zhou, X.; Liu, J.; Sun, C.; Vogler, A.P.; Cai, W. Capturing the phylogeny of Holometabola with mitochondrial genome data and Bayesian site-heterogeneous mixture models. Genome Biol. Evol. 2016, 8, 1411–1426. [Google Scholar] [CrossRef]
- Lightowlers, R.N.; Chinnery, P.F.; Turnbull, D.M.; Howell, N. Mammalian mitochondrial genetics: Heredity, heteroplasmy and disease. Trends Genet. 1997, 13, 450–455. [Google Scholar] [CrossRef]
- Tang, C.; Du, X. Complete mitochondrial genomes of two moths in the tribe Trichaeini (Lepidoptera: Crambidae) and their phylogenetic implications. Ecol. Evol. 2023, 13, e10188. [Google Scholar] [CrossRef]
- Wang, W. Studies on Mitochondrial Genomes and Phylogeny of Lepidoptera Insects. Master’s Thesis, Nanjing Agricultural University, Nanjing, China, 2014. [Google Scholar]
- Chen, L. Analysis of Structural Characteristics of Mitochondrial Genomes in Lepidoptera Insects. Master’s Thesis, Hunan Agricultural University, Changsha, China, 2021. [Google Scholar]
- Zaspel, J.M.; Weller, S.J.; Wardwell, C.T.; Zahiri, R.; Wahlberg, N. Phylogeny and Evolution of Pharmacophagy in Tiger Moths (Lepidoptera: Erebidae: Arctiinae). PLoS ONE 2014, 9, e101975. [Google Scholar] [CrossRef]
- Zenker, M.M.; Wahlberg, N.; Brehm, G.; Teston, J.A.; Przybylowicz, L.; Pie, M.R.; Freitas, A.V.L. Systematics and origin of moths in the subfamily Arctiinae (Lepidoptera, Erebidae) in the Neotropical region. Zool. Scr. 2017, 46, 348–362. [Google Scholar] [CrossRef]
- Dowdy, N.J.; Keating, S.; Lemmon, A.R.; Lemmon, E.M.; Conner, W.E.; Chialvo, C.H.S.; Weller, S.J.; Simmons, R.B.; Sisson, M.S.; Zaspel, J.M. A deeper meaning for shallow-level phylogenomic studies: Nested anchored hybrid enrichment offers great promise for resolving the tiger moth tree of life (Lepidoptera: Erebidae: Arctiinae). Syst. Entomol. 2020, 45, 874–893. [Google Scholar] [CrossRef]
- Przybylowicz, L.; Lees, D.C.; Zenker, M.M.; Wahlberg, N. Molecular systematics of the arctiine tribe Syntomini (Lepidoptera, Erebidae). Syst. Entomol. 2019, 44, 624–637. [Google Scholar] [CrossRef]
- Barker, K. Phenol-chloroform isoamyl alcohol (PCI) DNA extraction. Bench 1998, 31, 735. [Google Scholar]
- Guo, Y.; Peng, D.; Han, L.; Liu, T.; Li, G.; Garber, P.A.; Zhou, J. Mitochondrial DNA control region sequencing of the critically endangered Hainan gibbon (Nomascus hainanus) reveals two female origins and extremely low genetic diversity. Mitochondrial DNA B Resour. 2021, 6, 1355–1359. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Freudenthal, J.A.; Pfaff, S.; Terhoeven, N.; Korte, A.; Ankenbrand, M.J.; Förster, F. A systematic comparison of chloroplast genome assembly tools. Genome Biol. 2020, 21, 254. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.D.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, 181–184. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11 Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Juehling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Puetz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Liao, F.; Wang, L.; Wu, S.; Li, Y.P.; Zhao, L.; Huang, G.M.; Niu, C.J.; Liu, Y.Q.; Li, M.G. The complete mitochondrial genome of the fall webworm, Hyphantria cunea (Lepidoptera: Arctiidae). Int. J. Biol. Sci. 2010, 6, 172–186. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.F.; Su, T.J.; Luo, A.R.; Zhu, C.D.; Wu, C.S. Characterization of the Complete Mitochondrion Genome of Diurnal Moth Amata emma (Butler) (Lepidoptera: Erebidae) and Its Phylogenetic Implications. PLoS ONE 2013, 8, e72410. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Tian, S.; Qian, C.; Sun, Y.X.; Abbas, M.N.; Kausar, S.; Wang, L.; Wei, G.; Zhu, B.J.; Liu, C.L. Characterization of the complete mitochondrial genome of Spilarctia robusta (Lepidoptera: Noctuoidea: Erebidae) and its phylogenetic implications. Eur. J. Entomol. 2016, 113, 558–570. [Google Scholar] [CrossRef][Green Version]
- Yang, J.; Kong, W. The complete mitochondrial genome of Lemyra melli (Daniel) (Lepidoptera: Erebidae) and a comparative analysis within the Noctuoidea. Zool. Syst. 2016, 41, 366–378. [Google Scholar]
- Liu, N.; Li, N.; Yang, P.; Sun, C.; Fang, J.; Wang, S. The complete mitochondrial genome of Damora sagana and phylogenetic analyses of the family Nymphalidae. Genes Genom. 2018, 40, 109–122. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Macgregor, C.J.; Saccheri, I.; Fox, B.; Boyes, D. The genome sequence of the Rosy Footman, Miltochrista miniata (Forster, 1771) [version 1; peer review: Awaiting peer review]. Wellcome Open Res. 2023, 8, 582. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Darriba, D.; Posada, D. jModelTest 2.0 Manual v0. 1.1. 2014. Available online: https://www.phylo.org/pdf_docs/jmodeltest-2.1.6-manual.pdf (accessed on 4 July 2024).
- Lemey, P.; Rambaut, A.; Drummond, A.J.; Suchard, M.A. Bayesian Phylogeography Finds Its Roots. PLoS Comput. Biol. 2009, 5, e1000520. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Tan, M.; Meng, G.; Yang, S.; Su, X.; Liu, S.; Song, W.; Li, Y.; Wu, Q.; Zhang, A.; et al. Multiplex sequencing of pooled mitochondrial genomes—A crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 2014, 42, e166. [Google Scholar] [CrossRef] [PubMed]
- Galarza, J.A.; Mappes, J. The complete mitochondrial genome of the wood tiger moth (Arctia plantaginis) and phylogenetic analyses within Arctiinae. Mitochondrial DNA Part B-Resour. 2021, 6, 2171–2173. [Google Scholar] [CrossRef]
- Peng, X.Y.; Duan, X.Y.; Qiang, Y. Characterization of the complete mitochondrial genome of the Scarlet Tiger moth Callimorpha dominula (Insecta: Lepidoptera: Arctiidae). Mitochondrial DNA Part A 2016, 27, 3749–3750. [Google Scholar] [CrossRef]
- Wang, Z.; Jiang, W.; Ye, J.; Wu, H.; Wang, Y.; Xiong, F. Decoding the Mitochondrial Genome of the Tiger Shrimp: Comparative Genomics and Phylogenetic Placement Within Caridean Shrimps. Genes 2025, 16, 457. [Google Scholar] [CrossRef]
- Shi, Q.; Wang, X.; Xing, J.; Xu, X.; Sun, G.; Zhang, J. Complete Mitochondrial Genome of Two Amathusiini Species (Lepidoideae: Nymphalidae: Satyrinae): Characterization, Comparative Analyses, and Phylogenetic Implications. Genes 2025, 16, 447. [Google Scholar] [CrossRef]
- Wei, S.J.; Chen, X.X. Progress in research on the comparative mitogenomics of insects. Chin. J. Appl. Entomol. 2011, 48, 1573–1585. [Google Scholar]
- Vila, M.; Bjorklund, M. The utility of the neglected mitochondrial control region for evolutionary studies in Lepidoptera (Insecta). J. Mol. Evol. 2004, 58, 280–290. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Zhang, D.; Hao, J.; Huang, D.; Cameron, S.; Zhu, C. The complete mitochondrial genome of the yellow coaster, Acraea issoria (Lepidoptera: Nymphalidae: Heliconiinae: Acraeini): Sequence, gene organization and a unique tRNA translocation event. Mol. Biol. Rep. 2010, 37, 3431–3438. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.F. Systematic Taxonomic Study of Amata (Lepidoptera: Ctenuchidae). Natl. Inst. Health J. 2013, 8, e72410. (In Chinese) [Google Scholar]
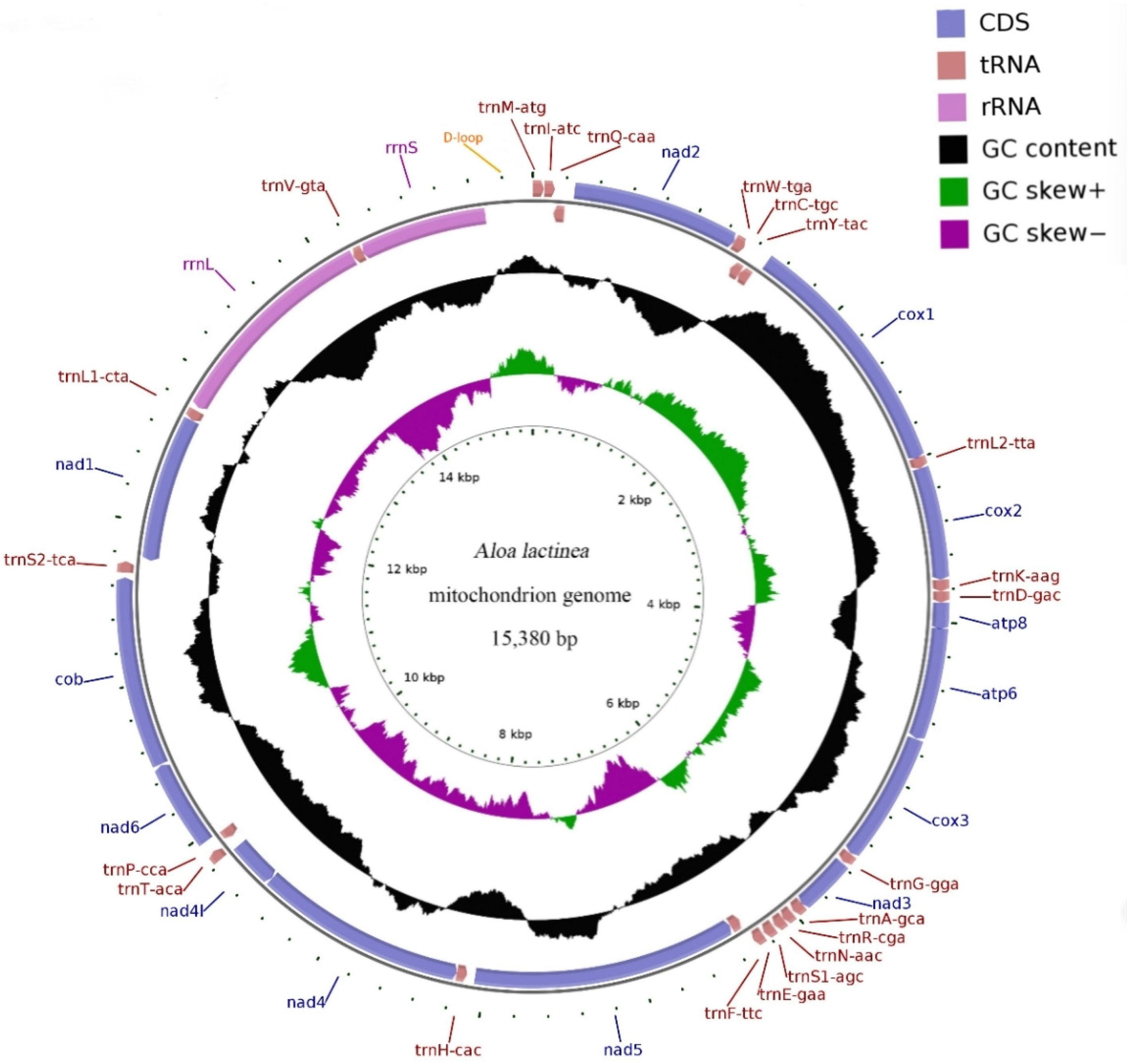
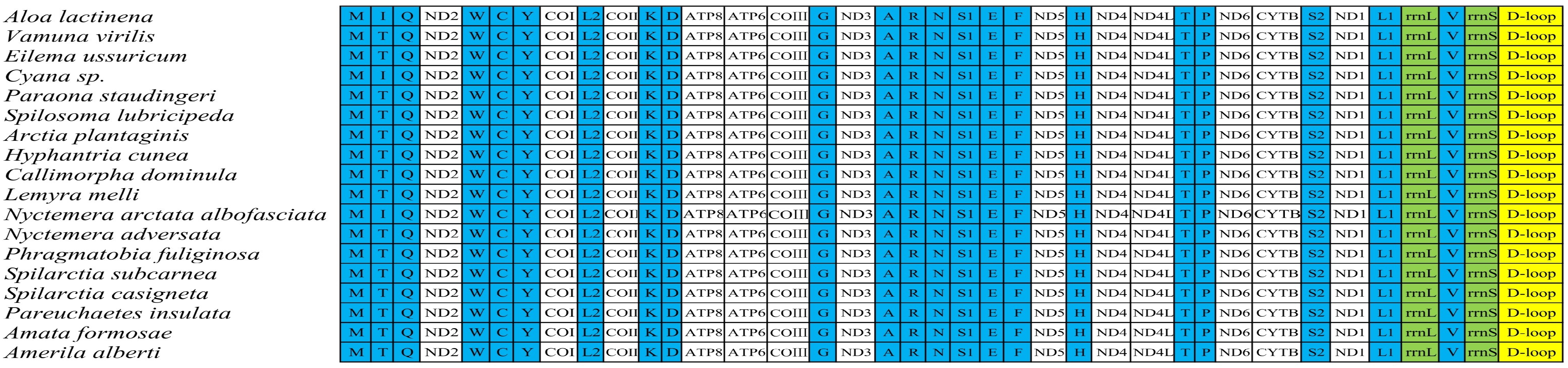
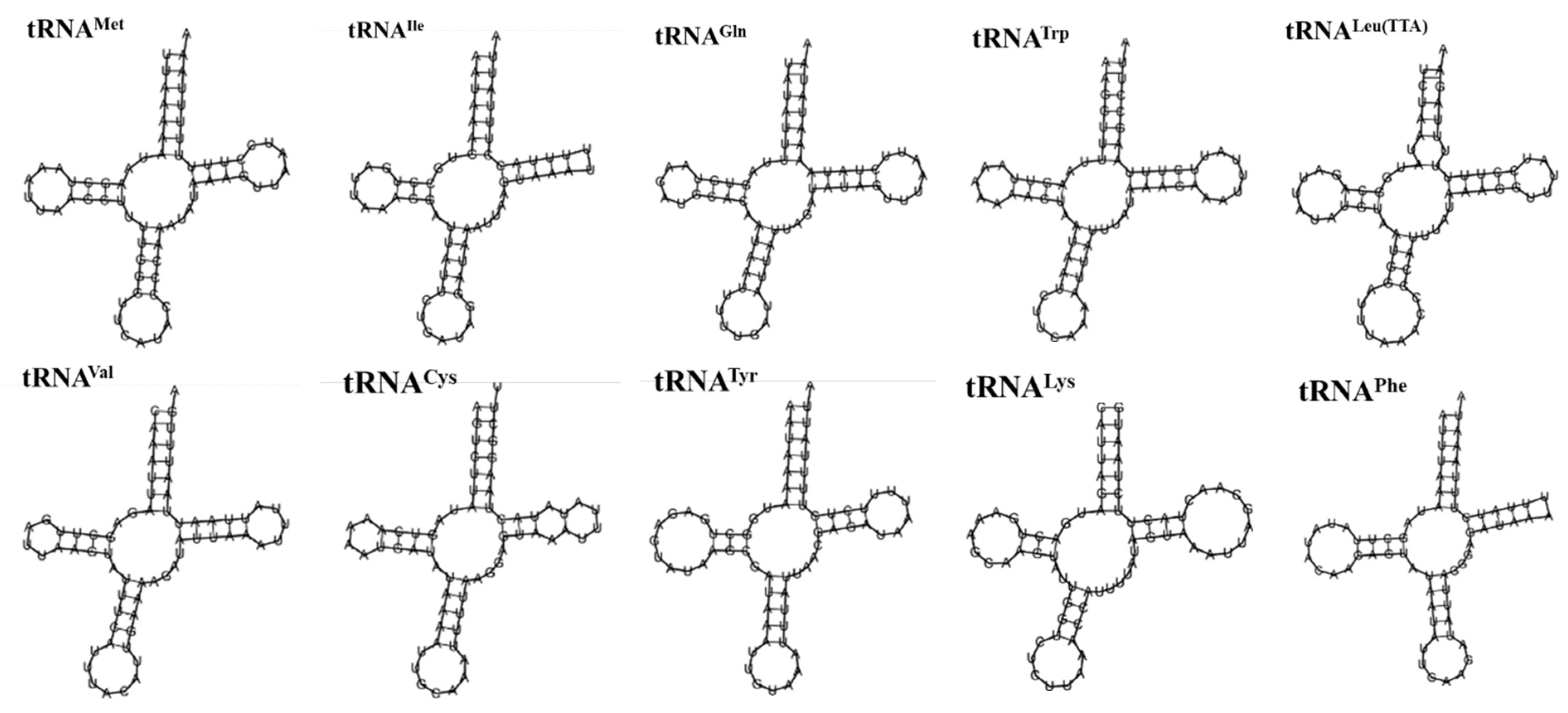


| Gene | Strand | Position | Length | Start Codon | Stop Codon | Anti Codons | Intergenic Nucleotides | Overlapping Nucleotide |
|---|---|---|---|---|---|---|---|---|
| tRNAMet | H | 1–67 | 67 | - | - | CAT | - | |
| tRNAIle | H | 70–133 | 64 | - | - | GAT | 2 | |
| tRNAGln | L | 131–199 | 69 | - | - | TTG | 3 | |
| ND2 | H | 252–1262 | 1011 | ATT | TAA | 52 | ||
| tRNATrp | H | 1264–1332 | 69 | - | - | TCA | 1 | |
| tRNACys | L | 1325–1389 | 65 | - | - | GCA | 8 | |
| tRNATyr | L | 1390–1457 | 68 | - | - | GTA | ||
| COI | H | 1495–3000 | 1506 | ATT | TAA | 37 | ||
| tRNALeu(TTA) | H | 2996–3062 | 67 | - | - | TAA | 5 | |
| COII | H | 3063–3744 | 682 | ATG | T | |||
| tRNALys | H | 3745–3814 | 70 | - | - | CTT | ||
| tRNAAsp | H | 3815–3881 | 67 | - | - | GTC | ||
| ATP8 | H | 3882–4043 | 162 | ATT | TAA | |||
| ATP6 | H | 4037–4714 | 678 | ATG | TAA | 7 | ||
| COIII | H | 4719–5510 | 792 | ATG | TAA | 4 | ||
| tRNAGly | H | 5514–5578 | 65 | - | - | TCC | 3 | |
| ND3 | H | 5588–5932 | 345 | ATA | TAA | 9 | ||
| tRNAAla | H | 5932–5999 | 68 | - | - | TGC | 1 | |
| tRNAArg | H | 6012–6074 | 63 | - | - | TCG | 12 | |
| tRNAAsn | H | 6076–6140 | 65 | - | - | GTT | 1 | |
| tRNASer(AGC) | H | 6151–6216 | 66 | - | - | GCT | 10 | |
| tRNAGlu | H | 6229–6294 | 66 | - | - | TTC | 12 | |
| tRNAPhe | L | 6301–6367 | 67 | - | - | GAA | 6 | |
| ND5 | L | 6371–8059 | 1689 | ATA | TAA | 3 | ||
| tRNAHis | L | 8111–8177 | 67 | - | - | GTG | 51 | |
| ND4 | L | 8178–9516 | 1339 | ATG | T | |||
| ND4L | L | 9519–9815 | 297 | ATA | TAA | 2 | ||
| tRNAThr | H | 9822–9886 | 65 | - | - | TGT | 6 | |
| tRNAPro | L | 9887–9953 | 67 | - | - | TGG | ||
| ND6 | H | 9961–10,491 | 531 | ATA | TAA | 7 | ||
| CYTB | H | 10,499–11,650 | 1152 | ATG | TAA | 7 | ||
| tRNASer(TCA) | H | 11,680–11,747 | 68 | - | - | TGA | 29 | |
| ND1 | L | 11,765–12,703 | 939 | ATG | TAA | 17 | ||
| tRNALeu(CTA) | L | 12,705–12,772 | 68 | - | - | TAG | 1 | |
| 18S rRNA | L | 12,795–14,190 | 1396 | - | - | 22 | ||
| tRNAVal | L | 14,194–14,259 | 66 | - | - | TAC | 3 | |
| 12S rRNA | L | 14,259–15,077 | 819 | - | - | 1 | ||
| D-loop | H | 15,078–15,380 | 303 | - | - |
| Gene/Region | Base Composition (%) | AT Skew | GC Skew | |||||
|---|---|---|---|---|---|---|---|---|
| A | T | C | G | A+T | G+C | |||
| Genome | 39.95 | 41.05 | 11.37 | 7.62 | 81.01 | 18.99 | −0.0136 | −0.1975 |
| Protein-coding genes (total) | 34.48 | 45.44 | 9.65 | 10.44 | 79.92 | 20.08 | −0.1371 | 0.0393 |
| ND2 | 36.30 | 48.66 | 9.20 | 5.84 | 84.96 | 15.04 | −0.1455 | −0.2234 |
| COI | 31.54 | 41.10 | 13.75 | 13.61 | 72.64 | 27.36 | −0.1316 | −0.0051 |
| COII | 35.78 | 41.50 | 12.46 | 10.26 | 77.28 | 22.72 | −0.0740 | −0.0968 |
| ATP8 | 45.06 | 48.77 | 4.32 | 1.85 | 93.83 | 6.17 | −0.0395 | −0.4003 |
| ATP6 | 35.55 | 43.81 | 12.83 | 7.82 | 79.36 | 20.65 | −0.1041 | −0.2426 |
| COIII | 33.08 | 41.54 | 13.51 | 11.87 | 74.62 | 25.38 | −0.1134 | −0.0646 |
| ND3 | 35.65 | 46.96 | 11.30 | 6.09 | 82.61 | 17.39 | −0.1369 | −0.2996 |
| ND5 | 35.70 | 46.12 | 6.10 | 12.08 | 81.82 | 18.18 | −0.1274 | 0.3289 |
| ND4 | 35.18 | 47.27 | 5.90 | 11.65 | 82.45 | 17.55 | −0.1466 | 0.3276 |
| ND4L | 31.99 | 54.55 | 3.03 | 10.44 | 86.54 | 13.47 | −0.2607 | 0.5501 |
| ND6 | 37.10 | 50.09 | 8.10 | 4.71 | 87.19 | 12.81 | −0.1490 | −0.2646 |
| CYTB | 33.33 | 44.18 | 12.67 | 9.81 | 77.51 | 22.48 | −0.1400 | −0.1272 |
| ND1 | 31.95 | 47.28 | 7.24 | 13.53 | 79.23 | 20.77 | −0.1935 | 0.3028 |
| First site | 36.57 | 45.66 | 8.41 | 9.33 | 82.24 | 17.75 | −0.1105 | 0.0518 |
| Secondary site | 35.06 | 43.42 | 10.44 | 11.06 | 78.49 | 21.5 | −0.1065 | 0.0288 |
| Tertiary site | 31.75 | 47.23 | 10.09 | 10.92 | 78.97 | 21.01 | −0.1960 | 0.0395 |
| tRNA gene | 41.38 | 40.49 | 7.57 | 10.57 | 81.87 | 18.13 | 0.0109 | 0.1655 |
| rRNA gene | 43.12 | 42.17 | 4.74 | 9.98 | 85.28 | 14.72 | 0.0111 | 0.3559 |
| D-loop zone | 32.01 | 42.24 | 11.88 | 13.86 | 74.25 | 25.75 | −0.1378 | 0.0769 |
| Species | Whole Genome | PCGs | tRNA Gene | rRNA Gene | D-Loop | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Size | A+T | GC/AT Skew | Size | A+T | GC/AT Skew | Size | A+T | GC/AT Skew | Size | A+T | GC/AT Skew | Size | A+T | GC/AT Skew | |
| (bp) | (%) | (bp) | (%) | (bp) | (%) | (bp) | (%) | (bp) | (%) | ||||||
| Aloa lactinea | 15,380 | 81.01 | 14.522059 | 11,123 | 79.92 | 0.0393/−0.1371 | 1467 | 81.87 | 0.1655/0.0109 | 2215 | 85.28 | 0.3559/0.0111 | 303 | 74.25 | 0.0769/−0.1378 |
| Vamuna virilis | 15,417 | 80.4 | 458.2 | 10,755 | 78.14 | 0.0467/−0.1564 | 1456 | 81.59 | 0.1793/0.0219 | 2176 | 84.6 | 0.3727/0.0310 | 362 | 95.03 | 8.7214397 |
| Eilema | 15,344 | 80.46 | 452.4 | 11,199 | 78.75 | 0.0118/−0.1512 | 1465 | 81.91 | 0.1852/0.0383 | 2034 | 84.27 | 16.299145 | 329 | 94.22 | 1.5237173 |
| ussuricum | |||||||||||||||
| Cyana sp. | 15,494 | 81.2 | 15.622378 | 10,767 | 79.18 | 0.0634/−0.1510 | 1475 | 81.69 | 0.1628/0.0224 | 2192 | 84.67 | 0.3810/0.0464 | \ | \ | \ |
| MT-2014 | |||||||||||||||
| Paraona staudingeri | 15,427 | 80.19 | −29.17442 | 10,821 | 78.03 | 0.0073/−0.1464 | 1462 | 81.12 | 0.1525/0.0270 | 2181 | 84.46 | 0.3925/0.0000 | 362 | 94.48 | 28.409091 |
| Spilosoma | 15,369 | 81.38 | 13.305732 | 10,734 | 79.54 | 0.0323/−0.1433 | 1463 | 81.68 | 0.1714/0.0159 | 2187 | 85.05 | 0.3271/0.0140 | 361 | 95.01 | 4.8993363 |
| lubricipeda | |||||||||||||||
| Arctia plantaginis | 15,479 | 80.78 | 14.15528 | 10,731 | 78.7 | 0.0413/−0.1422 | 1464 | 81.56 | 0.1779/0.0267 | 2204 | 84.53 | 18.743719 | 401 | 96.01 | 8.2881806 |
| Hyphantria cunea | 15,481 | 80.38 | −23.75258 | 10,752 | 78.18 | 0.0339/−0.1480 | 1473 | 81.74 | 0.1599/0.0250 | 2212 | 84.67 | 0.3509/0.0017 | 357 | 94.96 | 14.631579 |
| Callimorpha dominula | 15,496 | 81.02 | 18.46789 | 10,785 | 79.85 | 0.0491/−0.1456 | 1462 | 82.08 | 0.1607/0.0217 | 2154 | 84.54 | 15.012195 | 486 | 75.1 | −2.231746 |
| Lemyra melli | 15,418 | 78.67 | −204.1818 | 10,749 | 76.2 | 0.0361/−0.1512 | 1468 | 80.65 | 0.1829/0.0068 | 2233 | 84.19 | −117.5938 | 338 | 94.38 | 5.5945626 |
| Nyctemera | 15,432 | 80.05 | 15.489796 | 10,761 | 78.94 | 0.0228/−0.1430 | 1445 | 81.11 | 0.1941/0.0359 | 2058 | 84.25 | 13.5 | \ | \ | \ |
| albofasciata | |||||||||||||||
| Nyctemera | 15,429 | 79.21 | 2670 | 10,761 | 76.67 | 0.0212014 | 1460 | 81.16 | 0.1783/0.0261 | 2033 | 84.55 | 14.264 | 292 | 94.86 | 129.69444 |
| adversata | |||||||||||||||
| Phragmatobia fuliginosa | 15,469 | 80.98 | 26.785714 | 10,767 | 78.88 | 0.0208/−0.1496 | 1463 | 81.68 | 0.1567/0.0260 | 2181 | 85.1 | 0.3597/0.0172 | 181 | 96.69 | 0.0000/−0.0057 |
| Codon | Amino Acid | n | % | RSCU | Codon | Amino Acid | n | % | RSCU |
|---|---|---|---|---|---|---|---|---|---|
| GCA | Ala | 48 | 1.30 | 1.51 | AAA | Lys | 98 | 2.65 | 1.90 |
| GCC | Ala | 3 | 0.08 | 0.09 | AAG | Lys | 5 | 0.14 | 0.10 |
| GCG | Ala | 2 | 0.05 | 0.06 | AUA | Met | 277 | 7.49 | 1.87 |
| GCU | Ala | 74 | 2.00 | 2.33 | AUG | Met | 20 | 0.54 | 0.13 |
| CGA | Arg | 39 | 1.06 | 3.00 | UUC | Phe | 21 | 0.57 | 0.12 |
| CGC | Arg | 0 | 0.00 | 0.00 | UUU | Phe | 323 | 8.74 | 1.88 |
| CGG | Arg | 2 | 0.05 | 0.15 | CCA | Pro | 31 | 0.84 | 0.98 |
| CGU | Arg | 11 | 0.30 | 0.85 | CCC | Pro | 7 | 0.19 | 0.22 |
| AAC | Asn | 10 | 0.27 | 0.08 | CCG | Pro | 2 | 0.05 | 0.06 |
| AAU | Asn | 238 | 6.44 | 1.92 | CCU | Pro | 86 | 2.33 | 2.73 |
| GAC | Asp | 1 | 0.03 | 0.03 | AGA | Ser | 88 | 2.38 | 2.23 |
| GAU | Asp | 62 | 1.68 | 1.97 | AGC | Ser | 0 | 0.00 | 0.00 |
| UGC | Cys | 0 | 0.00 | 0.00 | AGG | Ser | 0 | 0.00 | 0.00 |
| UGU | Cys | 29 | 0.78 | 2.00 | AGU | Ser | 26 | 0.70 | 0.66 |
| CAA | Gln | 62 | 1.68 | 2.00 | UCA | Ser | 78 | 2.11 | 1.98 |
| CAG | Gln | 0 | 0.00 | 0.00 | UCC | Ser | 9 | 0.24 | 0.23 |
| GAA | Glu | 73 | 1.98 | 1.90 | UCG | Ser | 0 | 0.00 | 0.00 |
| GAG | Glu | 4 | 0.11 | 0.10 | UCU | Ser | 114 | 3.08 | 2.90 |
| GGA | Gly | 111 | 3.00 | 2.24 | ACA | Thr | 61 | 1.65 | 1.64 |
| GGC | Gly | 0 | 0.00 | 0.00 | ACC | Thr | 14 | 0.38 | 0.37 |
| GGG | Gly | 25 | 0.68 | 0.51 | ACG | Thr | 1 | 0.03 | 0.03 |
| GGU | Gly | 62 | 1.68 | 1.25 | ACU | Thr | 73 | 1.98 | 1.96 |
| CAC | His | 9 | 0.24 | 0.28 | UGG | Trp | 1 | 0.03 | 0.02 |
| CAU | His | 56 | 1.52 | 1.72 | UGA | Trp | 95 | 2.57 | 1.98 |
| AUC | Ile | 16 | 0.43 | 0.07 | UAC | Tyr | 10 | 0.27 | 0.10 |
| AUU | Ile | 451 | 12.2 | 1.93 | UAU | Tyr | 187 | 5.06 | 1.90 |
| CUA | Leu | 16 | 0.43 | 0.18 | GUA | Val | 68 | 1.84 | 2.05 |
| CUC | Leu | 2 | 0.05 | 0.02 | GUC | Val | 2 | 0.05 | 0.06 |
| CUG | Leu | 0 | 0.00 | 0.00 | GUG | Val | 0 | 0.00 | 0.00 |
| CUU | Leu | 43 | 1.16 | 0.47 | GUU | Val | 63 | 1.70 | 1.89 |
| UUA | Leu | 475 | 12.85 | 5.20 | UAG * | - | - | - | - |
| UUG | Leu | 12 | 0.32 | 0.13 | UAA * | - | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, C.; Xu, S.; Shu, Y.; Fang, J. The Complete Mitochondrial Genome of Red Costate Tiger Moth (Aloa lactinea [Cramer, 1777]), and Phylogenetic Analyses of the Subfamily Arctiinae. Genes 2025, 16, 554. https://doi.org/10.3390/genes16050554
Pan C, Xu S, Shu Y, Fang J. The Complete Mitochondrial Genome of Red Costate Tiger Moth (Aloa lactinea [Cramer, 1777]), and Phylogenetic Analyses of the Subfamily Arctiinae. Genes. 2025; 16(5):554. https://doi.org/10.3390/genes16050554
Chicago/Turabian StylePan, Chengrong, Sheng Xu, Yu Shu, and Jie Fang. 2025. "The Complete Mitochondrial Genome of Red Costate Tiger Moth (Aloa lactinea [Cramer, 1777]), and Phylogenetic Analyses of the Subfamily Arctiinae" Genes 16, no. 5: 554. https://doi.org/10.3390/genes16050554
APA StylePan, C., Xu, S., Shu, Y., & Fang, J. (2025). The Complete Mitochondrial Genome of Red Costate Tiger Moth (Aloa lactinea [Cramer, 1777]), and Phylogenetic Analyses of the Subfamily Arctiinae. Genes, 16(5), 554. https://doi.org/10.3390/genes16050554





