Advancing Non-Invasive Prenatal Screening: A Targeted 1069-Gene Panel for Comprehensive Detection of Monogenic Disorders and Copy Number Variations
Abstract
1. Introduction
2. Materials and Methods
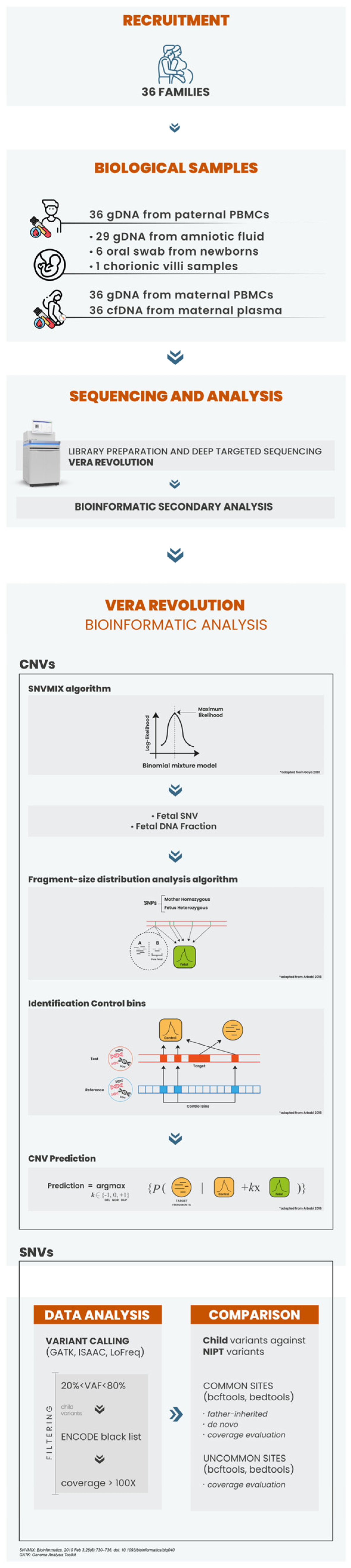
2.1. Patient Recruitment and Sample Processing
2.2. DNA Isolation and NIPS Analysis
2.3. VERA Revolution Gene Panel: Design and Selection Criteria
2.4. VERA Revolution Library Preparation
2.5. CNV Calling
2.6. Follow-Up Diagnostic Testing
3. Results
3.1. VERA Revolution Gene Panel and Depth of Analysis
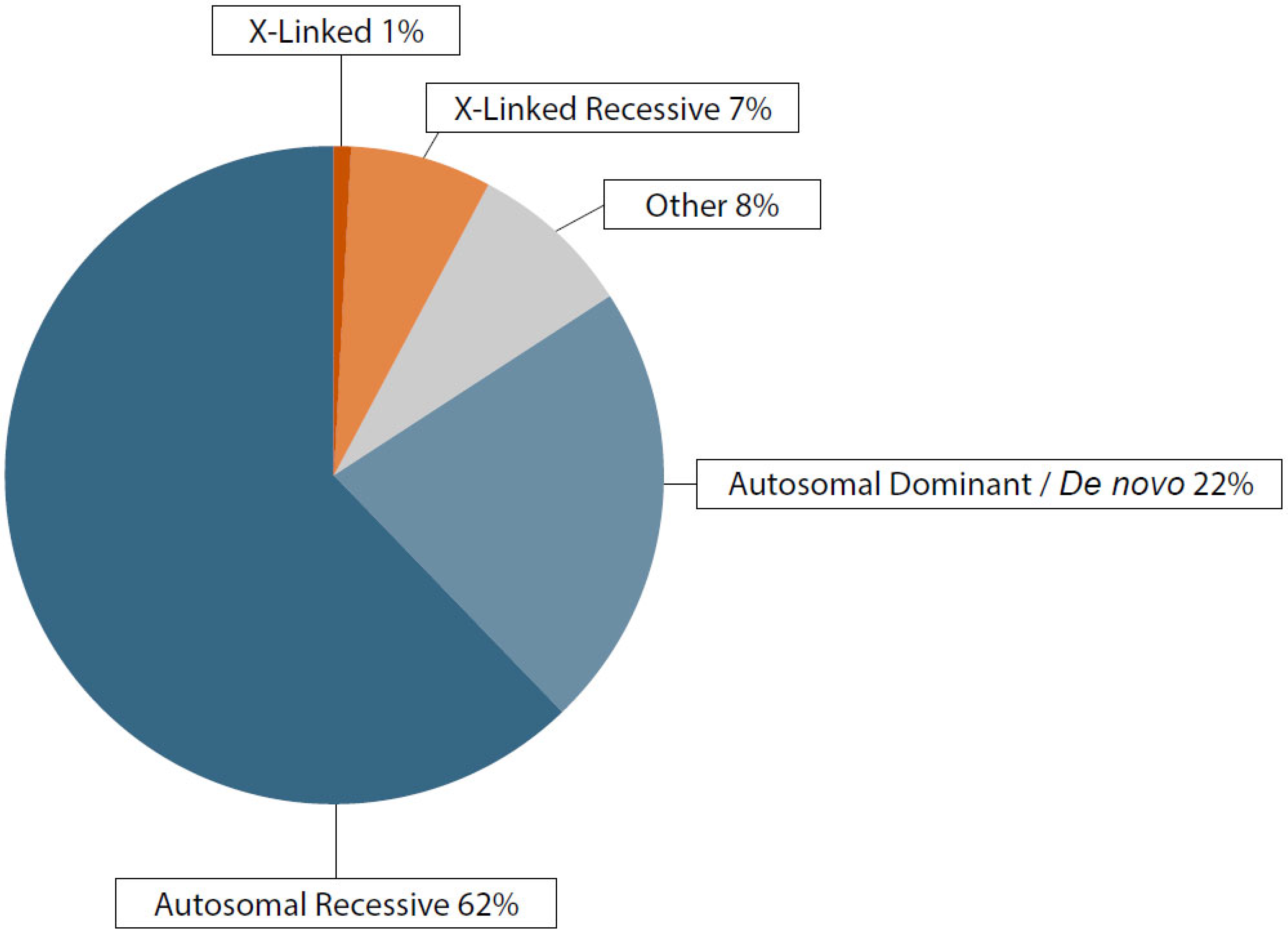
3.2. Genetic Findings
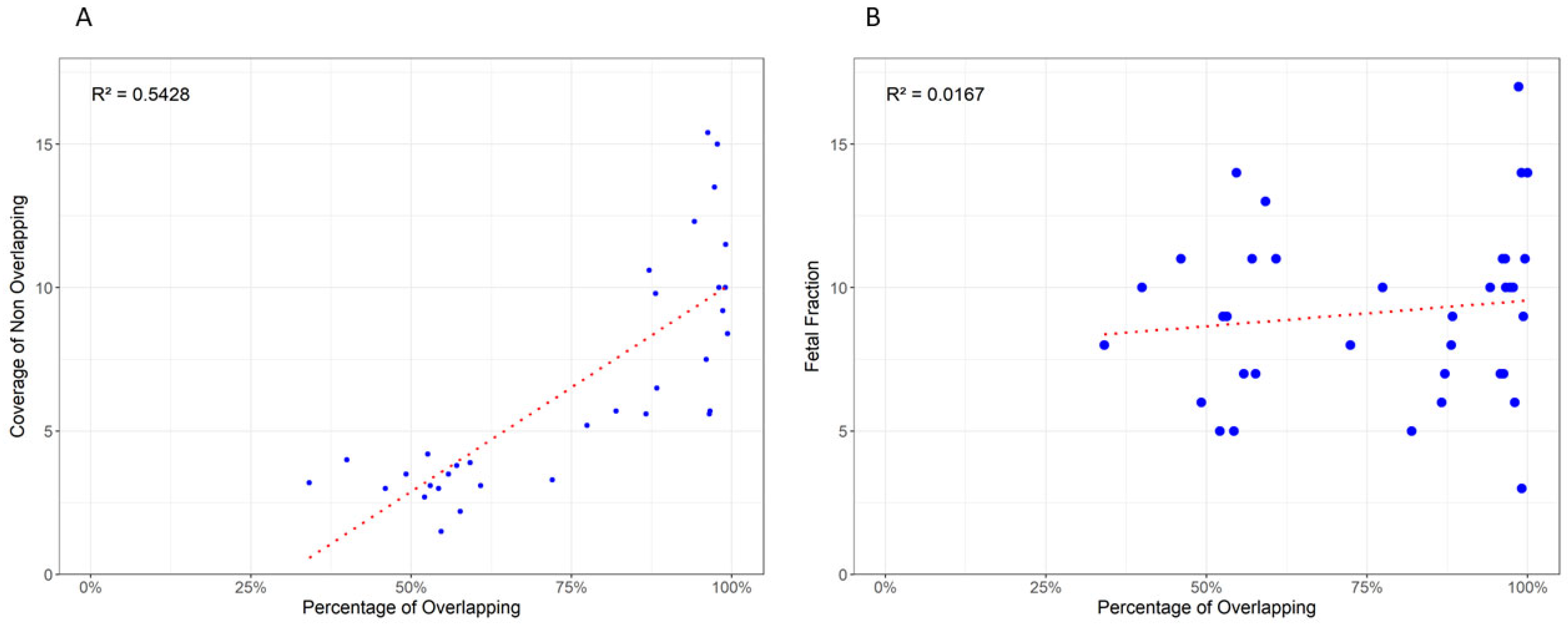
3.3. Variant Overlap and Coverage Influence
4. Discussion
4.1. Broad-Spectrum Analysis
4.2. Clinical Implications in Early Pregnancy
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Atkins, J.C.; Padgett, C.R. Living with a Rare Disease: Psychosocial Impacts for Parents and Family Members—A Systematc Review. J. Child Fam. Stud. 2024, 33, 617–636. [Google Scholar] [CrossRef]
- Alfirevic, Z.; Navaratnam, K.; Mujezinovic, F. Amniocentesis and Chorionic Villus Sampling for Prenatal Diagnosis. Cochrane Database Syst. Rev. 2017, 2017, CD003252. [Google Scholar] [CrossRef]
- Lo, Y.M.D.; Corbetta, N.; Chamberlain, P.F.; Rai, V.; Sargent, I.L.; Redman, C.W.; Wainscoat, J.S. Presence of Fetal DNA in Maternal Plasma and Serum. Lancet 1997, 350, 485–487. [Google Scholar] [CrossRef] [PubMed]
- Nectoux, J. Current, Emerging, and Future Applications of Digital PCR in Non-Invasive Prenatal Diagnosis. Mol. Diagn. Ther. 2018, 22, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, J.; Saucier, J.B.; Feng, Y.; Jiang, Y.; Sinson, J.; McCombs, A.K.; Schmitt, E.S.; Peacock, S.; Chen, S.; et al. Non-Invasive Prenatal Sequencing for Multiple Mendelian Monogenic Disorders Using Circulating Cell-Free Fetal DNA. Nat. Med. 2019, 25, 439–447. [Google Scholar] [CrossRef]
- Chiu, R.W.K.; Chan, K.C.A.; Gao, Y.; Lau, V.Y.M.; Zheng, W.; Leung, T.Y.; Foo, C.H.F.; Xie, B.; Tsui, N.B.Y.; Lun, F.M.F.; et al. Noninvasive Prenatal Diagnosis of Fetal Chromosomal Aneuploidy by Massively Parallel Genomic Sequencing of DNA in Maternal Plasma. Proc. Natl. Acad. Sci. USA 2008, 105, 20458–20463. [Google Scholar] [CrossRef]
- Chen, C.; Li, R.; Sun, J.; Zhu, Y.; Jiang, L.; Li, J.; Fu, F.; Wan, J.; Guo, F.; An, X.; et al. Noninvasive Prenatal Testing of α-Thalassemia and β-Thalassemia through Population-Based Parental Haplotyping. Genome Med. 2021, 13, 18. [Google Scholar] [CrossRef]
- Hui, L.; Bianchi, D.W. Recent Advances in the Prenatal Interrogation of the Human Fetal Genome. Trends Genet. 2013, 29, 84–91. [Google Scholar] [CrossRef]
- Gadsbøll, K.; Petersen, O.B.; Gatinois, V.; Strange, H.; Jacobsson, B.; Wapner, R.; Vermeesch, J.R.; The NIPT-map Study Group; Vogel, I. Current Use of Noninvasive Prenatal Testing in Europe, Australia and the USA: A Graphical Presentation. Acta Obstet. Gynecol. Scand 2020, 99, 722–730. [Google Scholar] [CrossRef]
- Vora, N.L.; Langlois, S.; Chitty, L.S. Current Controversy in Prenatal Diagnosis: The Use of cfDNA to Screen for Monogenic Conditions in Low Risk Populations Is Ready for Clinical Use. Prenat. Diagn. 2024, 44, 389–397. [Google Scholar] [CrossRef]
- Rafalko, J.; Soster, E.; Caldwell, S.; Almasri, E.; Westover, T.; Weinblatt, V.; Cacheris, P. Genome-Wide Cell-Free DNA Screening: A Focus on Copy-Number Variants. Genet. Med. 2021, 23, 1847–1853. [Google Scholar] [CrossRef] [PubMed]
- Dungan, J.S.; Klugman, S.; Darilek, S.; Malinowski, J.; Akkari, Y.M.N.; Monaghan, K.G.; Erwin, A.; Best, R.G.; ACMG Board of Directors. Noninvasive Prenatal Screening (NIPS) for Fetal Chromosome Abnormalities in a General-Risk Population: An Evidence-Based Clinical Guideline of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2023, 25, 100336. [Google Scholar] [CrossRef]
- Amemiya, H.M.; Kundaje, A.; Boyle, A.P. The ENCODE Blacklist: Identification of Problematic Regions of the Genome. Sci. Rep. 2019, 9, 9354. [Google Scholar] [CrossRef]
- Goya, R.; Sun, M.G.F.; Morin, R.D.; Leung, G.; Ha, G.; Wiegand, K.C.; Senz, J.; Crisan, A.; Marra, M.A.; Hirst, M.; et al. SNVMix: Predicting Single Nucleotide Variants from next-Generation Sequencing of Tumors. Bioinformatics 2010, 26, 730–736. [Google Scholar] [CrossRef] [PubMed]
- Arbabi, A.; Rampášek, L.; Brudno, M. Cell-Free DNA Fragment-Size Distribution Analysis for Non-Invasive Prenatal CNV Prediction. Bioinformatics 2016, 32, 1662–1669. [Google Scholar] [CrossRef]
- De Falco, L.; Savarese, G.; Savarese, P.; Petrillo, N.; Ianniello, M.; Ruggiero, R.; Suero, T.; Barbato, C.; Mori, A.; Ramiro, C.; et al. Clinical Experience with Genome-Wide Noninvasive Prenatal Screening in a Large Cohort of Twin Pregnancies. Genes 2023, 14, 982. [Google Scholar] [CrossRef] [PubMed]
- De Falco, L.; Vitiello, G.; Savarese, G.; Suero, T.; Ruggiero, R.; Savarese, P.; Ianniello, M.; Petrillo, N.; Bruno, M.; Legnante, A.; et al. A Case Report of a Feto-Placental Mosaicism Involving a Segmental Aneuploidy: A Challenge for Genome Wide Screening by Non-Invasive Prenatal Testing of Cell-Free DNA in Maternal Plasma. Genes 2023, 14, 668. [Google Scholar] [CrossRef]
- Scotchman, E.; Shaw, J.; Paternoster, B.; Chandler, N.; Chitty, L.S. Non-Invasive Prenatal Diagnosis and Screening for Monogenic Disorders. Eur. J. Obstet. Gynecol. Reprod. Biol. 2020, 253, 320–327. [Google Scholar] [CrossRef]
- Hanson, B.; Scotchman, E.; Chitty, L.S.; Chandler, N.J. Non-Invasive Prenatal Diagnosis (NIPD): How Analysis of Cell-Free DNA in Maternal Plasma Has Changed Prenatal Diagnosis for Monogenic Disorders. Clin. Sci. 2022, 136, 1615–1629. [Google Scholar] [CrossRef]
- Hill, M.; Twiss, P.; Verhoef, T.I.; Drury, S.; McKay, F.; Mason, S.; Jenkins, L.; Morris, S.; Chitty, L.S. Non-Invasive Prenatal Diagnosis for Cystic Fibrosis: Detection of Paternal Mutations, Exploration of Patient Preferences and Cost Analysis. Prenat. Diagn. 2015, 35, 950–958. [Google Scholar] [CrossRef]
- Sims, D.; Sudbery, I.; Ilott, N.E.; Heger, A.; Ponting, C.P. Sequencing Depth and Coverage: Key Considerations in Genomic Analyses. Nat. Rev. Genet. 2014, 15, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Kotsopoulou, I.; Tsoplou, P.; Mavrommatis, K.; Kroupis, C. Non-Invasive Prenatal Testing (NIPT): Limitations on the Way to Become Diagnosis. Diagnosis 2015, 2, 141–158. [Google Scholar] [CrossRef] [PubMed]
| ID | Gesta-tional Week | FF | Indic-ation for NIPS | NIPS Results | Vera Revolution Panel Results | Confirmatory Test | Disease | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Biological Sample | NGS Results | Karyotype | Array Results | ACMG Classification | Maternal DNA | Paternal DNA | |||||||
| 1 | 10 + 4 | 11 | MA | WT,XY | GJB2 NM_004004.6: c.35del p.(Gly12ValfsTer2) | AF | GJB2 NM_004004.6: c.35del p.(Gly12ValfsTer2), c.35del p.(Gly12ValfsTer2) | 46,XY | ND | P (PVS1, PP5, PM2) | GJB2 NM_004004.6: c.35del p.(Gly12ValfsTer2) | GJB2 NM_004004.6: c.35del p.(Gly12ValfsTer2) | Deafness, autosomal recessive 1A |
| 2 | 11 + 5 | 7 | MA | WT,XY | WT | AF | WT | 46,XY | arr(X,Y)x1,(1-22)x2 | NA | WT | WT | NA |
| 3 | 15 + 2 | 11 | AMA | dup(8)(p22q22.1), XY | WT | AF | WT | 46, XY | arr(X,Y)x1,(1-22)x2, negative for UPD | NA | WT | WT | NA |
| 4 | 14 | 10 | MA | WT,XX | g.16(222,899_223,019dup);g.16(223,093_223, 625dup) | BS | WT | 46,XX | arr[GRCh37] 16p13.3 (105,320-507,904)x3 pat 410kb; arr[GRCh37] 22q11.21-q11.22 (21,806,401-22,903,788)x1 dn 1.1 Mb | VUS/P | arr(X,1-22)x2 | arr[GRCh37] 16p13.3 (105,320-507,904)x3 | NA |
| 5 | 13 + 6 | 9 | MA | WT,XY | CFTR c.1521_1523del, p.Phe508del | AF | CFTR c.1521_1523del, p.Phe508del, c.1521_1523del, p.Phe508del | 46,XY | ND | P (PS3, PM1, PM4, PP5) | c.1521_1523del, p.Phe508del | c.1521_1523del, p.Phe508del | Cystc fibrosis |
| 6 | 12 | 8 | AMA | WT, XY | g.22(18,222,123_18,222,243del);g.22(18,222,8 5_18,222,955del) | AF | NA | 46,XY | arr[GRCh37] 22q11.21(18,877,787-21,462,353)x1 dn, 2.6 Mb | P | arr(X,1-22)x2 | WT | DiGeorge syndrome |
| 7 | 10 + 3 | 11 | Previous pregnancy with IUGR | dup(4)(p16.3p12), XX | g.4(499476_499744del);g.4(500494_500614del) | AF | WT | arr[GRCh37] 4p16.3 (48,283- 1,243,573)x1 dn, 1.2Mb | P | arr(X,1-22)x2 | arr(X,Y)x1,(1-22)x2 | Wolf-Hirschhorn syndrome | |
| 8 | 12 + 6 | 14 | MA | border trisomy 21, XY | WT | AF | NA | 46,XX | arr(X,1-22)x2 | NA | arr(X,1-22)x2 | arr(X,Y)x1,(1-22)x2 | |
| 9 | 11 + 2 | 10 | Congenital heart disease, suspicion of DiGeorge Syndrome | WT,XX | WT | AF | WT | 46,XX | arr(1-22,X)x2, negative for UPD | NA | arr(X,1-22)x2 | arr(X,Y)x1,(1-22)x2 | NA |
| 10 | 12 + 5 | 5 | MA | dup(5)(p15.31p15.2), XX | g.5(6,602,533_6,602,653dup); g.5(6,604,260_6,60 | AF | WT | 46,XX,dup(5)(p15.32p15.1) | arr[GRCh37] 5p15.32 - p15.1(5.320.850-15.346.106)x3 dn 10Mb | P | arr(X,1-22)x2 | arr(X,Y)x1,(1-22)x2 | 5p duplication syndrome |
| 11 | 10 + 3 | 10 | MA | WT,XY | GJB2 NM_004004.6: c.35del p.(Gly12ValfsTer2) | AF | GJB2 NM_004004.6: c.35del p.(Gly12ValfsTer2) | NA | NA | P (PVS1, PP5, PM2) | WT | GJB2 NM_004004.6: c.35del p.(Gly12ValfsTer2) | Deafness, autosomal recessive 1A |
| 12 | 13 + 5 | 9 | cystic igroma | WT,XY | RIT1 NM_006912.6: c.229G>T (p.Ala77Ser) | AF | RIT1 NM_006912.6: c.229G>T (p.Ala77Ser) | 46,XY | arr(X,Y)x1,(1-22)x2 | P (PM5, PP5, PM1, PP3, PM2) | WT | WT | Noonan Symdrome |
| 13 | 11 + 1 | 6 | AMA | dup(22)(q11.21q12.1), XY | WT | AF | WT | 46,XY | arr(X,Y)x1,(1-22)x2 | arr(X,1-22)x2 | arr(X,Y)x1,(1-22)x2 | NA | |
| 14 | 12 | 6 | MA | WT,XX | PAH NM_000277.3: c.533A>G (p.Glu178Gly), c.1208C>T (p.Ala403Val) | AF | PAH NM_000277.3: c.533A>G (p.Glu178Gly), c.1208C>T (p.Ala403Val) | 46,XX | arr(X,1-22)x2 | P (PP5, PM1, PM5, PP3, PM2)/P (PP5, PM1, PS3, BP4) | PAH NM_000277.1: c.533A>G (p.Glu178Gly) | PAH NM_000277.1: c.1208C>T (p.Ala403Val) | Phenylchetonuria |
| 15 | 11 | 14 | Increased NT, previous pregnancy with 15q11.2 deletion | WT,XX | WT | AF | WT | 46,XX | arr(X,1-22)x2 | NA | arr(X,1-22)x2 | arr(X,Y)x1,(1-22)x2 | NA |
| 16 | 13 + 1 | 7 | Increased NT | WT, XX | WT | AF | WT | 46,XX | arr(X,1-22)x2 | NA | arr(X,1-22)x2 | arr(X,Y)x1,(1-22)x2 | |
| 17 | 12 + 5 | 17 | Increased NT | WT,XX | DHCR7 NM_001360.2 c.964-3C>G | CVS | DHCR7 NM_001360.2 c.964-3C>G | 46,XX | arr(X,1-22)x2 | VUS (PP3, PM2) | WT | DHCR7 NM_001360.2 c.964-3C>G | Smith-Lemli-Opitz Syndrome |
| 18 | 12 + 2 | 14 | Increased NT | WT,XX | WT | AF | NA | 46,XX | arr(X,1-22)x2 | NA | arr(X,1-22)x2 | arr(X,Y)x1,(1-22)x2 | NA |
| 19 | 11 | 11 | MA | Border trisomy 18, XX | WT | AF | NA | 46,XX | arr(X,1-22)x2 | NA | arr(X,1-22)x2 | arr(X,Y)x1,(1-22)x2 | NA |
| 20 | 12 | 5 | Increased NT | WT | WT | AF | NA | NA | NA | ||||
| 21 | 12 + 3 | 7 | MA | del(13)(q21.2q31.2), XX | WT | AF | NA | 46,XX,del(13)(q21.2q31.2) | arr[GRCh37] 13q21.2 - q31.1(60.932.642-81.657.010)x1 dn | NA | arr(X,1-22)x2 | arr(X,Y)x1,(1-22)x2 | NA |
| 22 | 12 + 4 | 7 | MA | WT,XY | GJB2 NM_004004.6: c.109G>A p.(Val37Ile); c.583A>G, p.(Met195Val) | AF | NA | 46,XY | GJB2 NM_004004.6: c.109G>A p.(Val37Ile); c.583A>G, p.(Met195Val) | P (PM1, PM5, PP3, PP5, PM2)/P (PM5, PM1, PP3, PM2, PP5) | GJB2 NM_004004.6: c.109G>A p.(Val37Ile) | GJB2 NM_004004.6: c.583A>G, p.(Met195Val) | Deafness, autosomal recessive 1A |
| 23 | 12 | 5 | PMA | trisomy 3, XX | WT | AF | NA | 46,XX | arr(X,1-22)x2 | NA | arr(X,1-22)x2 | arr(X,Y)x1,(1-22)x2 | NA |
| 24 | 9 | 10 | WT | WT | WT | BS | NA | 46,XY | arr(X,Y)x1,(1-22)x2 | NA | arr(X,1-22)x2 | arr(X,Y)x1,(1-22)x2 | NA |
| 25 | 14 + 5 | 10 | NA | WT | TCOF1 NM_001371623.1:c.1999dup, (p.Arg667fs) | AF | TCOF1 NM_001371623.1: c.1999dup, (p.Arg667fs) | 46,XY | arr(X,Y)x1,(1-22)x2 | P (PVS1, PM2, PP5) | arr(X,1-22)x2 | arr(X,Y)x1,(1-22)x2 | Treacher Collins |
| 26 | 10 + 3 | 8 | AMA | WT,XY | PTPN11 NM_002834.5 c.1471C>T p.(Pro491Ser) | AF | PTPN11 NM_001330437.2 c.1483C>T p.(Pro495Ser) | 46,XY | arr(X,Y)x1,(1-22)x2 | P (PP5, PM5, PM1, PP3, PM2) | WT | WT | RASopathy |
| 27 | 12 + 3 | 5 | MA | WT,XY | MCCC2 NM_022132.5 c.1015G>A, p.(Val339Met) | AF | MCCC2 NM_022132.5 c.1015G>A, p.(Val339Met) | 46,XY | arr(X,Y)x1,(1-22)x2 | P (PP5, PM1) | MCCC2 NM_022132.5 c.1015G>A, p.(Val339Met) | MCCC2 NM_022132.5 c.1015G>A, p.(Val339Met) | 3-Methylcrotonyl-CoA carboxylase 2 deficiency |
| 28 | 11 + 1 | 12 | MA | WT,XY | GJB2 NM_004004.6: c.35del p.(Gly12ValfsTer2) | BS | GJB2 NM_004004.6: c.35del p.(Gly12ValfsTer2) | 46,XY | arr(X,Y)x1,(1-22)x2 | p (PVS1, PP5, PM2) | WT | GJB2 NM_004004.6: c.35del p.(Gly12ValfsTer2) | Deafness, autosomal recessive 1A |
| 29 | 13 + 3 | 20 | MA | WT,XY | GBA1 NM_001005741.3: c.1226A>G (p.Asn409Ser) | BS | GBA1 NM_001005741.3: c.1226A>G (p.Asn409Ser) | 46,XY | arr(X,Y)x1,(1-22)x2 | P (PS3, PM1, BP4) | GBA1 NM_001005741.3: c.1226A>G (p.Asn409Ser) | WT | Gaucher disease type I (GD1) |
| 30 | 12 + 2 | 6 | WT,XY | PTPN11 NM_002834.5 c.5C>T, p.(Thr2Ile) | AF | PTPN11 NM_002834.5 c.5C>T, p.(Thr2Ile) | 46,XY | arr(X,Y)x1,(1-22)x2 | P (PP5, PM2, PP2, BP3) | WT | WT | RASopathy | |
| 31 | 12 + 6 | 11 | AMA | dup(2)(q31.1q32.1), XX | ABCA4 NM_000350.3 c.286A>G, p.(Asn96Asp), GBA1 NM_001005741.3 c.1226A>G, p.(Asn409Ser) | AF | ABCA4 NM_000350.3 c.286A>G, p.(Asn96Asp), GBA1 NM_001005741.3 c.1226A>G, p.(Asn409Ser) | 46,XX | arr[GRCh37] 2q31.1 - q32.1 (173.278.777-188.970.013)x3 dn, 15.7 Mb | P/PL (PM5, PP5, PM1, PP3, PM2), P/PL (PS3, PM1, BP4) | WT | ABCA4 NM_000350.3 c.286A>G, p.(Asn96Asp), GBA1 NM_001005741.3 c.1226A>G, p.(Asn409Ser) | Stargardt disease 1, Gaucher disease type I (GD1) |
| 32 | 11 + 3 | 12 | AMA | WT,XY | PAH NM_000277.3, c.143T>C p.(Leu48Ser) | BS | PAH NM_000277.3, c.143T>C p.(Leu48Ser) | 46,XY | arr(X,Y)x1,(1-22)x2 | P (PP5, PP3, PM1, PS3, PM2) | WT | PAH NM_000277.1, c.143T>C p.(Leu48Ser) | Phenylchetonuria |
| 33 | 10 + 4 | 9 | AMA | WT,XY | PAH NM_000277.1, c.688G>A p.(Val230Ile) | BS | PAH NM_000277.1, c.688G>A p.(Val230Ile) | 46,XY | arr(X,Y)x1,(1-22)x2 | LP (PP5, PM1, PM5, PS3, PM2, BP4 | PAH NM_000277.1, c.688G>A p.(Val230Ile) | WT | Phenylchetonuria |
| 34 | 11 + 4 | 15 | AMA | WT,XY | PTPN11 NM_001330437.2:c.1504C>T, p.(Arg502Trp) | AF | PTPN11 NM_001330437.2:c.1504C>T, p.Arg502Trp | 46,XY | arr(X,Y)x1,(1-22)x2 | P (PM1, PM5, PP3, PM2) | WT | WT | RASopathy |
| 35 | 12 + 0 | 11 | MA | WT,XX | MYBPC3 NM_000256.3 c.927-9G>A | AF | MYBPC3 NM_000256.3 c.927-9G>A | 46,XX | arr(X,1-22)x2 | P (PP5, PP3, PM2) | WT | MYBPC3 NM_000256.3 c.927-9G>A | Hypertrophic cardiomyopathy |
| 36 | 12 + 0 | 18 | MA | WT,XX | GBA1 NM_001005741.3 c.1448T>C, p.(Leu483Pro) | AF | GBA1 NM_001005741.3 c.1448T>C, p.(Leu483Pro) | 46,XX | arr(X,1-22)x2 | P (PP3. PP5, PM1, PM5, PM2) | GBA1 NM_001005741.3 c.1448T>C, p.(Leu483Pro) | GBA1 NM_001005741.3 c.1448T>C, p.(Leu483Pro) | Gaucher disease type I (GD1) |
| Father-Inherited | De Novo | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ID | Total Variants | Common Variants | Uncommon Variants | % of Common | Coverage of Uncommon | Total Variants | Common Variants | Uncommon Variants | % of Common | Coverage of Uncommon |
| 1 | 3448 | 1633 | 1815 | 47% | 3.8 | 6754 | 2446 | 4308 | 36% | 2.2 |
| 2 | 559 | 487 | 72 | 87% | 10.6 | 309 | 309 | 0 | 100% | 19.3 |
| 3 | 460 | 444 | 16 | 96% | 5.6 | 272 | 272 | 0 | 100% | 15.8 |
| 4 | 2682 | 1583 | 1099 | 59% | 4.0 | 4038 | 1613 | 2425 | 40% | 1.6 |
| 5 | 3652 | 1920 | 1732 | 52% | 4.2 | 6139 | 2018 | 4121 | 33% | 1.6 |
| 6 | 2700 | 1256 | 1444 | 46% | 3.2 | 4656 | 1587 | 3069 | 34% | 1.7 |
| 7 | 1921 | 1379 | 542 | 72% | 3.1 | 1838 | 1118 | 720 | 61% | 1.4 |
| 8 | 530 | 525 | 5 | 99% | 11.5 | 343 | 341 | 2 | 99% | 7.0 |
| 9 | 448 | 438 | 10 | 98% | 15.0 | 320 | 320 | 0 | 100% | 12.8 |
| 10 | 164 | 89 | 75 | 54% | 3.0 | 680 | 476 | 204 | 70% | 3.3 |
| 11 | 500 | 483 | 17 | 96% | 5.7 | 204 | 204 | 0 | 100% | 8.8 |
| 12 | 468 | 465 | 3 | 99% | 8.4 | 301 | 258 | 43 | 86% | 6.9 |
| 13 | 464 | 402 | 62 | 87% | 5.6 | 288 | 288 | 0 | 100% | 4.5 |
| 14 | 2720 | 1605 | 1115 | 59% | 3.5 | 4383 | 1535 | 2848 | 35% | 1.4 |
| 15 | 426 | 233 | 193 | 55% | 1.5 | 1832 | 231 | 1601 | 13% | 0.9 |
| 16 | 458 | 441 | 17 | 96% | 15.4 | 405 | 287 | 118 | 71% | 3.3 |
| 17 | 498 | 491 | 7 | 99% | 9.2 | 290 | 290 | 0 | 100% | 7.2 |
| 18 | 478 | 478 | 0 | 100% | 21.0 | 304 | 304 | 0 | 100% | 3.7 |
| 19 | 490 | 488 | 2 | 99% | 18.4 | 258 | 258 | 0 | 100% | 24.4 |
| 20 | 410 | 336 | 74 | 82% | 5.7 | 3222 | 262 | 2960 | 8% | 0.8 |
| 21 | 470 | 271 | 199 | 56% | 2.2 | 221 | 147 | 74 | 67% | 2.5 |
| 22 | 6586 | 1958 | 4628 | 30% | 3.5 | 3214 | 2112 | 1102 | 66% | 1.5 |
| 23 | 478 | 249 | 229 | 52% | 2.7 | 315 | 240 | 75 | 76% | 3.7 |
| 24 | 1952 | 1511 | 441 | 77% | 5.2 | 1952 | 1511 | 441 | 77% | 1.9 |
| 25 | 481 | 453 | 28 | 94% | 12.3 | 318 | 318 | 0 | 100% | 9.9 |
| 26 | 610 | 535 | 75 | 88% | 9.8 | 355 | 350 | 5 | 99% | 17.1 |
| 27 | 2900 | 1700 | 1200 | 59% | 3.9 | 5000 | 1800 | 3200 | 36% | 2.0 |
| 28 | 430 | 380 | 50 | 88% | 6.5 | 260 | 255 | 5 | 98% | 5.2 |
| 29 | 580 | 565 | 15 | 97% | 13.5 | 330 | 330 | 0 | 100% | 11.0 |
| 30 | 2500 | 1150 | 1350 | 46% | 3.0 | 4500 | 1500 | 3000 | 33% | 1.8 |
| 31 | 1800 | 1300 | 500 | 72% | 3.3 | 1700 | 1050 | 650 | 62% | 1.3 |
| 32 | 495 | 490 | 5 | 99% | 10.0 | 310 | 308 | 2 | 99% | 6.8 |
| 33 | 450 | 440 | 10 | 98% | 16.0 | 300 | 300 | 0 | 100% | 13.0 |
| 34 | 150 | 80 | 70 | 53% | 3.1 | 700 | 490 | 210 | 70% | 3.5 |
| 35 | 510 | 490 | 20 | 96% | 7.5 | 210 | 210 | 0 | 100% | 9.0 |
| 36 | 470 | 460 | 10 | 98% | 10.0 | 295 | 250 | 45 | 85% | 7.1 |
| Father-Inherited | De Novo | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ID | Total Variants | Common Variants | Uncommon Variants | % of Common | Coverage of Uncommon | Total Variants | Common Variants | Uncommon Variants | % of Common | Coverage of Uncommon |
| 1 | 3448 | 1633 | 1815 | 47% | 3.8 | 6754 | 2446 | 4308 | 36% | 2.2 |
| 2 | 559 | 487 | 72 | 87% | 10.6 | 309 | 309 | 0 | 100% | 19.3 |
| 3 | 460 | 444 | 16 | 96% | 5.6 | 272 | 272 | 0 | 100% | 15.8 |
| 4 | 2682 | 1583 | 1099 | 59% | 4.0 | 4038 | 1613 | 2425 | 40% | 1.6 |
| 5 | 3652 | 1920 | 1732 | 52% | 4.2 | 6139 | 2018 | 4121 | 33% | 1.6 |
| 6 | 2700 | 1256 | 1444 | 46% | 3.2 | 4656 | 1587 | 3069 | 34% | 1.7 |
| 7 | 1921 | 1379 | 542 | 72% | 3.1 | 1838 | 1118 | 720 | 61% | 1.4 |
| 8 | 530 | 525 | 5 | 99% | 11.5 | 343 | 341 | 2 | 99% | 7.0 |
| 9 | 448 | 438 | 10 | 98% | 15.0 | 320 | 320 | 0 | 100% | 12.8 |
| 10 | 164 | 89 | 75 | 54% | 3.0 | 680 | 476 | 204 | 70% | 3.3 |
| 11 | 500 | 483 | 17 | 96% | 5.7 | 204 | 204 | 0 | 100% | 8.8 |
| 12 | 468 | 465 | 3 | 99% | 8.4 | 301 | 258 | 43 | 86% | 6.9 |
| 13 | 464 | 402 | 62 | 87% | 5.6 | 288 | 288 | 0 | 100% | 4.5 |
| 14 | 2720 | 1605 | 1115 | 59% | 3.5 | 4383 | 1535 | 2848 | 35% | 1.4 |
| 15 | 426 | 233 | 193 | 55% | 1.5 | 1832 | 231 | 1601 | 13% | 0.9 |
| 16 | 458 | 441 | 17 | 96% | 15.4 | 405 | 287 | 118 | 71% | 3.3 |
| 17 | 498 | 491 | 7 | 99% | 9.2 | 290 | 290 | 0 | 100% | 7.2 |
| 18 | 478 | 478 | 0 | 100% | 21.0 | 304 | 304 | 0 | 100% | 3.7 |
| 19 | 490 | 488 | 2 | 99% | 18.4 | 258 | 258 | 0 | 100% | 24.4 |
| 20 | 410 | 336 | 74 | 82% | 5.7 | 3222 | 262 | 2960 | 8% | 0.8 |
| 21 | 470 | 271 | 199 | 56% | 2.2 | 221 | 147 | 74 | 67% | 2.5 |
| 22 | 6586 | 1958 | 4628 | 30% | 3.5 | 3214 | 2112 | 1102 | 66% | 1.5 |
| 23 | 478 | 249 | 229 | 52% | 2.7 | 315 | 240 | 75 | 76% | 3.7 |
| 24 | 1952 | 1511 | 441 | 77% | 5.2 | 1952 | 1511 | 441 | 77% | 1.9 |
| 25 | 481 | 453 | 28 | 94% | 12.3 | 318 | 318 | 0 | 100% | 9.9 |
| 26 | 610 | 535 | 75 | 88% | 9.8 | 355 | 350 | 5 | 99% | 17.1 |
| 27 | 2900 | 1700 | 1200 | 59% | 3.9 | 5000 | 1800 | 3200 | 36% | 2.0 |
| 28 | 430 | 380 | 50 | 88% | 6.5 | 260 | 255 | 5 | 98% | 5.2 |
| 29 | 580 | 565 | 15 | 97% | 13.5 | 330 | 330 | 0 | 100% | 11.0 |
| 30 | 2500 | 1150 | 1350 | 46% | 3.0 | 4500 | 1500 | 3000 | 33% | 1.8 |
| 31 | 1800 | 1300 | 500 | 72% | 3.3 | 1700 | 1050 | 650 | 62% | 1.3 |
| 32 | 495 | 490 | 5 | 99% | 10.0 | 310 | 308 | 2 | 99% | 6.8 |
| 33 | 450 | 440 | 10 | 98% | 16.0 | 300 | 300 | 0 | 100% | 13.0 |
| 34 | 150 | 80 | 70 | 53% | 3.1 | 700 | 490 | 210 | 70% | 3.5 |
| 35 | 510 | 490 | 20 | 96% | 7.5 | 210 | 210 | 0 | 100% | 9.0 |
| 36 | 470 | 460 | 10 | 98% | 10.0 | 295 | 250 | 45 | 85% | 7.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sirica, R.; Ottaiano, A.; D’Amore, L.; Ianniello, M.; Petrillo, N.; Ruggiero, R.; Castiello, R.; Mori, A.; Evangelista, E.; De Falco, L.; et al. Advancing Non-Invasive Prenatal Screening: A Targeted 1069-Gene Panel for Comprehensive Detection of Monogenic Disorders and Copy Number Variations. Genes 2025, 16, 427. https://doi.org/10.3390/genes16040427
Sirica R, Ottaiano A, D’Amore L, Ianniello M, Petrillo N, Ruggiero R, Castiello R, Mori A, Evangelista E, De Falco L, et al. Advancing Non-Invasive Prenatal Screening: A Targeted 1069-Gene Panel for Comprehensive Detection of Monogenic Disorders and Copy Number Variations. Genes. 2025; 16(4):427. https://doi.org/10.3390/genes16040427
Chicago/Turabian StyleSirica, Roberto, Alessandro Ottaiano, Luigi D’Amore, Monica Ianniello, Nadia Petrillo, Raffaella Ruggiero, Rosa Castiello, Alessio Mori, Eloisa Evangelista, Luigia De Falco, and et al. 2025. "Advancing Non-Invasive Prenatal Screening: A Targeted 1069-Gene Panel for Comprehensive Detection of Monogenic Disorders and Copy Number Variations" Genes 16, no. 4: 427. https://doi.org/10.3390/genes16040427
APA StyleSirica, R., Ottaiano, A., D’Amore, L., Ianniello, M., Petrillo, N., Ruggiero, R., Castiello, R., Mori, A., Evangelista, E., De Falco, L., Santorsola, M., Misasi, M., Savarese, G., & Fico, A. (2025). Advancing Non-Invasive Prenatal Screening: A Targeted 1069-Gene Panel for Comprehensive Detection of Monogenic Disorders and Copy Number Variations. Genes, 16(4), 427. https://doi.org/10.3390/genes16040427






