Whole-Genome Resequencing in Sheep: Applications in Breeding, Evolution, and Conservation
Abstract
1. Introduction
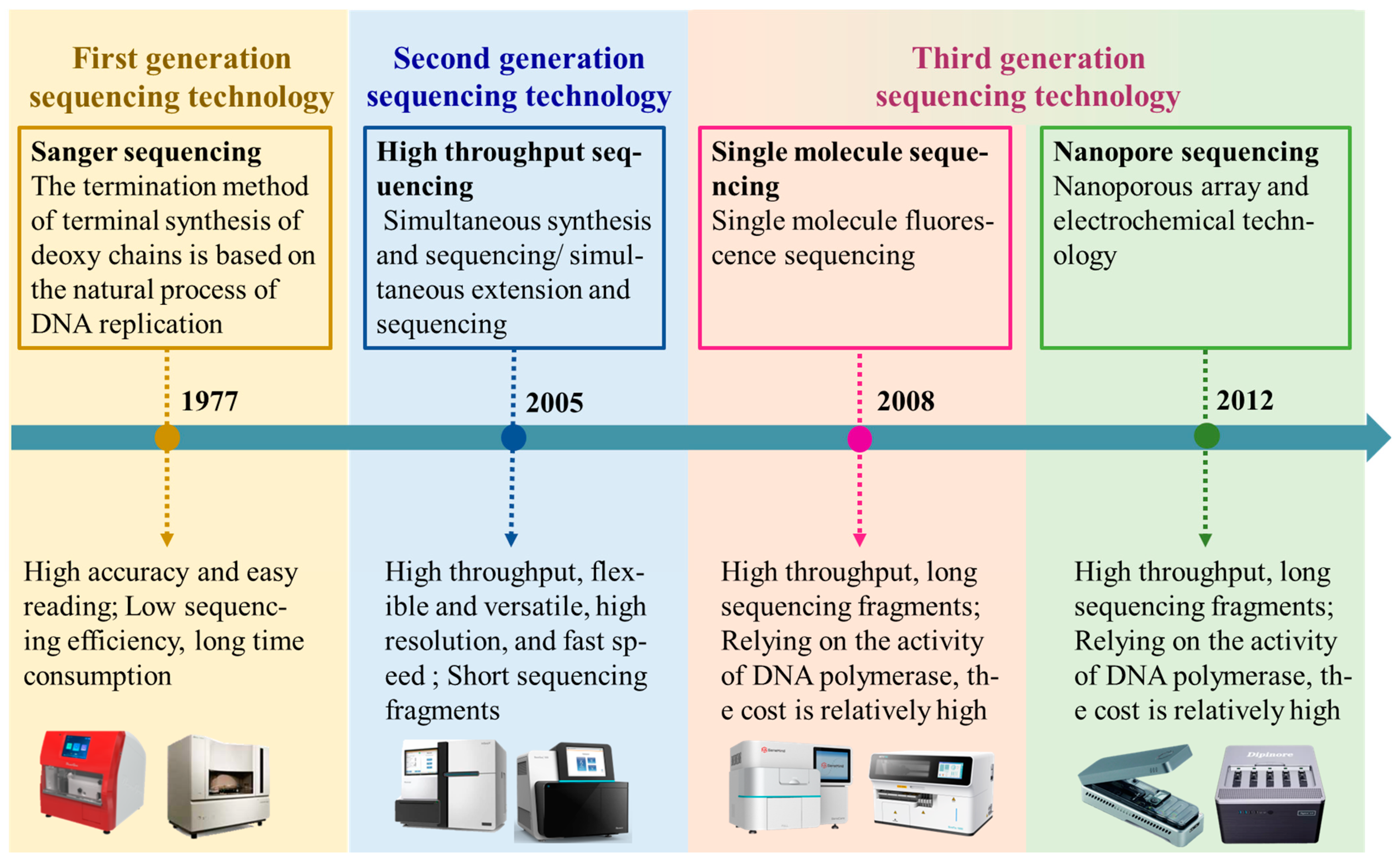
2. Whole-Genome Resequencing
3. Origin and Evolution of Sheep
4. Construction of Sheep Genetic Maps
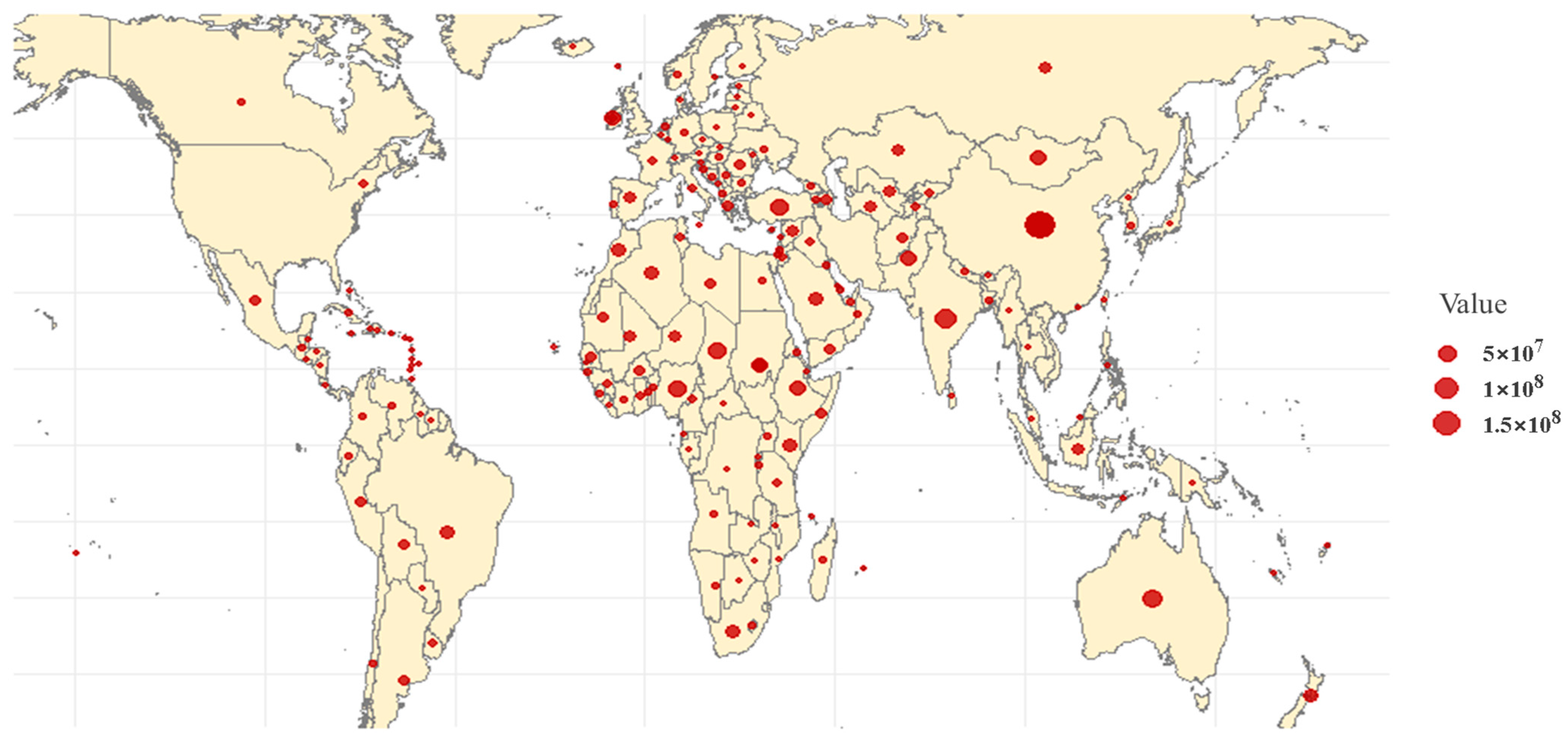
5. Genetic Structure Differentiation in Sheep Populations
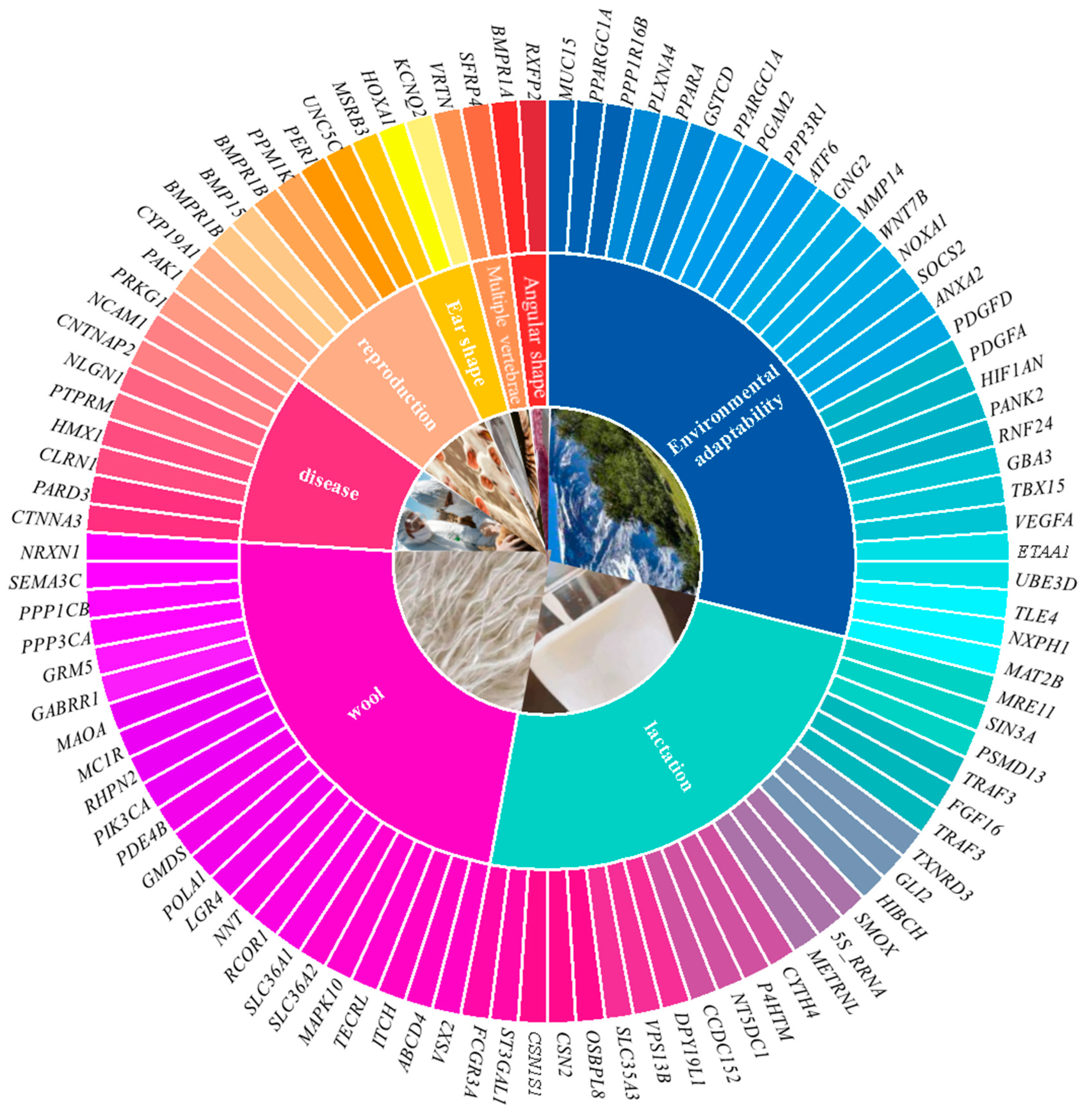
6. Sheep Trait Association Analysis
6.1. Wool Fineness and Color
6.2. Lactation Traits
6.3. Reproductive Traits
6.4. Multi-Vertebrae Features
6.5. Ear Shape and Horn Shape
6.6. Disease Resistance Traits
6.7. Environmental Adaptability Traits
7. Development, Utilization, and Conservation of Genetic Resource Diversity
8. Summary and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Larsson, M.N.A.; Morell Miranda, P.; Pan, L.; Başak Vural, K.; Kaptan, D.; Rodrigues Soares, A.E.; Kivikero, H.; Kantanen, J.; Somel, M.; Özer, F.; et al. Ancient Sheep Genomes Reveal Four Millennia of North European Short-Tailed Sheep in the Baltic Sea Region. Genome Biol. Evol. 2024, 16, evae114. [Google Scholar] [CrossRef]
- Kaptan, D.; Atağ, G.; Vural, K.B.; Morell Miranda, P.; Akbaba, A.; Yüncü, E.; Buluktaev, A.; Abazari, M.F.; Yorulmaz, S.; Kazancı, D.D.; et al. The Population History of Domestic Sheep Revealed by Paleogenomes. Mol. Biol. Evol. 2024, 41, msae158. [Google Scholar] [CrossRef]
- Saadatabadi, L.M.; Mohammadabadi, M.; Ghanatsaman, Z.A.; Babenko, O.; Stavetska, R.V.; Kalashnik, O.M.; Afanasenko, V.; Kochuk-Yashchenko, O.A.; Kucher, D.M.; Nanaei, H.A. Data of Whole-Genome Sequencing of Karakul, Zel, and Kermani Sheep Breeds. BMC Res. Notes 2023, 16, 353. [Google Scholar] [CrossRef]
- Zhang, D.; Cheng, J.; Li, X.; Huang, K.; Yuan, L.; Zhao, Y.; Xu, D.; Zhang, Y.; Zhao, L.; Yang, X.; et al. Comprehensive Multi-tissue Epigenome Atlas in Sheep: A Resource for Complex Traits, Domestication, and Breeding. Imeta 2024, 3, e254. [Google Scholar] [CrossRef] [PubMed]
- Aydin, K.B.; Bi, Y.; Brito, L.F.; Ulutaş, Z.; Morota, G. Review of Sheep Breeding and Genetic Research in Türkiye. Front. Genet. 2024, 15, 1308113. [Google Scholar] [CrossRef] [PubMed]
- Notter, D.R. Genetic Improvement of Reproductive Efficiency of Sheep and Goats. Anim. Reprod. Sci. 2012, 130, 147–151. [Google Scholar] [CrossRef]
- Bai, L.; Zhou, H.; Li, W.; Tao, J.; Hickford, J.G.H. Exploring Variation in Ovine KRTAP19-5 and Its Effect on Fine Wool Fibre Curvature in Chinese Tan Sheep. Animals 2024, 14, 2155. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, N.; Su, Y.; Long, Q.; Peng, Y.; Shangguan, L.; Zhang, F.; Cao, S.; Wang, X.; Ge, M.; et al. Grapevine Pangenome Facilitates Trait Genetics and Genomic Breeding. Nat. Genet. 2024, 56, 2804–2814. [Google Scholar] [CrossRef]
- Rubin, C.-J.; Zody, M.C.; Eriksson, J.; Meadows, J.R.S.; Sherwood, E.; Webster, M.T.; Jiang, L.; Ingman, M.; Sharpe, T.; Ka, S.; et al. Whole-Genome Resequencing Reveals Loci under Selection during Chicken Domestication. Nature 2010, 464, 587–591. [Google Scholar] [CrossRef]
- Zhao, X.; Zheng, T.; Gao, T.; Song, N. Whole-Genome Resequencing Reveals Genetic Diversity and Selection Signals in Warm Temperate and Subtropical Sillago Sinica Populations. BMC Genom. 2023, 24, 547. [Google Scholar] [CrossRef]
- Xu, L.; Zhou, K.; Huang, X.; Chen, H.; Dong, H.; Chen, Q. Whole-Genome Resequencing Provides Insights into the Diversity and Adaptation to Desert Environment in Xinjiang Mongolian Cattle. BMC Genom. 2024, 25, 176. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, H.; Luo, Y.; Li, J.; Sun, A.; Ahmed, Z.; Zhang, B.; Lei, C.; Yi, K. Comprehensive Whole-Genome Resequencing Unveils Genetic Diversity and Selective Signatures of the Xiangdong Black Goat. Front. Genet. 2024, 15, 1326828. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, S.; Terao, C. Comparative Evaluation of SNVs, Indels, and Structural Variations Detected with Short- and Long-Read Sequencing Data. Hum. Genome Var. 2024, 11, 18. [Google Scholar] [CrossRef] [PubMed]
- Saada, B.; Zhang, T.; Siga, E.; Zhang, J.; Magalhães Muniz, M.M. Whole-Genome Alignment: Methods, Challenges, and Future Directions. Appl. Sci. 2024, 14, 4837. [Google Scholar] [CrossRef]
- Dorado, G.; Gálvez, S.; Rosales, T.E.; Vásquez, V.F.; Hernández, P. Analyzing Modern Biomolecules: The Revolution of Nucleic-Acid Sequencing—Review. Biomolecules 2021, 11, 1111. [Google Scholar] [CrossRef]
- Hu, T.; Chitnis, N.; Monos, D.; Dinh, A. Next-Generation Sequencing Technologies: An Overview. Hum. Immunol. 2021, 82, 801–811. [Google Scholar] [CrossRef]
- Yao, Z.; Zhang, S.; Wang, X.; Guo, Y.; Xin, X.; Zhang, Z.; Xu, Z.; Wang, E.; Jiang, Y.; Huang, Y. Genetic Diversity and Signatures of Selection in BoHuai Goat Revealed by Whole-Genome Sequencing. BMC Genom. 2023, 24, 116. [Google Scholar] [CrossRef]
- Ahmed, Z.; Xiang, W.; Wang, F.; Nawaz, M.; Kuthu, Z.H.; Lei, C.; Xu, D. Whole-Genome Resequencing Deciphers Patterns of Genetic Diversity, Phylogeny, and Evolutionary Dynamics in Kashmir Cattle. Anim. Genet. 2024, 55, 511–526. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, Y.; Guo, Y.; Chen, Q.; Zhang, M.; Chen, H.; Geng, J.; Huang, X. Whole-Genome Resequencing Uncovers Diversity and Selective Sweep in Kazakh Cattle. Anim. Genet. 2024, 55, 377–386. [Google Scholar] [CrossRef]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of Age: Ten Years of next-Generation Sequencing Technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef]
- Xiong, X.; Liu, J.; Rao, Y. Whole Genome Resequencing Helps Study Important Traits in Chickens. Genes 2023, 14, 1198. [Google Scholar] [CrossRef] [PubMed]
- Dementieva, N.V.; Shcherbakov, Y.S.; Stanishevskaya, O.I.; Vakhrameev, A.B.; Larkina, T.A.; Dysin, A.P.; Nikolaeva, O.A.; Ryabova, A.E.; Azovtseva, A.I.; Mitrofanova, O.V.; et al. Large-Scale Genome-Wide SNP Analysis Reveals the Rugged (and Ragged) Landscape of Global Ancestry, Phylogeny, and Demographic History in Chicken Breeds. J. Zhejiang Univ. Sci. B 2024, 25, 324–340. [Google Scholar] [CrossRef]
- Zhang, L.; Li, H.; Zhao, X.; Wu, Y.; Li, J.; Yao, Y.; Yao, Y.; Wang, L. Whole Genome Resequencing Reveals the Adaptability of Native Chickens to Drought, Tropical and Frigid Environments in Xinjiang. Poult. Sci. 2024, 103, 103947. [Google Scholar] [CrossRef] [PubMed]
- Chong, Y.; Xiong, H.; Gao, Z.; Lu, Y.; Hong, J.; Wu, J.; He, X.; Xi, D.; Tu, X.; Deng, W. Genomic and Transcriptomic Landscape to Decipher the Genetic Basis of Hyperpigmentation in Lanping Black-Boned Sheep (Ovis Aries). BMC Genom. 2024, 25, 845. [Google Scholar] [CrossRef]
- Alberto, F.J.; Boyer, F.; Orozco-terWengel, P.; Streeter, I.; Servin, B.; de Villemereuil, P.; Benjelloun, B.; Librado, P.; Biscarini, F.; Colli, L.; et al. Convergent Genomic Signatures of Domestication in Sheep and Goats. Nat. Commun. 2018, 9, 813. [Google Scholar] [CrossRef]
- Ciani, E.; Mastrangelo, S.; Da Silva, A.; Marroni, F.; Ferenčaković, M.; Ajmone-Marsan, P.; Baird, H.; Barbato, M.; Colli, L.; Delvento, C.; et al. On the Origin of European Sheep as Revealed by the Diversity of the Balkan Breeds and by Optimizing Population-Genetic Analysis Tools. Genet. Sel. Evol. 2020, 52, 25. [Google Scholar] [CrossRef]
- Hu, X.-J.; Yang, J.; Xie, X.-L.; Lv, F.-H.; Cao, Y.-H.; Li, W.-R.; Liu, M.-J.; Wang, Y.-T.; Li, J.-Q.; Liu, Y.-G.; et al. The Genome Landscape of Tibetan Sheep Reveals Adaptive Introgression from Argali and the History of Early Human Settlements on the Qinghai-Tibetan Plateau. Mol. Biol. Evol. 2019, 36, 283–303. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.-X.; Yang, J.; Lv, F.-H.; Hu, X.-J.; Xie, X.-L.; Zhang, M.; Li, W.-R.; Liu, M.-J.; Wang, Y.-T.; Li, J.-Q.; et al. Genomic Reconstruction of the History of Native Sheep Reveals the Peopling Patterns of Nomads and the Expansion of Early Pastoralism in East Asia. Mol. Biol. Evol. 2017, 34, 2380–2395. [Google Scholar] [CrossRef]
- Lv, F.-H.; Peng, W.-F.; Yang, J.; Zhao, Y.-X.; Li, W.-R.; Liu, M.-J.; Ma, Y.-H.; Zhao, Q.-J.; Yang, G.-L.; Wang, F.; et al. Mitogenomic Meta-Analysis Identifies Two Phases of Migration in the History of Eastern Eurasian Sheep. Mol. Biol. Evol. 2015, 32, 2515–2533. [Google Scholar] [CrossRef]
- Deng, J.; Xie, X.L.; Wang, D.F.; Zhao, C.; Lv, F.H.; Li, X.; Yang, J.; Yu, J.L.; Shen, M.; Gao, L.; et al. Paternal Origins and Migratory Episodes of Domestic Sheep. Curr. Biol. 2020, 30, 4085–4095.e6. [Google Scholar] [CrossRef]
- Cao, Y.-H.; Xu, S.-S.; Shen, M.; Chen, Z.-H.; Gao, L.; Lv, F.-H.; Xie, X.-L.; Wang, X.-H.; Yang, H.; Liu, C.-B.; et al. Historical Introgression from Wild Relatives Enhanced Climatic Adaptation and Resistance to Pneumonia in Sheep. Mol. Biol. Evol. 2021, 38, 838–855. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.H.; Xu, Y.X.; Xie, X.L.; Wang, D.F.; Aguilar-Gómez, D.; Liu, G.J.; Li, X.; Esmailizadeh, A.; Rezaei, V.; Kantanen, J.; et al. Whole-Genome Sequence Analysis Unveils Different Origins of European and Asiatic Mouflon and Domestication-Related Genes in Sheep. Commun. Biol. 2021, 4, 1307. [Google Scholar] [CrossRef]
- Kandoussi, A.; Boujenane, I.; Auger, C.; Serranito, B.; Germot, A.; Piro, M.; Maftah, A.; Badaoui, B.; Petit, D. The Origin of Sheep Settlement in Western Mediterranean. Sci. Rep. 2020, 10, 10225. [Google Scholar] [CrossRef]
- Rezaei, H.R.; Naderi, S.; Chintauan-Marquier, I.C.; Jordan, S.; Taberlet, P.; Virk, A.T.; Naghash, H.R.; Rioux, D.; Kaboli, M.; Luikart, G.; et al. Evolution and Taxonomy of the Wild Species of the Genus Ovis (Mammalia, Artiodactyla, Bovidae). Mol. Phylogenet. Evol. 2010, 54, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Mereu, P.; Pirastru, M.; Sanna, D.; Bassu, G.; Naitana, S.; Leoni, G.G. Phenotype Transition from Wild Mouflon to Domestic Sheep. Genet. Sel. Evol. 2024, 56, 1. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Zhang, Z.; Wen, J.; Lenstra, J.A.; Heller, R.; Cai, Y.; Guo, Y.; Li, M.; Li, R.; Li, W.; et al. Long Divergent Haplotypes Introgressed from Wild Sheep Are Associated with Distinct Morphological and Adaptive Characteristics in Domestic Sheep. PLoS Genet. 2023, 19, e1010615. [Google Scholar] [CrossRef]
- Taylor, W.T.T.; Pruvost, M.; Posth, C.; Rendu, W.; Krajcarz, M.T.; Abdykanova, A.; Brancaleoni, G.; Spengler, R.; Hermes, T.; Schiavinato, S.; et al. Evidence for Early Dispersal of Domestic Sheep into Central Asia. Nat. Hum. Behav. 2021, 5, 1169–1179. [Google Scholar] [CrossRef]
- Zhang, D.-Y.; Zhang, X.-X.; Li, F.-D.; Yuan, L.-F.; Li, X.-L.; Zhang, Y.-K.; Zhao, Y.; Zhao, L.-M.; Wang, J.-H.; Xu, D.; et al. Whole-Genome Resequencing Reveals Molecular Imprints of Anthropogenic and Natural Selection in Wild and Domesticated Sheep. Zool. Res. 2022, 43, 695–705. [Google Scholar] [CrossRef]
- Her, C.; Rezaei, H.-R.; Hughes, S.; Naderi, S.; Duffraisse, M.; Mashkour, M.; Naghash, H.-R.; Bălășescu, A.; Luikart, G.; Jordan, S.; et al. Broad Maternal Geographic Origin of Domestic Sheep in Anatolia and the Zagros. Anim. Genet. 2022, 53, 13191. [Google Scholar] [CrossRef]
- Wientjes, Y.C.J.; Bijma, P.; Calus, M.P.L.; Zwaan, B.J.; Vitezica, Z.G.; van den Heuvel, J. The Long-Term Effects of Genomic Selection: 1. Response to Selection, Additive Genetic Variance, and Genetic Architecture. Genet. Sel. Evol. 2022, 54, 19. [Google Scholar] [CrossRef]
- Littrell, J.; Tsaih, S.-W.; Baud, A.; Rastas, P.; Solberg-Woods, L.; Flister, M.J. A High-Resolution Genetic Map for the Laboratory Rat. G3 (Bethesda) 2018, 8, 2241–2248. [Google Scholar] [CrossRef] [PubMed]
- Portnoy, D.S.; Renshaw, M.A.; Hollenbeck, C.M.; Gold, J.R. A Genetic Linkage Map of Red Drum, Sciaenops Ocellatus. Anim. Genet. 2010, 41, 630–641. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.-Y.; Wu, H.; Zhao, L.-M.; Zhang, Y.-H.; Huang, J.-H.; Liu, Q.-Y.; Wang, H.-T.; Mo, D.-X.; EEr, H.-H.; Zhang, L.-Q.; et al. Telomere-to-Telomere Sheep Genome Assembly Reveals New Variants Associated with Wool Fineness Trait. bioRxiv 2024, 2024.07.21.604451. [Google Scholar]
- Crawford, A.M.; Dodds, K.G.; Ede, A.J.; Pierson, C.A.; Montgomery, G.W.; Garmonsway, H.G.; Beattie, A.E.; Davies, K.; Maddox, J.F.; Kappes, S.W. An Autosomal Genetic Linkage Map of the Sheep Genome. Genetics 1995, 140, 703–724. [Google Scholar] [CrossRef]
- de Gortari, M.J.; Freking, B.A.; Cuthbertson, R.P.; Kappes, S.M.; Keele, J.W.; Stone, R.T.; Leymaster, K.A.; Dodds, K.G.; Crawford, A.M.; Beattie, C.W. A Second-Generation Linkage Map of the Sheep Genome. Mamm. Genome 1998, 9, 204–209. [Google Scholar] [CrossRef]
- Maddox, J.F.; Davies, K.P.; Crawford, A.M.; Hulme, D.J.; Vaiman, D.; Cribiu, E.P.; Freking, B.A.; Beh, K.J.; Cockett, N.E.; Kang, N.; et al. An Enhanced Linkage Map of the Sheep Genome Comprising More than 1000 Loci. Genome Res 2001, 11, 1275–1289. [Google Scholar] [CrossRef]
- Jiang, Y.; Xie, M.; Chen, W.; Talbot, R.; Maddox, J.F.; Faraut, T.; Wu, C.; Muzny, D.M.; Li, Y.; Zhang, W.; et al. The Sheep Genome Illuminates Biology of the Rumen and Lipid Metabolism. Science 2014, 344, 1168–1173. [Google Scholar] [CrossRef]
- Yuan, C.; Lu, Z.; Guo, T.; Yue, Y.; Wang, X.; Wang, T.; Zhang, Y.; Hou, F.; Niu, C.; Sun, X.; et al. A Global Analysis of CNVs in Chinese Indigenous Fine-Wool Sheep Populations Using Whole-Genome Resequencing. BMC Genom. 2021, 22, 78. [Google Scholar] [CrossRef]
- Davenport, K.M.; Bickhart, D.M.; Worley, K.; Murali, S.C.; Salavati, M.; Clark, E.L.; Cockett, N.E.; Heaton, M.P.; Smith, T.P.L.; Murdoch, B.M.; et al. An Improved Ovine Reference Genome Assembly to Facilitate In-Depth Functional Annotation of the Sheep Genome. Gigascience 2022, 11, giab096. [Google Scholar] [CrossRef]
- Luo, L.-Y.; Wu, H.; Zhao, L.-M.; Zhang, Y.-H.; Huang, J.-H.; Liu, Q.-Y.; Wang, H.-T.; Mo, D.-X.; EEr, H.-H.; Zhang, L.-Q.; et al. Telomere-to-Telomere Sheep Genome Assembly Identifies Variants Associated with Wool Fineness. Nat. Genet. 2025, 57, 218–230. [Google Scholar] [CrossRef]
- Lv, F.-H.; Cao, Y.-H.; Liu, G.-J.; Luo, L.-Y.; Lu, R.; Liu, M.-J.; Li, W.-R.; Zhou, P.; Wang, X.-H.; Shen, M.; et al. Whole-Genome Resequencing of Worldwide Wild and Domestic Sheep Elucidates Genetic Diversity, Introgression, and Agronomically Important Loci. Mol. Biol. Evol. 2022, 39, msab353. [Google Scholar] [CrossRef] [PubMed]
- Woolley, S.A.; Salavati, M.; Clark, E.L. Recent Advances in the Genomic Resources for Sheep. Mamm. Genome 2023, 34, 545–558. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Wang, J.; Li, J.; Chen, Z.; Wang, N.; Li, M.; Wang, L.; Si, Y.; Lu, S.; Cui, Z.; et al. Decoding the Fish Genome Opens a New Era in Important Trait Research and Molecular Breeding in China. Sci. China Life Sci. 2024, 67, 2064–2083. [Google Scholar] [CrossRef]
- Schierenbeck, K.A. Population-Level Genetic Variation and Climate Change in a Biodiversity Hotspot. Ann. Bot. 2017, 119, 215–228. [Google Scholar] [CrossRef]
- Bourgeois, Y.X.C.; Warren, B.H. An Overview of Current Population Genomics Methods for the Analysis of Whole-Genome Resequencing Data in Eukaryotes. Mol. Ecol. 2021, 30, 6036–6071. [Google Scholar] [CrossRef] [PubMed]
- Deniskova, T.; Dotsev, A.; Lushihina, E.; Shakhin, A.; Kunz, E.; Medugorac, I.; Reyer, H.; Wimmers, K.; Khayatzadeh, N.; Sölkner, J.; et al. Population Structure and Genetic Diversity of Sheep Breeds in the Kyrgyzstan. Front. Genet. 2019, 10, 1311. [Google Scholar] [CrossRef]
- Cheng, J.; Zhao, H.; Chen, N.; Cao, X.; Hanif, Q.; Pi, L.; Hu, L.; Chaogetu, B.; Huang, Y.; Lan, X.; et al. Population Structure, Genetic Diversity, and Selective Signature of Chaka Sheep Revealed by Whole Genome Sequencing. BMC Genom. 2020, 21, 520. [Google Scholar] [CrossRef]
- Wang, R.; Wang, X.; Qi, Y.; Li, Y.; Na, Q.; Yuan, H.; Rong, Y.; Ao, X.; Guo, F.; Zhang, L.; et al. Genetic Diversity Analysis of Inner Mongolia Cashmere Goats (Erlangshan Subtype) Based on Whole Genome Re-Sequencing. BMC Genom. 2024, 25, 698. [Google Scholar] [CrossRef]
- Liu, Z.; Tan, X.; Wang, J.; Jin, Q.; Meng, X.; Cai, Z.; Cui, X.; Wang, K. Whole Genome Sequencing of Luxi Black Head Sheep for Screening Selection Signatures Associated with Important Traits. Anim. Biosci. 2022, 35, 1340–1350. [Google Scholar] [CrossRef]
- Kerr, E.; Marr, M.M.; Collins, L.; Dubarry, K.; Salavati, M.; Scinto, A.; Woolley, S.; Clark, E.L. Analysis of Genotyping Data Reveals the Unique Genetic Diversity Represented by the Breeds of Sheep Native to the United Kingdom. BMC Genom. Data 2024, 25, 82. [Google Scholar] [CrossRef]
- Sun, X.; Guo, J.; Li, R.; Zhang, H.; Zhang, Y.; Liu, G.E.; Emu, Q.; Zhang, H. Whole-Genome Resequencing Reveals Genetic Diversity and Wool Trait-Related Genes in Liangshan Semi-Fine-Wool Sheep. Animals 2024, 14, 444. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Wang, G.; Chen, M.; Pang, Q.; Jiang, S.; Zeng, J.; Du, D.; Yang, P.; Wu, W.; Zhao, H. Genetic Diversity and Population Structure of Sheep (Ovis Aries) in Sichuan, China. PLoS ONE 2021, 16, e0257974. [Google Scholar] [CrossRef]
- Montossi, F.; Font-i-Furnols, M.; del Campo, M.; San Julián, R.; Brito, G.; Sañudo, C. Sustainable Sheep Production and Consumer Preference Trends: Compatibilities, Contradictions, and Unresolved Dilemmas. Meat Sci. 2013, 95, 772–789. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, Q.; Zhang, X.; Li, C.; Zhang, D.; Li, G.; Zhang, Y.; Zhao, Y.; Shi, Z.; Wang, W.; et al. Whole-Genome Resequencing to Study Brucellosis Susceptibility in Sheep. Front. Genet. 2021, 12, 653927. [Google Scholar] [CrossRef]
- Zhang, W.; Jin, M.; Li, T.; Lu, Z.; Wang, H.; Yuan, Z.; Wei, C. Whole-Genome Resequencing Reveals Selection Signal Related to Sheep Wool Fineness. Animals 2023, 13, 2944. [Google Scholar] [CrossRef]
- Zhao, H.; Guo, T.; Lu, Z.; Liu, J.; Zhu, S.; Qiao, G.; Han, M.; Yuan, C.; Wang, T.; Li, F.; et al. Genome-Wide Association Studies Detects Candidate Genes for Wool Traits by Re-Sequencing in Chinese Fine-Wool Sheep. BMC Genom. 2021, 22, 127. [Google Scholar] [CrossRef]
- Cao, C.; Kang, Y.; Zhou, Q.; Nanaei, H.A.; Bo, D.; Liu, P.; Bai, Y.; Li, R.; Jiang, Y.; Lan, X.; et al. Whole-Genome Resequencing Reveals the Genomic Diversity and Signatures of Selection in Romanov Sheep. J. Anim. Sci. 2023, 101, skad291. [Google Scholar] [CrossRef]
- Zhang, W.; Jin, M.; Lu, Z.; Li, T.; Wang, H.; Yuan, Z.; Wei, C. Whole Genome Resequencing Reveals Selection Signals Related to Wool Color in Sheep. Animals 2023, 13, 3265. [Google Scholar] [CrossRef]
- Akers, R.M. A 100-Year Review: Mammary Development and Lactation. J. Dairy Sci. 2017, 100, 10332–10352. [Google Scholar] [CrossRef]
- Staiger, E.A.; Thonney, M.L.; Buchanan, J.W.; Rogers, E.R.; Oltenacu, P.A.; Mateescu, R.G. Effect of Prolactin, Beta-Lactoglobulin, and Kappa-Casein Genotype on Milk Yield in East Friesian Sheep. J. Dairy Sci. 2010, 93, 1736–1742. [Google Scholar] [CrossRef]
- Li, R.; Zhao, Y.; Liang, B.; Pu, Y.; Jiang, L.; Ma, Y. Genome-Wide Signal Selection Analysis Revealing Genes Potentially Related to Sheep-Milk-Production Traits. Animals 2023, 13, 1654. [Google Scholar] [CrossRef] [PubMed]
- Rezvannejad, E.; Asadollahpour Nanaei, H.; Esmailizadeh, A. Detection of Candidate Genes Affecting Milk Production Traits in Sheep Using Whole-Genome Sequencing Analysis. Vet. Med. Sci. 2022, 8, 1197–1204. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yuan, L.; Wang, W.; Zhang, D.; Zhao, Y.; Chen, J.; Xu, D.; Zhao, L.; Li, F.; Zhang, X. Whole Genome Re-Sequencing Reveals Artificial and Natural Selection for Milk Traits in East Friesian Sheep. Front. Vet. Sci. 2022, 9, 1034211. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Wang, H.; Li, J.; Lv, D.; Xu, J.; Liu, M.; Yin, G. Sheep Litter Size Heredity Basis Using Genome-Wide Selective Analysis. Reprod. Domest. Anim. 2024, 59, e14689. [Google Scholar] [CrossRef]
- Krupová, Z.; Wolfová, M.; Krupa, E.; Oravcová, M.; Daňo, J.; Huba, J.; Polák, P. Impact of Production Strategies and Animal Performance on Economic Values of Dairy Sheep Traits. Animal 2012, 6, 440–448. [Google Scholar] [CrossRef]
- Smołucha, G.; Gurgul, A.; Jasielczuk, I.; Kawęcka, A.; Miksza-Cybulska, A. A Genome-Wide Association Study for Prolificacy in Three Polish Sheep Breeds. J. Appl. Genet. 2021, 62, 323–326. [Google Scholar] [CrossRef]
- Wang, Y.; Chi, Z.; Jia, S.; Zhao, S.; Cao, G.; Purev, C.; Cang, M.; Yu, H.; Li, X.; Bao, S.; et al. Effects of Novel Variants in BMP15 Gene on Litter Size in Mongolia and Ujimqin Sheep Breeds. Theriogenology 2023, 198, 1–11. [Google Scholar] [CrossRef]
- Zhu, M.; Yang, Y.; Yang, H.; Zhao, Z.; Zhang, H.; Blair, H.T.; Zheng, W.; Wang, M.; Fang, C.; Yu, Q.; et al. Whole-Genome Resequencing of the Native Sheep Provides Insights into the Microevolution and Identifies Genes Associated with Reproduction Traits. BMC Genom. 2023, 24, 392. [Google Scholar] [CrossRef]
- Bao, J.; Xiong, J.; Huang, J.; Yang, P.; Shang, M.; Zhang, L. Genetic Diversity, Selection Signatures, and Genome-Wide Association Study Identify Candidate Genes Related to Litter Size in Hu Sheep. Int. J. Mol. Sci. 2024, 25, 9397. [Google Scholar] [CrossRef]
- Abuzahra, M.; Wijayanti, D.; Effendi, M.H.; Mustofa, I.; Lamid, M. Association of Melatonin Receptor 1 A with Litter Size in Sheep: A Review. F1000Research 2023, 12, 900. [Google Scholar] [CrossRef]
- Chantepie, L.; Bodin, L.; Sarry, J.; Woloszyn, F.; Plisson-Petit, F.; Ruesche, J.; Drouilhet, L.; Fabre, S. Genome-Wide Identification of a Regulatory Mutation in BMP15 Controlling Prolificacy in Sheep. Front. Genet. 2020, 11, 585. [Google Scholar]
- Zhong, T.; Hou, D.; Zhao, Q.; Zhan, S.; Wang, L.; Li, L.; Zhang, H.; Zhao, W.; Yang, S.; Niu, L. Comparative Whole-Genome Resequencing to Uncover Selection Signatures Linked to Litter Size in Hu Sheep and Five Other Breeds. BMC Genom. 2024, 25, 480. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhang, X.; Li, F.; Liu, T.; Hu, Z.; Gao, N.; Yuan, L.; Li, X.; Zhao, Y.; Zhao, L.; et al. Whole-Genome Resequencing Identified Candidate Genes Associated with the Number of Ribs in Hu Sheep. Genomics 2021, 113, 2077–2084. [Google Scholar] [CrossRef]
- Li, C.; Liu, K.; Dai, J.; Li, X.; Liu, X.; Ni, W.; Li, H.; Wang, D.; Qiao, J.; Wang, Y.; et al. Whole-Genome Resequencing to Investigate the Determinants of the Multi-Lumbar Vertebrae Trait in Sheep. Gene 2022, 809, 146020. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, M.; Li, X.; Ni, W.; Xu, Y.; Yao, R.; Wei, B.; Zhang, M.; Li, H.; Zhao, Y.; et al. Whole-Genome Resequencing Reveals Loci Associated With Thoracic Vertebrae Number in Sheep. Front. Genet. 2019, 10, 674. [Google Scholar] [CrossRef]
- Donaldson, C.L.; Lambe, N.R.; Maltin, C.A.; Knott, S.; Bunger, L. Between- and within-Breed Variations of Spine Characteristics in Sheep. J. Anim. Sci. 2013, 91, 995–1004. [Google Scholar] [CrossRef]
- Paris, J.M.; Letko, A.; Häfliger, I.M.; Ammann, P.; Drögemüller, C. Ear Type in Sheep Is Associated with the MSRB3 Locus. Anim. Genet. 2020, 51, 968–972. [Google Scholar] [CrossRef]
- Zhao, F.; Xie, R.; Fang, L.; Xiang, R.; Yuan, Z.; Liu, Y.; Wang, L. Analysis of 206 Whole-Genome Resequencing Reveals Selection Signatures Associated with Breed-Specific Traits in Hu Sheep. Evol. Appl. 2024, 17, e13697. [Google Scholar] [CrossRef]
- Tian, D.; Han, B.; Li, X.; Liu, D.; Zhou, B.; Zhao, C.; Zhang, N.; Wang, L.; Pei, Q.; Zhao, K. Genetic Diversity and Selection of Tibetan Sheep Breeds Revealed by Whole-Genome Resequencing. Anim. Biosci. 2023, 36, 991–1002. [Google Scholar] [CrossRef]
- Zhang, G.; Chu, M.; Yang, H.; Li, H.; Shi, J.; Feng, P.; Wang, S.; Pan, Z. Expression, Polymorphism, and Potential Functional Sites of the BMPR1A Gene in the Sheep Horn. Genes 2024, 15, 376. [Google Scholar] [CrossRef]
- Djokic, V.; Freddi, L.; de Massis, F.; Lahti, E.; van den Esker, M.H.; Whatmore, A.; Haughey, A.; Ferreira, A.C.; Garofolo, G.; Melzer, F.; et al. The Emergence of Brucella Canis as a Public Health Threat in Europe: What We Know and What We Need to Learn. Emerg. Microbes Infect. 2023, 12, 2249126. [Google Scholar] [CrossRef] [PubMed]
- Alasti, F.; Van Camp, G. Genetics of Microtia and Associated Syndromes. J. Med. Genet. 2009, 46, 361–369. [Google Scholar] [CrossRef]
- Mastrangelo, S.; Sottile, G.; Sutera, A.M.; Di Gerlando, R.; Tolone, M.; Moscarelli, A.; Sardina, M.T.; Portolano, B. Genome-Wide Association Study Reveals the Locus Responsible for Microtia in Valle Del Belice Sheep Breed. Anim. Genet. 2018, 49, 636–640. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Zhang, Z.; Sun, Y.; Ren, T.; Li, W.; Zhou, X.; Michal, J.J.; Jiang, Z.; Liu, M. Genome-Wide Association Study Shows That Microtia in Altay Sheep Is Caused by a 76 Bp Duplication of HMX1. Anim. Genet. 2020, 51, 132–136. [Google Scholar] [CrossRef] [PubMed]
- Cross, A.R.; Baldwin, V.M.; Roy, S.; Essex-Lopresti, A.E.; Prior, J.L.; Harmer, N.J. Zoonoses under Our Noses. Microbes Infect. 2019, 21, 10–19. [Google Scholar] [CrossRef]
- Elsohaby, I.; Villa, L. Zoonotic Diseases: Understanding the Risks and Mitigating the Threats. BMC Vet. Res. 2023, 19, 186. [Google Scholar] [CrossRef]
- Shi, S.; Shao, D.; Yang, L.; Liang, Q.; Han, W.; Xue, Q.; Qu, L.; Leng, L.; Li, Y.; Zhao, X.; et al. Whole Genome Analyses Reveal Novel Genes Associated with Chicken Adaptation to Tropical and Frigid Environments. J. Adv. Res. 2023, 47, 13–25. [Google Scholar] [CrossRef]
- Yang, J.; Li, W.-R.; Lv, F.-H.; He, S.-G.; Tian, S.-L.; Peng, W.-F.; Sun, Y.-W.; Zhao, Y.-X.; Tu, X.-L.; Zhang, M.; et al. Whole-Genome Sequencing of Native Sheep Provides Insights into Rapid Adaptations to Extreme Environments. Mol. Biol. Evol. 2016, 33, 2576–2592. [Google Scholar] [CrossRef]
- Barreiro, L.B.; Laval, G.; Quach, H.; Patin, E.; Quintana-Murci, L. Natural Selection Has Driven Population Differentiation in Modern Humans. Nat. Genet. 2008, 40, 340–345. [Google Scholar] [CrossRef]
- Zhang, C.-L.; Zhang, J.; Tuersuntuoheti, M.; Zhou, W.; Han, Z.; Li, X.; Yang, R.; Zhang, L.; Zheng, L.; Liu, S. Landscape Genomics Reveals Adaptive Divergence of Indigenous Sheep in Different Ecological Environments of Xinjiang, China. Sci. Total Environ. 2023, 904, 166698. [Google Scholar] [CrossRef]
- Kim, E.-S.; Elbeltagy, A.R.; Aboul-Naga, A.M.; Rischkowsky, B.; Sayre, B.; Mwacharo, J.M.; Rothschild, M.F. Multiple Genomic Signatures of Selection in Goats and Sheep Indigenous to a Hot Arid Environment. Heredity 2016, 116, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Edea, Z.; Dadi, H.; Dessie, T.; Kim, K.-S. Genomic Signatures of High-Altitude Adaptation in Ethiopian Sheep Populations. Genes. Genom. 2019, 41, 973–981. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.-X.; Wang, B.; Jing, J.-N.; Ma, R.; Luo, Y.-H.; Li, X.; Yan, Z.; Liu, Y.-J.; Gao, L.; Ren, Y.-L.; et al. Whole-Body Adipose Tissue Multi-Omic Analyses in Sheep Reveal Molecular Mechanisms Underlying Local Adaptation to Extreme Environments. Commun. Biol. 2023, 6, 159. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Han, Z.; Zhou, W.; Li, X.; Zhang, X.; Zhu, L.; Wang, J.; Li, X.; Zhang, C.-L.; Han, Y.; et al. Population Structure and Selective Signature of Kirghiz Sheep by Illumina Ovine SNP50 BeadChip. PeerJ 2024, 12, e17980. [Google Scholar] [CrossRef]
- Sweet-Jones, J.; Lenis, V.P.; Yurchenko, A.A.; Yudin, N.S.; Swain, M.; Larkin, D.M. Genotyping and Whole-Genome Resequencing of Welsh Sheep Breeds Reveal Candidate Genes and Variants for Adaptation to Local Environment and Socioeconomic Traits. Front. Genet. 2021, 12, 612492. [Google Scholar] [CrossRef]
- Jin, M.; Wang, H.; Liu, G.; Lu, J.; Yuan, Z.; Li, T.; Liu, E.; Lu, Z.; Du, L.; Wei, C. Whole-Genome Resequencing of Chinese Indigenous Sheep Provides Insight into the Genetic Basis Underlying Climate Adaptation. Genet. Sel. Evol. 2024, 56, 26. [Google Scholar] [CrossRef]
- Wiener, P.; Robert, C.; Ahbara, A.; Salavati, M.; Abebe, A.; Kebede, A.; Wragg, D.; Friedrich, J.; Vasoya, D.; Hume, D.A.; et al. Whole-Genome Sequence Data Suggest Environmental Adaptation of Ethiopian Sheep Populations. Genome Biol. Evol. 2021, 13, evab014. [Google Scholar] [CrossRef]
- Song, Y.; Yuan, C.; An, X.; Guo, T.; Zhang, W.; Lu, Z.; Liu, J. Genome-Wide Selection Signals Reveal Candidate Genes Associated with Plateau Adaptation in Tibetan Sheep. Animals 2024, 14, 3212. [Google Scholar] [CrossRef]
- Meyermans, R.; Gorssen, W.; Aerts, N.; Hooyberghs, K.; Chakkingal Bhaskaran, B.; Chapard, L.; Buys, N.; Janssens, S. Genomic Characterisation and Diversity Assessment of Eight Endangered Belgian Sheep Breeds. Animal 2024, 18, 101315. [Google Scholar] [CrossRef]
- Belabdi, I.; Ouhrouch, A.; Lafri, M.; Gaouar, S.B.S.; Ciani, E.; Benali, A.R.; Ould Ouelhadj, H.; Haddioui, A.; Pompanon, F.; Blanquet, V.; et al. Genetic Homogenization of Indigenous Sheep Breeds in Northwest Africa. Sci. Rep. 2019, 9, 7920. [Google Scholar] [CrossRef]
- Zhao, L.; Yuan, L.; Li, F.; Zhang, X.; Tian, H.; Ma, Z.; Zhang, D.; Zhang, Y.; Zhao, Y.; Huang, K.; et al. Whole-Genome Resequencing of Hu Sheep Identifies Candidate Genes Associated with Agronomic Traits. J. Genet. Genom. 2024, 51, 866–876. [Google Scholar] [CrossRef]
- Groeneveld, L.F.; Lenstra, J.A.; Eding, H.; Toro, M.A.; Scherf, B.; Pilling, D.; Negrini, R.; Finlay, E.K.; Jianlin, H.; Groeneveld, E.; et al. Genetic Diversity in Farm Animals—A Review. Anim. Genet. 2010, 41 (Suppl. 1), 6–31. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Li, T.; Su, M.; Wang, H.; Li, Q.; Lang, X.; Ma, Y. Identification of Copy Number Variation in Tibetan Sheep Using Whole Genome Resequencing Reveals Evidence of Genomic Selection. BMC Genom. 2023, 24, 555. [Google Scholar] [CrossRef]
- Yang, H.; Zhu, M.; Wang, M.; Zhou, H.; Zheng, J.; Qiu, L.; Fan, W.; Yang, J.; Yu, Q.; Yang, Y.; et al. Genome-Wide Comparative Analysis Reveals Selection Signatures for Reproduction Traits in Prolific Suffolk Sheep. Front. Genet. 2024, 15, 1404031. [Google Scholar] [CrossRef]
- Gudra, D.; Valdovska, A.; Kairisa, D.; Galina, D.; Jonkus, D.; Ustinova, M.; Viksne, K.; Kalnina, I.; Fridmanis, D. Genomic Diversity of the Locally Developed Latvian Darkheaded Sheep Breed. Heliyon 2024, 10, e31455. [Google Scholar] [CrossRef] [PubMed]
- Senczuk, G.; Di Civita, M.; Rillo, L.; Macciocchi, A.; Occidente, M.; Saralli, G.; D’Onofrio, V.; Galli, T.; Persichilli, C.; Di Giovannantonio, C.; et al. The Genome-Wide Relationships of the Critically Endangered Quadricorna Sheep in the Mediterranean Region. PLoS ONE 2023, 18, e0291814. [Google Scholar] [CrossRef]
- Barraclough, A.D.; Reed, M.G.; Måren, I.E.; Price, M.F.; Moreira-Muñoz, A.; Coetzer, K. Recognize 727 UNESCO Biosphere Reserves for Biodiversity COP15. Nature 2021, 598, 257. [Google Scholar] [CrossRef]
- Egoh, B.N.; Ntshotsho, P.; Maoela, M.A.; Blanchard, R.; Ayompe, L.M.; Rahlao, S. Setting the Scene for Achievable Post-2020 Convention on Biological Diversity Targets: A Review of the Impacts of Invasive Alien Species on Ecosystem Services in Africa. J. Environ. Manag. 2020, 261, 110171. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, R.; Lu, Y.; Li, M.; Gao, Z.; Li, D.; Gao, Y.; Deng, W.; Wang, B. Whole-Genome Resequencing in Sheep: Applications in Breeding, Evolution, and Conservation. Genes 2025, 16, 363. https://doi.org/10.3390/genes16040363
Ma R, Lu Y, Li M, Gao Z, Li D, Gao Y, Deng W, Wang B. Whole-Genome Resequencing in Sheep: Applications in Breeding, Evolution, and Conservation. Genes. 2025; 16(4):363. https://doi.org/10.3390/genes16040363
Chicago/Turabian StyleMa, Ruoshan, Ying Lu, Mengfei Li, Zhendong Gao, Dongfang Li, Yuyang Gao, Weidong Deng, and Bo Wang. 2025. "Whole-Genome Resequencing in Sheep: Applications in Breeding, Evolution, and Conservation" Genes 16, no. 4: 363. https://doi.org/10.3390/genes16040363
APA StyleMa, R., Lu, Y., Li, M., Gao, Z., Li, D., Gao, Y., Deng, W., & Wang, B. (2025). Whole-Genome Resequencing in Sheep: Applications in Breeding, Evolution, and Conservation. Genes, 16(4), 363. https://doi.org/10.3390/genes16040363




