Abundant β-Defensin Copy Number Variations in Pigs
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Tissues
2.2. Preparation of DNA
2.3. Semi-Quantitative PCR Using Genomic DNA
2.4. Real-Time Quantitative PCR Using Genomic DNA
2.5. Real-Time PCR Using Total RNA
2.6. Sequencing
2.7. Copy Number Estimation
2.8. Statistical Analysis
3. Results
3.1. Determination of Porcine β-Defensin Genes for the Analysis of Copy Number Variations
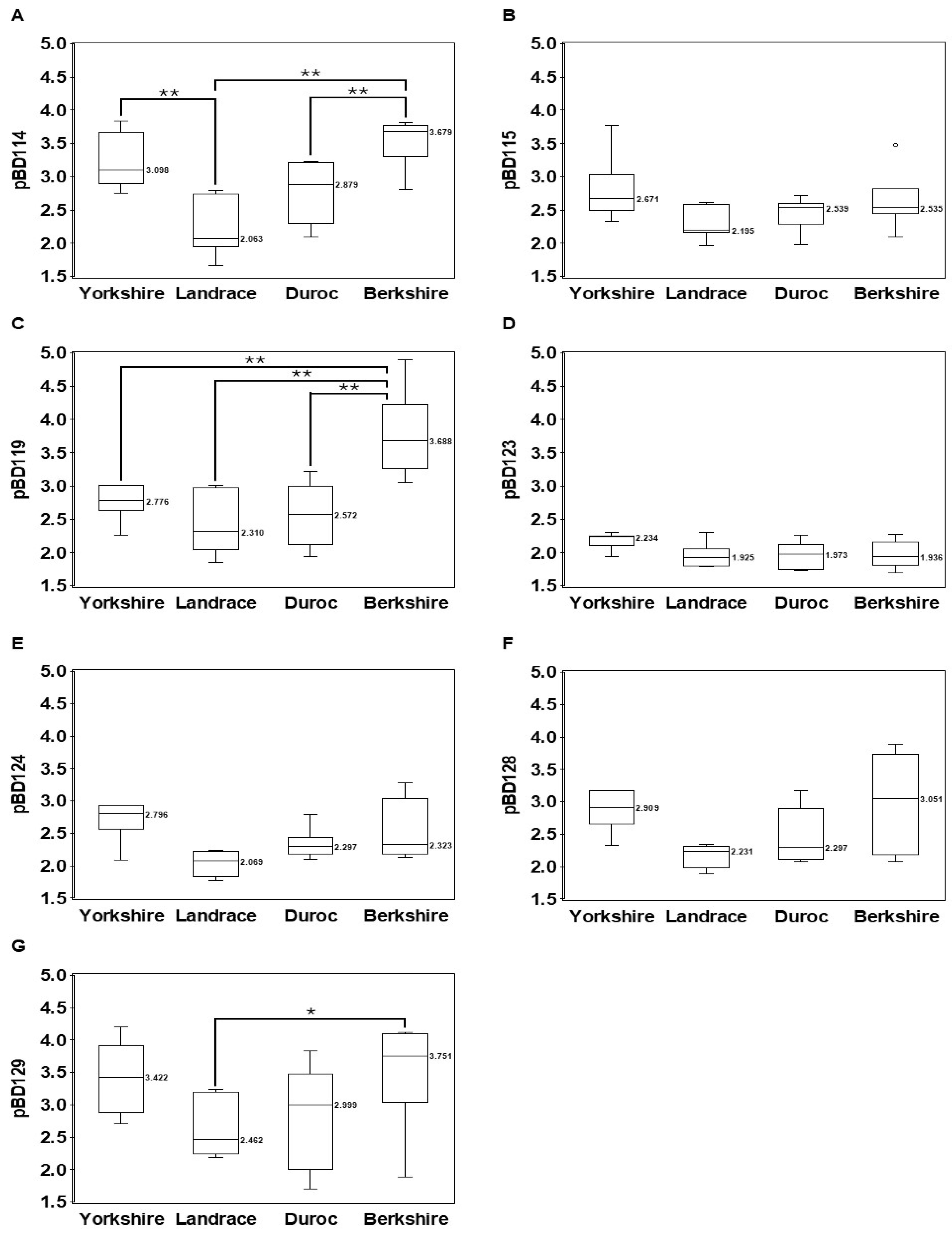
3.2. Breed Variations of CNVs for pBD114, pBD115, pBD119, pBD128, and pBD129
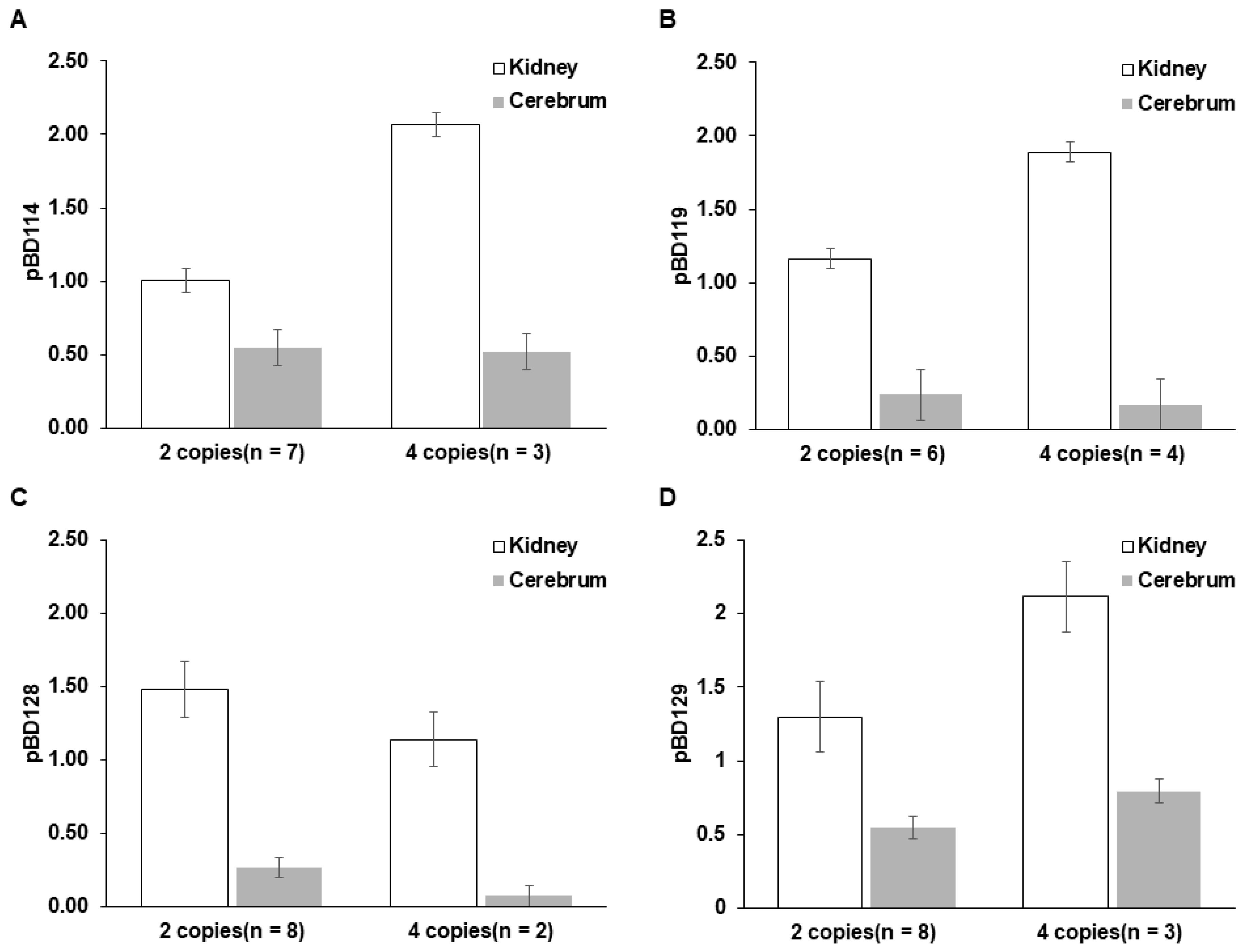
3.3. Significant Correlation Between pBD Copy Numbers and Expression in Kidneys
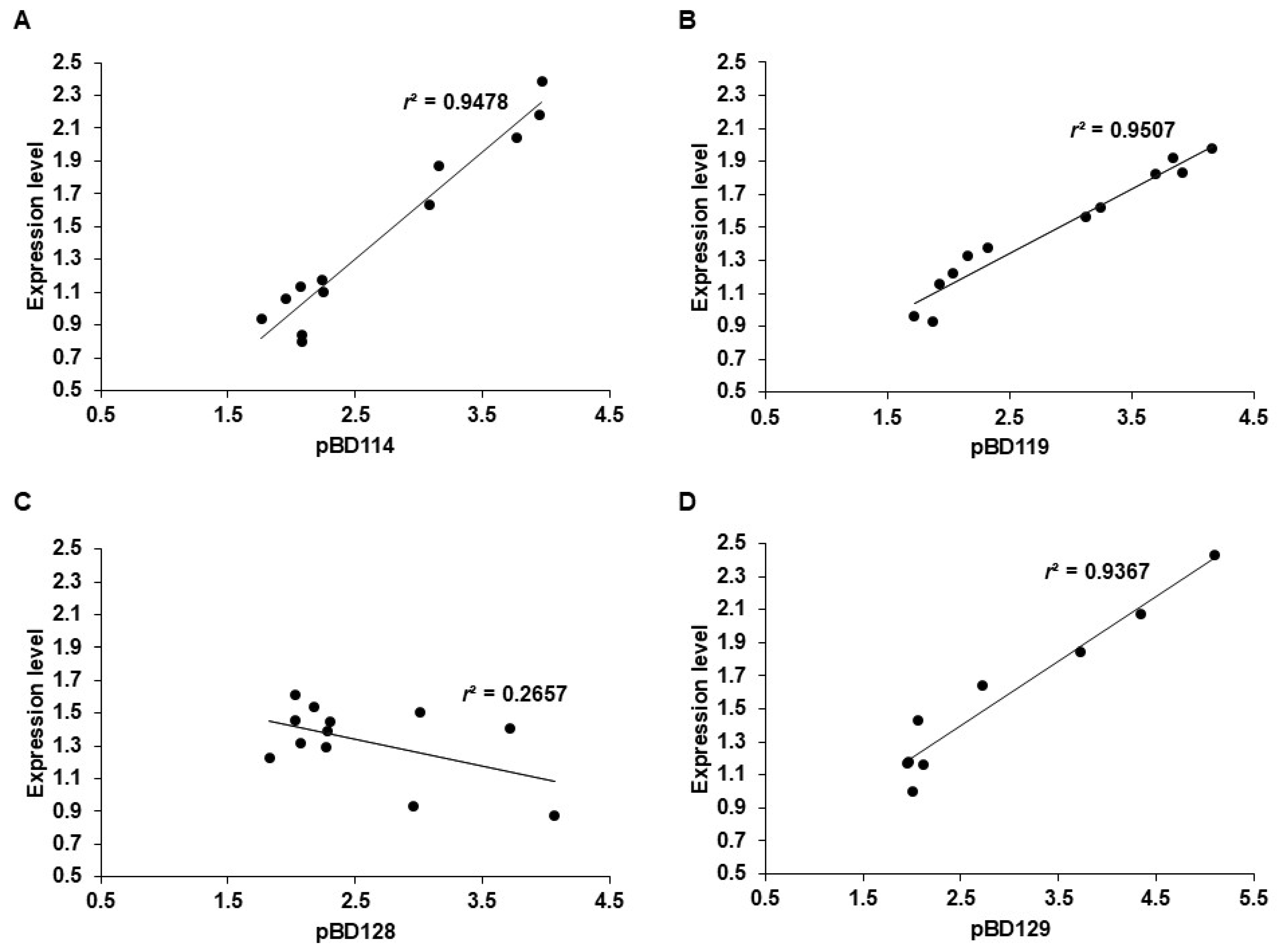
3.4. Correlation Between pBD Expression and Gene Copy Number
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AMPs | antimicrobial peptides |
| pBDs | porcine β-defensins |
| CNVs | copy number variations |
| RT-qPCR | real-time quantitative PCR |
| SSC | Sus scrofa chromosome |
| GAPDH | glyceraldehyde 3-phosphate dehydrogenase |
| GCG | glucagon |
| Ct | cycle threshold |
| LPS | lipopolysaccharide |
| TLRs | Toll-like receptors |
References
- Ganz, T.; Lehrer, R.I. Defensins. Pharmacol. Ther. 1995, 66, 191–205. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.; Ahn, B.; Yum, J.; Cho, H.s.; Choi, M.; Hong, K.; Choi, Y.; Kim, J.H.; Park, C. Influence of habitat change from land to sea on the evolution of antimicrobial peptide gene families, including β-defensin gene clusters, in mammals. J. Zool. Syst. Evol. Res. 2021, 59, 510–521. [Google Scholar] [CrossRef]
- Zou, J.; Mercier, C.; Koussounadis, A.; Secombes, C. Discovery of multiple β-defensin like homologues in teleost fish. Mol. Immunol. 2007, 44, 638–647. [Google Scholar] [CrossRef] [PubMed]
- Belov, K.; Sanderson, C.E.; Deakin, J.E.; Wong, E.S.; Assange, D.; McColl, K.A.; Gout, A.; de Bono, B.; Barrow, A.D.; Speed, T.P. Characterization of the opossum immune genome provides insights into the evolution of the mammalian immune system. Genome Res. 2007, 17, 982–991. [Google Scholar] [CrossRef]
- Choi, M.-K.; Le, M.T.; Nguyen, D.T.; Choi, H.; Kim, W.; Kim, J.-H.; Chun, J.; Hyeon, J.; Seo, K.; Park, C. Genome-level identification, gene expression, and comparative analysis of porcine β-defensin genes. BMC Genet. 2012, 13, 98. [Google Scholar] [CrossRef]
- Bauer, F.; Schweimer, K.; Klüver, E.; Conejo-Garcia, J.R.; Forssmann, W.G.; Rösch, P.; Adermann, K.; Sticht, H. Structure determination of human and murine β-defensins reveals structural conservation in the absence of significant sequence similarity. Protein Sci. 2001, 10, 2470–2479. [Google Scholar]
- Das, S.; Nikolaidis, N.; Goto, H.; McCallister, C.; Li, J.; Hirano, M.; Cooper, M.D. Comparative genomics and evolution of the α-defensin multigene family in primates. Mol. Biol. Evol. 2010, 27, 2333–2343. [Google Scholar] [CrossRef][Green Version]
- Patil, A.A.; Cai, Y.; Sang, Y.; Blecha, F.; Zhang, G. Cross-species analysis of the mammalian β-defensin gene family: Presence of syntenic gene clusters and preferential expression in the male reproductive tract. Physiol. Genom. 2005, 23, 5–17. [Google Scholar] [CrossRef]
- Meade, K.G.; O’Farrelly, C. β-Defensins: Farming the microbiome for homeostasis and health. Front. Immunol. 2019, 9, 3072. [Google Scholar] [CrossRef]
- Ganz, T. The role of antimicrobial peptides in innate immunity. Integr. Comp. Biol. 2003, 43, 300–304. [Google Scholar] [CrossRef]
- Lehrer, R.I. Primate defensins. Nat. Rev. Microbiol. 2004, 2, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Zanetti, M. Cathelicidins, multifunctional peptides of the innate immunity. J. Leucoc. Biol. 2004, 75, 39–48. [Google Scholar]
- Yang, D.; Chertov, O.; Bykovskaia, S.; Chen, Q.; Buffo, M.; Shogan, J.; Anderson, M.; Schroder, J.; Wang, J.; Howard, O. β-defensins: Linking innate and adaptive immunity through dendritic and T cell CCR6. Science 1999, 286, 525–528. [Google Scholar] [CrossRef]
- Ganz, T. Defensins: Antimicrobial peptides of vertebrates. Comptes Rendus Biol. 2004, 327, 539–549. [Google Scholar]
- Selsted, M.E.; Ouellette, A.J. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 2005, 6, 551–557. [Google Scholar]
- Semple, C.A.; Rolfe, M.; Dorin, J.R. Duplication and selection in the evolution of primate β-defensin genes. Genome Biol. 2003, 4, R31. [Google Scholar]
- Yuan, H.; Wei, W.; Zhang, Y.; Li, C.; Zhao, S.; Chao, Z.; Xia, C.; Quan, J.; Gao, C. Unveiling the Influence of Copy Number Variations on Genetic Diversity and Adaptive Evolution in China’s Native Pig Breeds via Whole-Genome Resequencing. Int. J. Mol. Sci. 2024, 25, 5843. [Google Scholar] [CrossRef]
- Stankiewicz, P.; Lupski, J.R. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 2010, 61, 437–455. [Google Scholar] [CrossRef]
- Durán Aguilar, M.; Román Ponce, S.; Ruiz López, F.; González Padilla, E.; Vásquez Peláez, C.; Bagnato, A.; Strillacci, M.G. Genome-wide association study for milk somatic cell score in holstein cattle using copy number variation as markers. J. Anim. Breed. Genet. 2017, 134, 49–59. [Google Scholar]
- Wang, Y.; Ma, J.; Wang, J.; Zhang, L.; Xu, L.; Chen, Y.; Zhu, B.; Wang, Z.; Gao, H.; Li, J. Genome-Wide Detection of Copy Number Variations and Their Potential Association with Carcass and Meat Quality Traits in Pingliang Red Cattle. Int. J. Mol. Sci. 2024, 25, 5626. [Google Scholar] [CrossRef]
- Chen, C.; Liu, C.; Xiong, X.; Fang, S.; Yang, H.; Zhang, Z.; Ren, J.; Guo, Y.; Huang, L. Copy number variation in the MSRB3 gene enlarges porcine ear size through a mechanism involving miR-584-5p. Genet. Sel. Evol. 2018, 50, 72. [Google Scholar] [PubMed]
- Perry, G.H.; Yang, F.; Marques-Bonet, T.; Murphy, C.; Fitzgerald, T.; Lee, A.S.; Hyland, C.; Stone, A.C.; Hurles, M.E.; Tyler-Smith, C. Copy number variation and evolution in humans and chimpanzees. Genome Res. 2008, 18, 1698–1710. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, M.; da Silva, V.H.; Megens, H.-J.; Visker, M.H.; Ajmone-Marsan, P.; Bâlteanu, V.A.; Dunner, S.; Garcia, J.F.; Ginja, C.; Kantanen, J. Distribution and functionality of copy number variation across European cattle populations. Front. Genet. 2017, 8, 108. [Google Scholar]
- Shi, H.; Li, T.; Su, M.; Wang, H.; Li, Q.; Lang, X.; Ma, Y. Identification of copy number variation in Tibetan sheep using whole genome resequencing reveals evidence of genomic selection. BMC Genom. 2023, 24, 555. [Google Scholar]
- Panda, S.; Kumar, A.; Gaur, G.K.; Ahmad, S.F.; Chauhan, A.; Mehrotra, A.; Dutt, T. Genome wide copy number variations using Porcine 60K SNP Beadchip in Landlly pigs. Anim. Biotechnol. 2023, 34, 1891–1899. [Google Scholar]
- Lin, S.; Lin, X.; Zhang, Z.; Jiang, M.; Rao, Y.; Nie, Q.; Zhang, X. Copy number variation in SOX6 contributes to chicken muscle development. Genes 2018, 9, 42. [Google Scholar] [CrossRef]
- Binversie, E.E.; Baker, L.A.; Engelman, C.D.; Hao, Z.; Moran, J.J.; Piazza, A.M.; Sample, S.J.; Muir, P. Analysis of copy number variation in dogs implicates genomic structural variation in the development of anterior cruciate ligament rupture. PLoS ONE 2020, 15, e0244075. [Google Scholar] [CrossRef]
- Machado, L.R.; Ottolini, B. An evolutionary history of defensins: A role for copy number variation in maximizing host innate and adaptive immune responses. Front. Immunol. 2015, 6, 115. [Google Scholar]
- Lee, M.O.; Bornelöv, S.; Andersson, L.; Lamont, S.J.; Chen, J.; Womack, J.E. Duplication of chicken defensin7 gene generated by gene conversion and homologous recombination. Proc. Natl. Acad. Sci. 2016, 113, 13815–13820. [Google Scholar] [CrossRef]
- Leonard, B.C.; Marks, S.L.; Outerbridge, C.A.; Affolter, V.K.; Kananurak, A.; Young, A.; Moore, P.F.; Bannasch, D.L.; Bevins, C.L. Activity, expression and genetic variation of canine β-defensin 103: A multifunctional antimicrobial peptide in the skin of domestic dogs. J. Innate Immun. 2012, 4, 248–259. [Google Scholar] [CrossRef]
- Hardwick, R.J.; Machado, L.R.; Zuccherato, L.W.; Antolinos, S.; Xue, Y.; Shawa, N.; Gilman, R.H.; Cabrera, L.; Berg, D.E.; Tyler-Smith, C. A worldwide analysis of β-defensin copy number variation suggests recent selection of a high-expressing DEFB103 gene copy in East Asia. Hum. Mutat. 2011, 32, 743–750. [Google Scholar] [PubMed]
- Wain, L.V.; Odenthal-Hesse, L.; Abujaber, R.; Sayers, I.; Beardsmore, C.; Gaillard, E.A.; Chappell, S.; Dogaru, C.M.; McKeever, T.; Guetta-Baranes, T. Copy number variation of the β-defensin genes in europeans: No supporting evidence for association with lung function, chronic obstructive pulmonary disease or asthma. PLoS ONE 2014, 9, e84192. [Google Scholar]
- Lee, A.S.; Gutiérrez-Arcelus, M.; Perry, G.H.; Vallender, E.J.; Johnson, W.E.; Miller, G.M.; Korbel, J.O.; Lee, C. Analysis of copy number variation in the rhesus macaque genome identifies candidate loci for evolutionary and human disease studies. Hum. Mol. Genet. 2008, 17, 1127–1136. [Google Scholar] [PubMed]
- Ottolini, B. Evolution of Copy Number Variation in the Rhesus Macaque β-Defensin Region. Ph.D. Thesis, University of Leicester, Leicester, UK, 2014. [Google Scholar]
- Sidekli, O.; Oketch, J.; Fair, S.; Meade, K.G.; Hollox, E.J. β-Defensin gene copy number variation in cattle. R. Soc. Open Sci. 2024, 11, 241154. [Google Scholar] [CrossRef]
- James, C.P.; Bajaj-Elliott, M.; Abujaber, R.; Forya, F.; Klein, N.; David, A.L.; Hollox, E.J.; Peebles, D.M. Human β defensin (HBD) gene copy number affects HBD2 protein levels: Impact on cervical bactericidal immunity in pregnancy. Eur. J. Hum. Genet. 2018, 26, 434–439. [Google Scholar]
- Stuart, P.E.; Hüffmeier, U.; Nair, R.P.; Palla, R.; Tejasvi, T.; Schalkwijk, J.; Elder, J.T.; Reis, A.; Armour, J.A. Association of β-defensin copy number and psoriasis in three cohorts of European origin. J. Investig. Dermatol. 2012, 132, 2407–2413. [Google Scholar]
- Sambrook, J. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Habor, NY, USA, 1989. [Google Scholar]
- Sang, Y.; Patil, A.A.; Zhang, G.; Ross, C.R.; Blecha, F. Bioinformatic and expression analysis of novel porcine β-defensins. Mamm. Genome 2006, 17, 332–339. [Google Scholar] [CrossRef]
- Ballester, M.; Castelló, A.; Ibáñez, E.; Sánchez, A.; Folch, J.M. Real-time quantitative PCR-based system for determining transgene copy number in transgenic animals. Biotechniques 2004, 37, 610–613. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar]
- Ramayo-Caldas, Y.; Castelló, A.; Pena, R.N.; Alves, E.; Mercadé, A.; Souza, C.A.; Fernández, A.I.; Perez-Enciso, M.; Folch, J.M. Copy number variation in the porcine genome inferred from a 60 k SNP BeadChip. BMC Genom. 2010, 11, 593. [Google Scholar] [CrossRef]
- Lee, J.; Le, M.T.; Choi, M.K.; Quy Le, V.C.; Choi, H.; Lee, H.; Song, H.; Kim, J.H.; Park, C. Development of a simple SLA-1 copy-number-variation typing and the comparison of typing accuracy between real-time quantitative and droplet digital PCR. Anim. Genet. 2019, 50, 315. [Google Scholar] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Lüdecke, D.; Ben-Shachar, M.S.; Patil, I.; Waggoner, P.; Makowski, D. performance: An R package for assessment, comparison and testing of statistical models. J. Open Source Softw. 2021, 6, 3139. [Google Scholar]
- Sedgwick, P. Pearson’s correlation coefficient. BMJ 2012, 345, e4483. [Google Scholar] [CrossRef]
- Villanueva, R.A.M.; Chen, Z.J. ggplot2: Elegant graphics for data analysis. Meas. Interdiscip. Res. Perspect. 2019, 17, 160–167. [Google Scholar] [CrossRef]
- Abe, S.; Miura, K.; Kinoshita, A.; Mishima, H.; Miura, S.; Yamasaki, K.; Hasegawa, Y.; Higashijima, A.; Jo, O.; Sasaki, K. Copy number variation of the antimicrobial-gene, defensin β 4, is associated with susceptibility to cervical cancer. J. Hum. Genet. 2013, 58, 250–253. [Google Scholar]
- Hollox, E.; Armour, J.; Barber, J. Extensive normal copy number variation of a β-defensin antimicrobial-gene cluster. Am. J. Hum. Genet. 2003, 73, 591–600. [Google Scholar] [CrossRef]
- Hollox, E.J.; Barber, J.C.; Brookes, A.J.; Armour, J.A. Defensins and the dynamic genome: What we can learn from structural variation at human chromosome band 8p23. 1. Genome Res. 2008, 18, 1686–1697. [Google Scholar]
- Ottolini, B.; Hornsby, M.J.; Abujaber, R.; MacArthur, J.A.; Badge, R.M.; Schwarzacher, T.; Albertson, D.G.; Bevins, C.L.; Solnick, J.V.; Hollox, E.J. Evidence of convergent evolution in humans and macaques supports an adaptive role for copy number variation of the β-defensin-2 gene. Genome Biol. Evol. 2014, 6, 3025–3038. [Google Scholar] [CrossRef][Green Version]
- Xie, K.; Su, G.; Chen, D.; Yu, B.; Huang, Z.; Yu, J.; Zheng, P.; Luo, Y.; Yan, H.; Li, H. The immunomodulatory function of the porcine β-defensin 129: Alleviate inflammatory response induced by LPS in IPEC-J2 cells. Int. J. Biol. Macromol. 2021, 188, 473–481. [Google Scholar]
- Zeng, F.; Wang, M.; Li, J.; Li, C.; Pan, X.; Meng, L.; Li, L.; Wei, H.; Zhang, S. Involvement of porcine β-defensin 129 in sperm capacitation and rescue of poor sperm in genital tract infection. Int. J. Mol. Sci. 2022, 23, 9441. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Zhang, Y.; Gao, H.; Fu, Y.; Chen, Y.; Yin, Y.; Xu, K. Differences in Immune Characteristics and Related Gene Expression in Spleen among Ningxiang, Berkshire Breeds and Their Hybrid Pigs. Genes 2024, 15, 205. [Google Scholar] [CrossRef] [PubMed]
- Su, G.; Luo, Y.; Chen, D.; Yu, B.; He, J. NF-κB-dependent induction of porcine β-defensin 114 regulates intestinal epithelium homeostasis. Int. J. Biol. Macromol. 2021, 192, 241–249. [Google Scholar] [CrossRef]
- Wan, M.L.-Y.; Woo, C.-S.J.; Allen, K.J.; Turner, P.C.; El-Nezami, H. Modulation of porcine β-defensins 1 and 2 upon individual and combined Fusarium toxin exposure in a swine jejunal epithelial cell line. Appl. Environ. Microbiol. 2013, 79, 2225–2232. [Google Scholar] [CrossRef]
- Tang, P.C.-T.; Zhang, Y.-Y.; Chan, M.K.-K.; Lam, W.W.-Y.; Chung, J.Y.-F.; Kang, W.; To, K.-F.; Lan, H.-Y.; Tang, P.M.-K. The Emerging Role of Innate Immunity in Chronic Kidney Diseases. Int. J. Mol. Sci. 2020, 21, 4018. [Google Scholar] [CrossRef]
- Ahn, B.; Jeon, H.; Cho, H.-s.; Nagasundarapandian, S.; Park, C. Sequence polymorphisms of PR39 cathelicidins and extensive copy variations in commercial pig breeds. Gene 2022, 822, 146323. [Google Scholar] [CrossRef]
- Choi, M.K.; Le, M.T.; Cho, H.; Soundrarajan, N.; Jeon, H.; Park, C.K.; Cha, S.Y.; Kim, J.H.; Seo, K.; Park, C. Defining the genetic relationship of protegrin-related sequences and the in vivo expression of protegrins. FEBS J. 2014, 281, 5420–5431. [Google Scholar] [CrossRef]
- Kościuczuk, E.M.; Lisowski, P.; Jarczak, J.; Strzałkowska, N.; Jóźwik, A.; Horbańczuk, J.; Krzyżewski, J.; Zwierzchowski, L.; Bagnicka, E. Cathelicidins: Family of antimicrobial peptides. A review. Mol. Biol. Rep. 2012, 39, 10957–10970. [Google Scholar] [CrossRef]
- Zhang, L.; Jie, H.; Xiao, Y.; Zhou, C.; Lyu, W.; Bai, W. Genomic identification and expression analysis of the cathelicidin gene family of the forest musk deer. Animals 2019, 9, 481. [Google Scholar] [CrossRef]
- Jungi, T.W.; Farhat, K.; Burgener, I.A.; Werling, D. Toll-like receptors in domestic animals. Cell Tissue Res. 2011, 343, 107–120. [Google Scholar] [CrossRef]
- Govoni, G.; Gros, P. Macrophage NRAMP1 and its role in resistance to microbial infections. Inflamm. Res. 1998, 47, 277–284. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, D.; Cho, H.-s.; Kang, M.; Ahn, B.; Shin, J.; Park, C. Abundant β-Defensin Copy Number Variations in Pigs. Genes 2025, 16, 430. https://doi.org/10.3390/genes16040430
Kim D, Cho H-s, Kang M, Ahn B, Shin J, Park C. Abundant β-Defensin Copy Number Variations in Pigs. Genes. 2025; 16(4):430. https://doi.org/10.3390/genes16040430
Chicago/Turabian StyleKim, Dohun, Hye-sun Cho, Mingue Kang, Byeongyong Ahn, Jaeyeol Shin, and Chankyu Park. 2025. "Abundant β-Defensin Copy Number Variations in Pigs" Genes 16, no. 4: 430. https://doi.org/10.3390/genes16040430
APA StyleKim, D., Cho, H.-s., Kang, M., Ahn, B., Shin, J., & Park, C. (2025). Abundant β-Defensin Copy Number Variations in Pigs. Genes, 16(4), 430. https://doi.org/10.3390/genes16040430




