Phylogenomic and Evolutionary Insights into Lipoprotein Lipase (LPL) Genes in Tambaqui: Gene Duplication, Tissue-Specific Expression and Physiological Implications
Abstract
1. Introduction
2. Materials and Methods
2.1. In Silico Analysis
2.1.1. Identification of lpl Sequences in Tambaqui and Other Teleost
2.1.2. Phylogenetic Analysis of lpl Genes
2.1.3. Synteny Analysis of lpl Gene Copies Among Teleosts
2.1.4. Predicted Structures and Properties of Tambaqui Lipoprotein Lipases
2.2. Gene Expression Analysis of Tambaqui lpl Gene Copies
2.2.1. Sample Collection and RNA Extraction
2.2.2. cDNA Synthesis and qPCR
2.2.3. Statistical Analysis
2.3. Comparative Gene Expression Analysis of lpl Gene Copies Across Teleosts
3. Results
3.1. Tambaqui lpl Genes
3.2. Phylogenetic Analysis of Teleost’s lpl Gene Copies
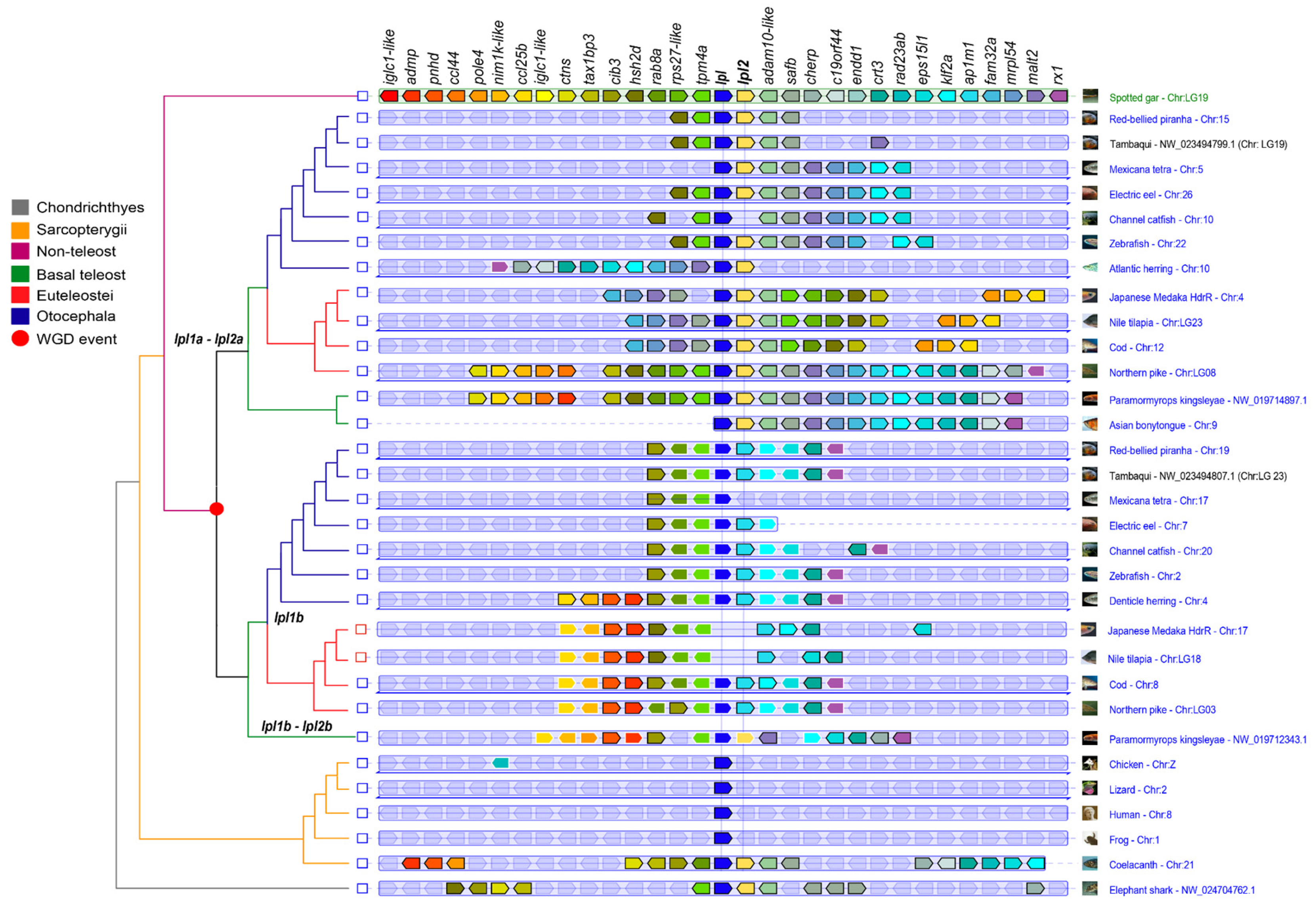
3.3. Synteny Analysis
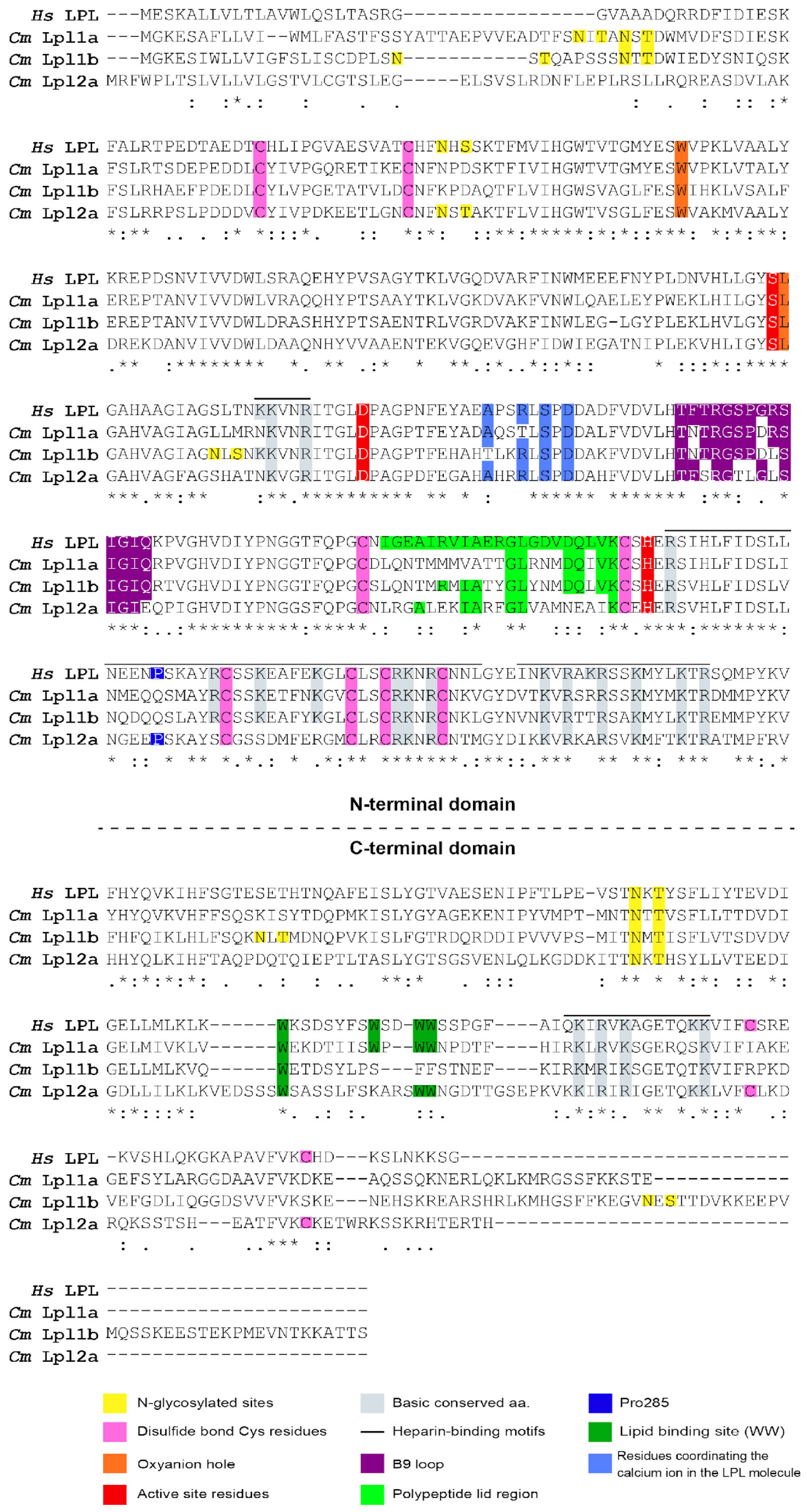
3.4. Characterization of Tambaqui Lpl1a, Lpl1b, and Lpl2a Protein Sequences
3.5. Comparisons of Lpls with Respective Orthologous
3.6. Comparative Analysis of Glycosylation Sites Across Vertebrate LPLs
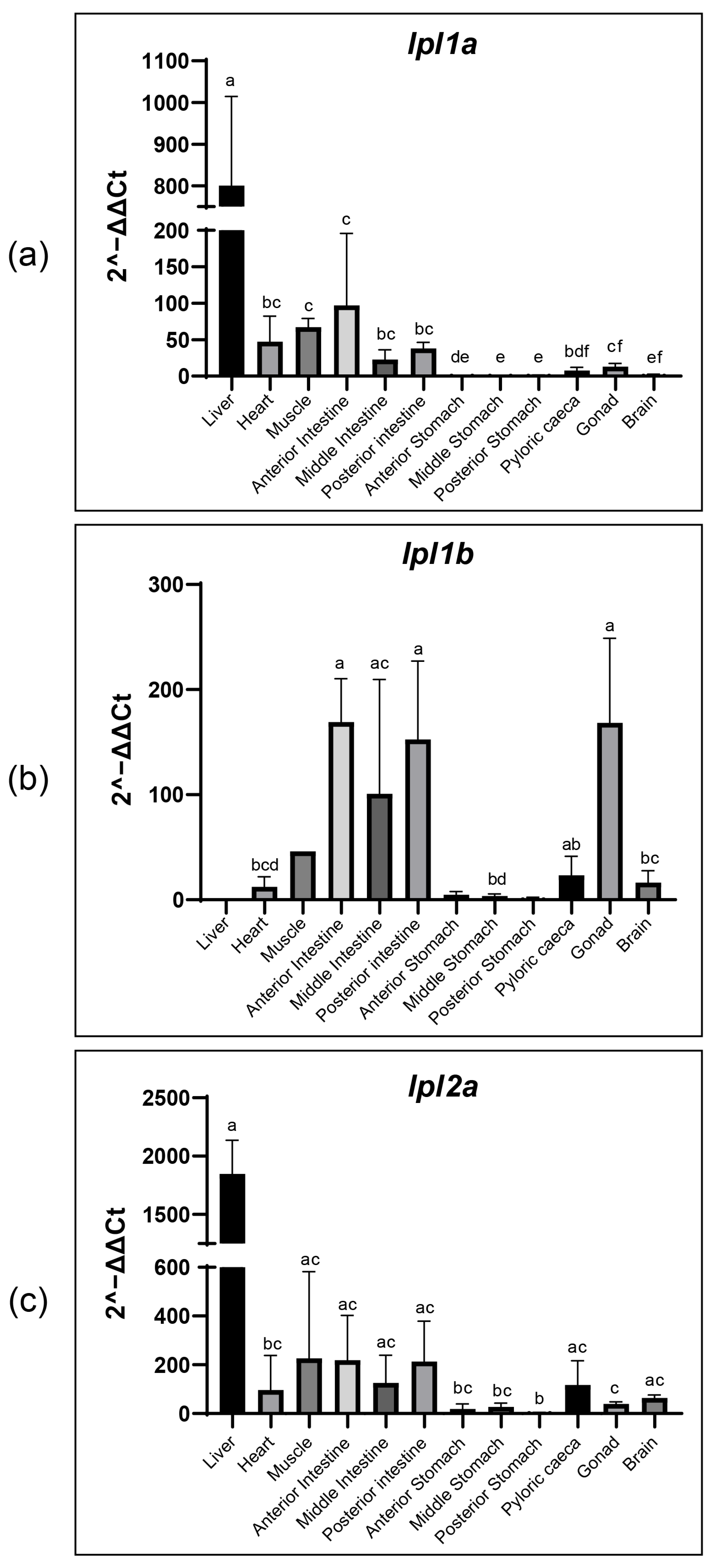
3.7. Tissue Distribution of Tambaqui lpl1a, lpl1b and lpl2a mRNA
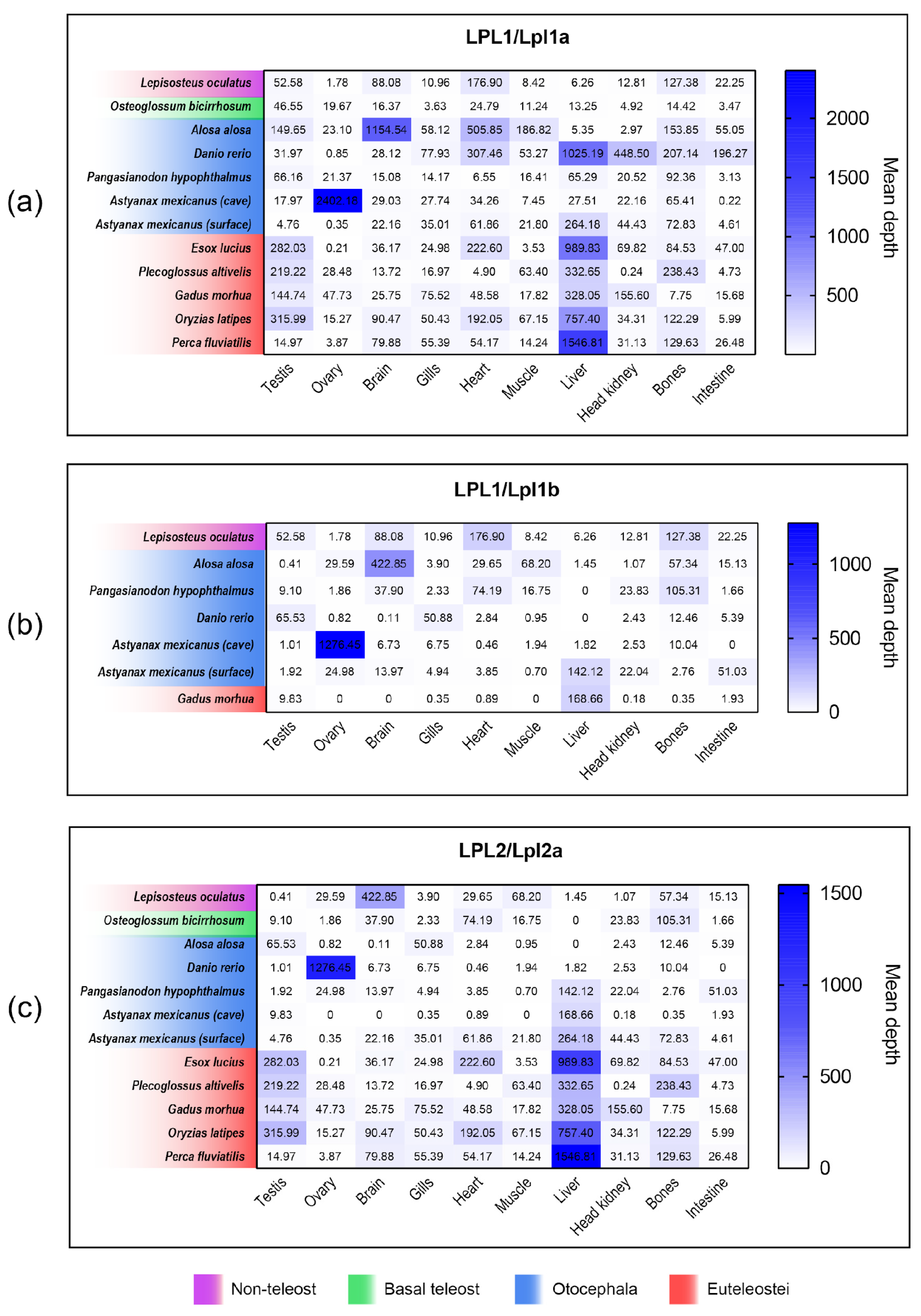
3.8. Comparison of lpl1a, lpl1b and lpl2a mRNA Tissue Distribution Between Non-Teleost and Teleost Fishes
4. Discussion
4.1. Evolution and Phylogeny of LPL Genes
4.2. Gene Retention, Loss, and Functional Diversification
4.3. Structural and Functional Insights of Lpl Proteins
4.4. Tissue-Specific Expression and Ecological Adaptations
4.5. Implications for Aquaculture, Conservation, and Future Research
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LPL | Lipoprotein lipase |
| VLDL | Very low-density lipoprotein |
| LIPC | Hepatic lipase |
| LIPG | Endothelial lipase |
| 3R | Teleost-specific whole genome duplication |
| qPCR | Real-time quantitative PCR |
| CDS | Coding sequence |
| BLAST | Basic Local Alignment Search Tool |
| MEGA | Molecular Evolutionary Genetic Analysis |
| JTT | Jones-Taylor Thornton (Amino acid substitution model) |
| F | Frequency |
| I | Invariant sites |
| G4 | Gamma distribution |
| IQ-Tree | Phylogenetic interference software |
| MrBayes | Bayesian phylogenetic analysis software |
| NCBI | National Center for Biotechnology Information |
| RNA-seq | RNA sequencing |
| CEUA | Animal Use Ethics Committee |
| TRIzol | RNA extraction reagent |
| cDNA | Complementary DNA |
| IDT | Integrating DNA Technologies |
| SYBR Green | qPCR dye |
| ΔΔCt | Delta-Delta Ct (Method for gene expression analysis) |
| ROUT | Method for outlier identification |
| SD | Standart deviation |
| Phylofish | Phylogenetic database of fish |
| LG | Linkage group |
| Da | Dalton (Atomic mass unit) |
| pI | Isoelectric point |
References
- Mead, J.; Irvine, S.; Ramji, D. Lipoprotein lipase: Structure, function, regulation, and role in disease. J. Mol. Med. 2002, 80, 753–769. [Google Scholar] [CrossRef] [PubMed]
- Holmes, R.S.; VandeBerg, J.L.; Cox, L.A. Comparative studies of vertebrate lipoprotein lipase: A key enzyme of very low density lipoprotein metabolism. Comp. Biochem. Physiol. Part. D Genom. Proteom. 2011, 6, 224–234. [Google Scholar] [CrossRef] [PubMed]
- Brault, D.; Noé, L.; Etienne, J.; Hamelin, J.; Raisonnier, A.; Souli, A.; Chuat, J.C.; Dugail, I.; Quignard-Boulangé, A.; Lavau, M.; et al. Sequence of rat lipoprotein lipase-encoding cDNA. Gene 1992, 121, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Chuat, J.-C.; Raisonnier, A.; Etienne, J.; Galibert, F. The lipoprotein lipase-encoding human gene: Sequence from intron-6 to intron-9 and presence in intron-7 of a 40-million-year-old Alu sequence. Gene 1992, 110, 257–261. [Google Scholar] [CrossRef]
- Zechner, R. The tissue-specific expression of lipoprotein lipase: Implications for energy and lipoprotein metabolism. Curr. Opin. Lipidol. 1997, 8, 77–88. [Google Scholar] [CrossRef]
- van Tilbeurgh, H.; Roussel, A.; Lalouel, J.M.; Cambillau, C. Lipoprotein lipase. Molecular model based on the pancreatic lipase X-Ray structure: Consequences for heparin binding and catalysis. J. Biol. Chem. 1994, 269, 4626–4633. [Google Scholar] [CrossRef]
- Gunn, K.H.; Neher, S.B. Structure of dimeric lipoprotein lipase reveals a pore adjacent to the active site. Nat. Commun. 2023, 14, 2569. [Google Scholar] [CrossRef]
- Wittrup, H.H.; Tybjærg-Hansen, A.; Nordestgaard, B.G. Lipoprotein Lipase Mutations, Plasma Lipids and Lipoproteins, and Risk of Ischemic Heart Disease. Circulation 1999, 99, 2901–2907. [Google Scholar] [CrossRef]
- Pulinilkunnil, T.; Rodrigues, B. Cardiac lipoprotein lipase: Metabolic basis for diabetic heart disease. Cardiovasc. Res. 2006, 69, 329–340. [Google Scholar] [CrossRef]
- Sagoo, G.S.; Tatt, I.; Salanti, G.; Butterworth, A.S.; Sarwar, N.; van Maarle, M.; Jukema, J.W.; Wiman, B.; Kastelein, J.J.; Bennet, A.M.; et al. Seven Lipoprotein Lipase Gene Polymorphisms, Lipid Fractions, and Coronary Disease: A HuGE Association Review and Meta-Analysis. Am. J. Epidemiol. 2008, 168, 1233–1246. [Google Scholar] [CrossRef]
- He, C.; Weston, T.A.; Jung, R.S.; Heizer, P.; Larsson, M.; Hu, X.; Alan, C.M.; Tontonoz, P.; Reue, K.; Beigneux, A.P.; et al. NanoSIMS Analysis of Intravascular Lipolysis and Lipid Movement across Capillaries and into Cardiomyocytes. Cell Metab. 2018, 27, 1055–1066. [Google Scholar] [CrossRef] [PubMed]
- Preiss-Landl, K.; Zimmermann, R.; Hämmerle, G.; Zechner, R. Lipoprotein lipase: The regulation of tissue specific expression and its role in lipid and energy metabolism. Curr. Opin. Lipidol. 2002, 13, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Kersten, S. Physiological regulation of lipoprotein lipase. Biochim. Et Biophys. Acta-Mol. Cell Biol. Lipids 2014, 1841, 919–933. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Aryal, B.; Chaube, B.; Rotllan, N.; Varela, L.; Horvath, T.L.; Suárez, Y.; Fernández-Hernando, C. Brown adipose tissue derived ANGPTL4 controls glucose and lipid metabolism and regulates thermogenesis. Mol. Metab. 2018, 11, 59–69. [Google Scholar] [CrossRef]
- Wu, S.A.; Kersten, S.; Qi, L. Lipoprotein Lipase and Its Regulators: An Unfolding Story. Trends Endocrinol. Metab. 2021, 32, 48–61. [Google Scholar] [CrossRef]
- Kumari, A.; Kristensen, K.K.; Ploug, M.; Winther, A.-M.L. The Importance of Lipoprotein Lipase Regulation in Atherosclerosis. Biomedicines 2021, 9, 782. [Google Scholar] [CrossRef]
- Birsoy, K.; Festuccia, W.T.; Laplante, M. A comparative perspective on lipid storage in animals. J. Cell Sci. 2013, 126, 1541–1552. [Google Scholar] [CrossRef]
- Weil, C.; Lefèvre, F.; Bugeon, J. Characteristics and metabolism of different adipose tissues in fish. Rev. Fish Biol. Fish. 2013, 23, 157–173. [Google Scholar] [CrossRef]
- Ranganathan, G.; Ong, J.M.; Yukht, A.; Saghizadeh, M.; Simsolo, R.B.; Pauer, A.; Kern, P.A. Tissue-specific Expression of Human Lipoprotein Lipase. J. Biol. Chem. 1995, 270, 7149–7155. [Google Scholar] [CrossRef]
- Wang, M.; Xu, D.; Liu, K.; Yang, J.; Xu, P. Molecular cloning and expression analysis on LPL of Coilia nasus. Gene 2016, 583, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wu, X.; Lai, J.; Liu, Y.; Song, M.; Li, F.; Gong, Q. Characterization of two lipid metabolism-associated genes and their expression profiles under different feeding conditions in Acipenser dabryanus. Aquac. Rep. 2021, 21, 100780. [Google Scholar] [CrossRef]
- Lindberg, A.; Olivecrona, G. Lipoprotein lipase from rainbow trout differs in several respects from the enzyme in mammals. Gene 2002, 292, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Oku, H.; Koizumi, N.; Okumura, T.; Kobayashi, T.; Umino, T. Molecular characterization of lipoprotein lipase, hepatic lipase and pancreatic lipase genes: Effects of fasting and refeeding on their gene expression in red sea bream Pagrus major. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2006, 145, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Eckel, R.H. Lipoprotein lipase: From gene to obesity. Am. J. Physiol.-Endocrinol. Metab. 2009, 297, E271–E288. [Google Scholar] [CrossRef]
- Zhao, W.S.; Hu, S.L.; Yu, K.; Wang, H.; Wang, W.; Loor, J.; Luo, J. Lipoprotein Lipase, Tissue Expression and Effects on Genes Related to Fatty Acid Synthesis in Goat Mammary Epithelial Cells. Int. J. Mol. Sci. 2014, 15, 22757–22771. [Google Scholar] [CrossRef]
- Liang, X.-F.; Ogata, H.Y.; Oku, H. Effect of dietary fatty acids on lipoprotein lipase gene expression in the liver and visceral adipose tissue of fed and starved red sea bream Pagrus major. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2002, 132, 913–919. [Google Scholar] [CrossRef]
- Tian, J.; Wu, F.; Yang, C.-G.; Jiang, M.; Liu, W.; Wen, H. Dietary lipid levels impact lipoprotein lipase, hormone-sensitive lipase, and fatty acid synthetase gene expression in three tissues of adult GIFT strain of Nile tilapia, Oreochromis niloticus. Fish Physiol. Biochem. 2015, 41, 1–18. [Google Scholar] [CrossRef]
- Zheng, K.; Zhu, X.; Han, D.; Yang, Y.; Lei, W.; Xie, S. Effects of dietary lipid levels on growth, survival and lipid metabolism during early ontogeny of Pelteobagrus vachelli larvae. Aquaculture 2010, 299, 121–127. [Google Scholar] [CrossRef]
- Kleveland, E.J.; Ruyter, B.; Vegusdal, A.; Sundvold, H.; Berge, R.K.; Gjøen, T. Effects of 3-thia fatty acids on expression of some lipid related genes in Atlantic salmon (Salmo salar L.). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2006, 145, 239–248. [Google Scholar] [CrossRef]
- Wang, A.; Han, G.; Qi, Z.; Lv, F.; Yu, Y.; Huang, J.; Wang, T.; Xu, P. Cloning of lipoprotein lipase (LPL) and the effects of dietary lipid levels on LPL expression in GIFT tilapia (Oreochromis niloticus). Aquac. Int. 2013, 21, 1219–1232. [Google Scholar] [CrossRef]
- Qin, Y.; He, L.; Wang, Y.; Li, D.; Chen, W.; Ye, J. Growth performance, fatty acid composition, and lipid metabolism are altered in groupers (Epinephelus coioides) by dietary fish oil replacement with palm oil. Anim. Nutr. 2022, 8, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Ryu, Y.W.; Tanaka, R.; Kasahara, A.; Ito, Y.; Hiramatsu, N.; Todo, T.; Sullivan, C.V.; Hara, A. Molecular Cloning and Transcript Expression of Genes Encoding Two Types of Lipoprotein Lipase in the Ovary of Cutthroat trout, Oncorhynchus clarki. Zoolog. Sci. 2013, 30, 224–237. [Google Scholar] [CrossRef] [PubMed]
- Glasauer, S.M.K.; Neuhauss, S.C.F. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genom. 2014, 289, 1045–1060. [Google Scholar] [CrossRef]
- Pasquier, J.; Cabau, C.; Nguyen, T.; Jouanno, E.; Severac, D.; Braasch, I.; Journot, L.; Pontarotti, P.; Klopp, C.; Postlethwait, J.H.; et al. Gene evolution and gene expression after whole genome duplication in fish: The PhyloFish database. BMC Genom. 2016, 17, 368. [Google Scholar] [CrossRef]
- De Almeida, L.C.; Lundstedt, L.M.; Moraes, G. Digestive enzyme responses of tambaqui (Colossoma macropomum) fed on different levels of protein and lipid. Aquac. Nutr. 2006, 12, 443–450. [Google Scholar] [CrossRef]
- Rauber, R.G.; Strictar, L.; Gomes, L.C.; Suzuki, H.I.; Agostinho, A.A. Spatial segregation in the reproductive activity of Neotropical fish species as an indicator of the migratory trait. J. Fish Biol. 2021, 98, 694–706. [Google Scholar] [CrossRef]
- Araujo-Lima, C.; Goulding, M. So Fruitful a Fish: Ecology, Conservation, and Aquaculture of the AMAZON’S Tambaqui; Columbia University Press: New York, NY, USA, 1997; p. 191. [Google Scholar]
- Sandre, L.C.G.; Buzollo, H.; Nascimento, T.M.T.; Neira, L.M.; Jomori, R.K.; Carneiro, D.J. Productive performance and digestibility in the initial growth phase of tambaqui (Colossoma macropomum) fed diets with different carbohydrate and lipid levels. Aquac. Rep. 2017, 6, 28–34. [Google Scholar] [CrossRef]
- Varela, E.S.; Bekaert, M.; Ganeco-Kirschnik, L.N.; Torati, L.S.; Shiotsuki, L.; de Almeida, F.L.; Villela, L.C.V.; Rezende, F.P.; da Silva Barroso, A.; de Freitas, L.E.L.; et al. A high-density linkage map and sex-linked markers for the Amazon Tambaqui Colossoma macropomum. BMC Genom. 2021, 22, 709. [Google Scholar] [CrossRef]
- Agudelo, J.F.G.; Mastrochirico-Filho, V.A.; Borges, C.H.S.; Ariede, R.B.; Lira, L.V.G.; Neto, R.R.O.; de Freitas, M.V.; Sucerquia, G.A.L.; Vera, M.; Berrocal, M.H.M.; et al. Genomic selection signatures in farmed Colossoma macropomum from tropical and subtropical regions in South America. Evol. Appl. 2022, 15, 679–693. [Google Scholar] [CrossRef]
- Hilsdorf, A.W.S.; Uliano-Silva, M.; Coutinho, L.L.; Montenegro, H.; Almeida-Val, V.M.F.; Pinhal, D. Genome assembly and annotation of the tambaqui (Colossoma macropomum): An emblematic fish of the Amazon River Basin. GigaByte 2021, 2021, gigabyte29. [Google Scholar] [CrossRef]
- Nguyen, N.T.T.; Vincens, P.; Dufayard, J.F.; Roest, H.; Louis, A. Genomicus in 2022: Comparative tools for thousands of genomes and reconstructed ancestors. Nucleic Acids Res. 2022, 50, D1025–D1031. [Google Scholar] [CrossRef] [PubMed]
- Griffon, N.; Budreck, E.C.; Long, C.J.; Broedl, U.C.; Marchadier, D.H.; Glick, J.M.; Rader, D.J. Substrate specificity of lipoprotein lipase and endothelial lipase: Studies of lid chimeras. J. Lipid Res. 2006, 47, 1803–1811. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, A.R.; Silva, G.F.; Gualberto, G.F.; Almeida, F.L. SHORT-COMMUNICATION Validation of reference genes for real-time quantitative PCR in tambaqui (Colossoma macropomum). Genet. Mol. Res. 2016, 15, 4. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Kaneko, G.; Yamada, T.; Han, Y.; Hirano, Y.; Khieokhajonkhet, A.; Shirakami, H.; Nagasaka, R.; Kondo, H.; Hirono, I.; Ushio, H.; et al. Differences in lipid distribution and expression of peroxisome proliferator-activated receptor gamma and lipoprotein lipase genes in torafugu and red seabream. General. Comp. Endocrinol. 2013, 184, 51–60. [Google Scholar] [CrossRef]
- Wang, L.; Kaneko, G.; Takahashi, S.-I.; Watabe, S.; Ushio, H. Identification and gene expression profile analysis of a major type of lipoprotein lipase in adult medaka Oryzias latipes. Fish. Sci. 2015, 81, 163–173. [Google Scholar] [CrossRef]
- Sharma, V.; Hecker, N.; Roscito, J.G.; Foerster, L.; Langer, B.E.; Hiller, M. A genomics approach reveals insights into the importance of gene losses for mammalian adaptations. Nat. Commun. 2018, 9, 1215. [Google Scholar] [CrossRef]
- German, J.B. Dietary lipids from an evolutionary perspective: Sources, structures and functions. Matern. Child Nutr. 2011, 7, 2–16. [Google Scholar] [CrossRef]
- Luca, F.; Perry, G.H.; Di Rienzo, A. Evolutionary Adaptations to Dietary Changes. Annu. Rev. Nutr. 2010, 30, 291–314. [Google Scholar] [CrossRef]
- Garnås, E. Saturated fat in an evolutionary context. Lipids Health Dis. 2025, 24, 28. [Google Scholar] [CrossRef]
- Tocher, D.R. Metabolism and Functions of Lipids and Fatty Acids in Teleost Fish. Rev. Fish. Sci. 2003, 11, 107–184. [Google Scholar] [CrossRef]
- Lee, M.-C.; Park, J.C.; Lee, J.-S. Effects of environmental stressors on lipid metabolism in aquatic invertebrates. Aquat. Toxicol. 2018, 200, 83–92. [Google Scholar] [CrossRef]
- Liu, J.; Lu, L.; Song, H.; Liu, S.; Liu, G.; Lou, B.; Shi, W. Effects of triclosan on lipid metabolism and underlying mechanisms in the cyprinid fish Squalidus argentatus. Sci. Total Environ. 2024, 951, 175627. [Google Scholar] [CrossRef]
- Parrish, C.C. Production, Transport, Fate and Effects of Lipids in the Marine Environment. Mar. Drugs 2025, 23, 52. [Google Scholar] [CrossRef]
- Brunet, F.G.; Roest Crollius, H.; Paris, M.; Aury, J.M.; Gibert, P.; Jaillon, O.; Laudet, V.; Robinson-Rechavi, M. Gene Loss and Evolutionary Rates Following Whole-Genome Duplication in Teleost Fishes. Mol. Biol. Evol. 2006, 23, 1808–1816. [Google Scholar] [CrossRef]
- Tang, S.-L.; Liang, X.-F.; He, S.; Li, L.; Alam, M.S.; Wu, J. Comparative Study of the Molecular Characterization, Evolution, and Structure Modeling of Digestive Lipase Genes Reveals the Different Evolutionary Selection Between Mammals and Fishes. Front. Genet. 2022, 13, 909091. [Google Scholar] [CrossRef]
- Burns, M.D.; Sidlauskas, B.L. Ancient and contingent body shape diversification in a hyperdiverse continental fish radiation. Evolution 2019, 73, 569–587. [Google Scholar] [CrossRef]
- Kolmann, M.A.; Hughes, L.C.; Hernandez, L.P.; Arcila, D.; Betancur, R.R.; Sabaj, M.H.; López-Fernández, H.; Ortí, G. Phylogenomics of Piranhas and Pacus (Serrasalmidae) Uncovers How Dietary Convergence and Parallelism Obfuscate Traditional Morphological Taxonomy. Syst. Biol. 2021, 70, 576–592. [Google Scholar] [CrossRef]
- Soares, B.E.; Benone, N.L.; Leitão, R.P.; Leal, C.G.; Santos, L.L.; de Assis Montag, L.F.; Caramaschi, É.P. The ecomorphological diversity of Amazonian stream fishes is constrained by phylogenetic relationships. EcoEvoRxiv 2023. [Google Scholar] [CrossRef]
- De, S.; Teichmann, S.A.; Babu, M.M. The impact of genomic neighborhood on the evolution of human and chimpanzee transcriptome. Genome Res. 2009, 19, 785–794. [Google Scholar] [CrossRef]
- Feng, D.; Huang, Q.Y.; Liu, K.; Zhang, S.C.; Liu, Z.H. Comparative studies of zebrafish Danio rerio lipoprotein lipase (LPL) and hepatic lipase (LIPC) genes belonging to the lipase gene family: Evolution and expression pattern. J. Fish Biol. 2014, 85, 329–342. [Google Scholar] [CrossRef] [PubMed]
- Guruprasad, K.; Reddy, B.V.B.; Pandit, M.W. Correlation between stability of a protein and its dipeptide composition: A novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. Des. Sel. 1990, 4, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Kurtovic, I.; Marshall, S.N.; Zhao, X.; Simpson, B.K. Lipases from Mammals and Fishes. Rev. Fish. Sci. 2009, 17, 18–40. [Google Scholar] [CrossRef]
- Taniguchi, A.; Takano, K.; Kamoi, I. Purification and Properties of Lipase from Tilapia Intestine. Digestive Enzyme of Tilapia VI. Nippon. Suisan Gakkaishi 2001, 67, 78–84. [Google Scholar] [CrossRef]
- Tyagi, P. Isolation, Purification and Characterization of Secondary Structure and Kinetic Study of Lipase from Indian Major Carp, Catla catla (Catla). Enzyme Eng. 2014, 3, 1–8. [Google Scholar] [CrossRef]
- Concha-Frías, B.; Gaxiola-Cortes, M.G.; De la Cruz-Alvarado, F.J.; Jimenez Martinez, L.D.; Peña-Marin, E.S.; Oliva-Arriagada, M.A.; Arias-Moscoso, J.L.; Alvarez-González, C.A. Intestinal Lipase Characterization in Common Snook (Centropomus undecimalis) Juveniles. Fishes 2022, 7, 107. [Google Scholar] [CrossRef]
- Arora, R.; Nimonkar, A.V.; Baird, D.; Wang, C.; Chiu, C.-H.; Horton, P.A.; Hanrahan, S.; Cubbon, R.; Weldon, S.; Tschantz, W.R.; et al. Structure of lipoprotein lipase in complex with GPIHBP1. Proc. Natl. Acad. Sci. USA 2019, 116, 10360–10365. [Google Scholar] [CrossRef]
- Anderson, J.T.; Rojas, J.S.; Flecker, A.S. High-quality seed dispersal by fruit-eating fishes in Amazonian floodplain habitats. Oecologia 2009, 161, 279–290. [Google Scholar] [CrossRef]
- Anderson, J.T.; Nuttle, T.; Rojas, J.S.S.; Pendergast, T.H.; Flecker, A.S. Extremely long-distance seed dispersal by an overfished Amazonian frugivore. Proc. R. Soc. B Biol. Sci. 2011, 278, 3329–3335. [Google Scholar] [CrossRef]
- Sado, R.Y.; de Souza, F.C.; Behr, E.R.; Mocha, P.R.E.; Baldisserotto, B. Anatomy of Teleosts and elasmobranchs. In Biology and Physiology of Freshwater Neotropical Fish; Elsevier: Amsterdam, The Netherlands, 2020; pp. 21–47. [Google Scholar] [CrossRef]
- Khan, F.I.; Lan, D.; Durrani, R.; Huan, W.; Zhao, Z.; Wang, Y. The Lid Domain in Lipases: Structural and Functional Determinant of Enzymatic Properties. Front. Bioeng. Biotechnol. 2017, 5, 16. [Google Scholar] [CrossRef]
- Smichi, N.; Fendri, A.; Triki, S.; Arondel, V.; Rebai, A.; Gargouri, Y.; Miled, N. Biochemical characterization, cloning and molecular modeling of a digestive lipase from red seabream (Pagrus major): Structural explanation of the interaction deficiency with colipase and lipidic interface. Eng. Life Sci. 2017, 17, 664–677. [Google Scholar] [CrossRef] [PubMed]
- Heras, J.; Chakraborty, M.; Emerson, J.J.; German, D.P. Genomic and biochemical evidence of dietary adaptation in a marine herbivorous fish. Proc. R. Soc. B Biol. Sci. 2020, 287, 20192327. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, J.; Inadera, H.; Fujita, Y.; Talley, G.; Morisaki, N.; Yoshida, S.; Saito, Y.; Fojo, S.S.; Brewer, H.B., Jr. A Naturally Occurring Mutation at the Second Base of Codon Asparagine 43 in the Proposed N-Linked Glycosylation Site of Human Lipoprotein Lipase: In Vivo Evidence That Asparagine 43 Is Essential for Catalysis and Secretion. Biochem. Biophys. Res. Commun. 1994, 205, 506–515. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liang, X.-F.; He, S.; Sun, J.; Wen, Z.-Y.; Shen, D.; Tao, Y.-X. Genomic structure, tissue expression and single nucleotide polymorphisms of lipoprotein lipase and hepatic lipase genes in Chinese perch. Aquac. Nutr. 2016, 22, 786–800. [Google Scholar] [CrossRef]
- Ibáñez, A.J.; Peinado-Onsurbe, J.; Sánchez, E.; Cerdá-Reverter, J.M.; Prat, F. Lipoprotein lipase (LPL) is highly expressed and active in the ovary of European sea bass (Dicentrarchus labrax L.), during gonadal development. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2008, 150, 347–354. [Google Scholar] [CrossRef]
- Makrakis, M.C.; Miranda, L.E.; Makrakis, S.; Xavier, A.M.M.; Fontes, H.M.; Morlis, W.G. Migratory movements of pacu, Piaractus mesopotamicus, in the highly impounded Paraná River. J. Appl. Ichthyol. 2007, 23, 700–704. [Google Scholar] [CrossRef]
- Nguyen, P.; Leray, V.; Diez, M.; Serisier, S.; Le Bloc’h, J.; Siliart, B.; Dumon, H. Liver lipid metabolism. J. Anim. Physiol. Anim. Nutr. 2008, 92, 272–283. [Google Scholar] [CrossRef]
- Ibáñez, A.J.; Peinado-Onsurbe, J.; Sánchez, E.; Prat, F. The role of lipoprotein lipase (LPL) in the incorporation of neutral lipids into the oocytes of the European sea bass (Dicentrarchus labrax L.) during gonadal development. Fish. Physiol. Biochem. 2003, 28, 291–293. [Google Scholar] [CrossRef]



| Protein | Taxonomic Groups | Species | Site 1 | Site 2 | Site 3 | Site 4 | Site 5 | Site 6 | Site 7 | Site 8 | Site 9 | Site 10 | Site 11 | Site 12 | Site 13 | Site 14 | Site 15 | Site 16 | Site 17 | Site 18 | Site 19 | Site 20 | Site 21 | Site 22 | Site 23 | Site 24 | Site 25 | Site 26 | Site 27 | Site 28 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LPL1 | Chondrichthyes | C. milii | NTS | NIS | NRT | NET | NQT | 5 | |||||||||||||||||||||||
| Sarcopterygii | L. chalumnae | NRT | NST | NLT | NWT | NKT | 5 | ||||||||||||||||||||||||
| H. sapiens | NHS | NKT | 2 | ||||||||||||||||||||||||||||
| Non-teleosts | E. calabaricus | NLT | NTS | NKT | 3 | ||||||||||||||||||||||||||
| L. oculatus | NTT | NIS | NET | NKT | 4 | ||||||||||||||||||||||||||
| Lpl1a | Basal teleosts | M. cyprinoides | NTT | NTT | NGS | 3 | |||||||||||||||||||||||||
| P. kingsleyae | NST | NST | NKT | NAT | NES | 5 | |||||||||||||||||||||||||
| S. formosus | NTT | NTT | NTT | 4 | |||||||||||||||||||||||||||
| Otocephala | C. harengus | NNT | NTT | NGS | 3 | ||||||||||||||||||||||||||
| C. chanos | NST | NST | NKT | NTT | 4 | ||||||||||||||||||||||||||
| D. rerio | NAT | NST | 2 | ||||||||||||||||||||||||||||
| E. electricus | NIT | NST | NTT | 3 | |||||||||||||||||||||||||||
| P. hypophthalmus | NLS | NTT | NST | NIT | 4 | ||||||||||||||||||||||||||
| I. punctatus | NIT | NST | NTT | 3 | |||||||||||||||||||||||||||
| A. mexicanus | NIT | NST | NTT | 3 | |||||||||||||||||||||||||||
| P. nattereri | NIT | NST | NTT | 3 | |||||||||||||||||||||||||||
| C. macropomum | NIT | NST | NTT | 3 | |||||||||||||||||||||||||||
| Euteleostei | E. lucius | NST | NST | NLS | 3 | ||||||||||||||||||||||||||
| G. morhua | NTT | NST | NST | 3 | |||||||||||||||||||||||||||
| O. niloticus | NTT | NET | NTT | NTT | NQS | 5 | |||||||||||||||||||||||||
| Ö. Latipes | NIS | NTT | NST | NNT | NIS | 5 | |||||||||||||||||||||||||
| Lpl1b | Basal teleosts | M. cyprinoides | NST | NAT | NTT | NGS | 4 | ||||||||||||||||||||||||
| P. kingsleyae | NTT | NTT | NFS | NQT | NST | 5 | |||||||||||||||||||||||||
| Otocephala | C. harengus | NTT | NTT | NIS | NFS | NMS | NTT | 6 | |||||||||||||||||||||||
| C. chanos | NTT | NST | NLT | NVS | NTT | 5 | |||||||||||||||||||||||||
| D. rerio | NST | NFT | NDS | NLT | NMT | NST | NQS | NTS | 8 | ||||||||||||||||||||||
| E. electricus | NST | NHS | NLT | NVS | NTT | 5 | |||||||||||||||||||||||||
| I. punctatus | NST | NTT | NLS | NTT | 4 | ||||||||||||||||||||||||||
| P. hypophthalmus | NST | NLT | NTT | NET | 4 | ||||||||||||||||||||||||||
| A. mexicanus | NPT | NTT | NDS | NMT | NTT | NET | 6 | ||||||||||||||||||||||||
| P. nattereri | NST | NTT | NLS | NTT | NES | 5 | |||||||||||||||||||||||||
| C. macropomum | NST | NTT | NLS | NLT | NMT | NES | 6 | ||||||||||||||||||||||||
| Euteleostei | E. lucius | ||||||||||||||||||||||||||||||
| G. morhua | NTT | NTT | NTT | NHT | NQT | 5 | |||||||||||||||||||||||||
| O. niloticus | |||||||||||||||||||||||||||||||
| Ö. latipes |
| Protein | Taxonomic Groups | Species | Site 1 | Site 2 | Site 3 | Site 4 | Site 5 | Site 6 | Site 7 | Site 8 | Site 9 | Site 10 | Site 11 | Site 12 | Site 13 | Site 14 | Site 15 | Site 16 | Site 17 | Site 18 | Site 19 | Site 20 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LPL2 | Chondrichthyes | C. milii | NTT | NAT | NVT | NHS | NKT | NQT | 6 | ||||||||||||||
| Non-teleost | L. chalumnae | NTT | NRT | 2 | |||||||||||||||||||
| E. calabaricus | NLT | NTS | NKT | 3 | |||||||||||||||||||
| L. oculatus | NHT | NKT | 2 | ||||||||||||||||||||
| Lpl2b | Basal teleosts | P. kingsleyae | NHT | NPT | NAT | NTS | NRT | NSS | 6 | ||||||||||||||
| Lpl2a | Basal teleosts | A. anguilla | NTT | NVT | NQS | NKT | 4 | ||||||||||||||||
| M. cyprinoides | NTT | NQS | NKT | 3 | |||||||||||||||||||
| P. kingsleyae | NTT | NCS | NNT | NRS | 4 | ||||||||||||||||||
| S. formosus | NAT | NHS | NRT | 3 | |||||||||||||||||||
| Otocephala | C. harengus | NST | NQS | NRT | 3 | ||||||||||||||||||
| C. chanos | NKT | NST | NRT | NKT | 4 | ||||||||||||||||||
| D. rerio | NIT | NPS | NHT | NKT | 4 | ||||||||||||||||||
| E. electricus | NLS | NTT | NQS | NGS | NKT | 5 | |||||||||||||||||
| P. hypophthalmus | NST | NQS | NTT | 3 | |||||||||||||||||||
| I. punctatus | |||||||||||||||||||||||
| A. mexicanus | NNT | NGS | NKT | NLS | NWS | 5 | |||||||||||||||||
| P. nattereri | NST | NKT | 2 | ||||||||||||||||||||
| C. macropomum | NST | NKT | 2 | ||||||||||||||||||||
| Euteleostei | E. lucius | NKT | NWS | 2 | |||||||||||||||||||
| G. morhua | NAT | NST | NRS | NKT | 4 | ||||||||||||||||||
| O. niloticus | NVT | NAT | NST | NSS | NKT | NVT | 6 | ||||||||||||||||
| Ö. Latipes | NST | NRT | NTS | NKT | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paixão, R.V.; Bandeira, I.C.; Reis, V.R.; da Silva, G.F.; Almeida O’Sullivan, F.L.d.; Varela, E.S. Phylogenomic and Evolutionary Insights into Lipoprotein Lipase (LPL) Genes in Tambaqui: Gene Duplication, Tissue-Specific Expression and Physiological Implications. Genes 2025, 16, 548. https://doi.org/10.3390/genes16050548
Paixão RV, Bandeira IC, Reis VR, da Silva GF, Almeida O’Sullivan FLd, Varela ES. Phylogenomic and Evolutionary Insights into Lipoprotein Lipase (LPL) Genes in Tambaqui: Gene Duplication, Tissue-Specific Expression and Physiological Implications. Genes. 2025; 16(5):548. https://doi.org/10.3390/genes16050548
Chicago/Turabian StylePaixão, Rômulo Veiga, Izabel Correa Bandeira, Vanessa Ribeiro Reis, Gilvan Ferreira da Silva, Fernanda Loureiro de Almeida O’Sullivan, and Eduardo Sousa Varela. 2025. "Phylogenomic and Evolutionary Insights into Lipoprotein Lipase (LPL) Genes in Tambaqui: Gene Duplication, Tissue-Specific Expression and Physiological Implications" Genes 16, no. 5: 548. https://doi.org/10.3390/genes16050548
APA StylePaixão, R. V., Bandeira, I. C., Reis, V. R., da Silva, G. F., Almeida O’Sullivan, F. L. d., & Varela, E. S. (2025). Phylogenomic and Evolutionary Insights into Lipoprotein Lipase (LPL) Genes in Tambaqui: Gene Duplication, Tissue-Specific Expression and Physiological Implications. Genes, 16(5), 548. https://doi.org/10.3390/genes16050548








