Alternative Splicing Events and ABA Hormone Regulation in Drought Response of Hippophae gyantsensis L.
Abstract
1. Introduction
2. Materials and Method
2.1. Plant Materials and Treatments
2.2. Determination of Drought-Related Physiological Indices
2.3. Next-Generation Transcriptome Sequencing and Analysis
2.4. Hormone Quantification
2.5. Full-Length Transcriptome Sequencing and Analysis
3. Results
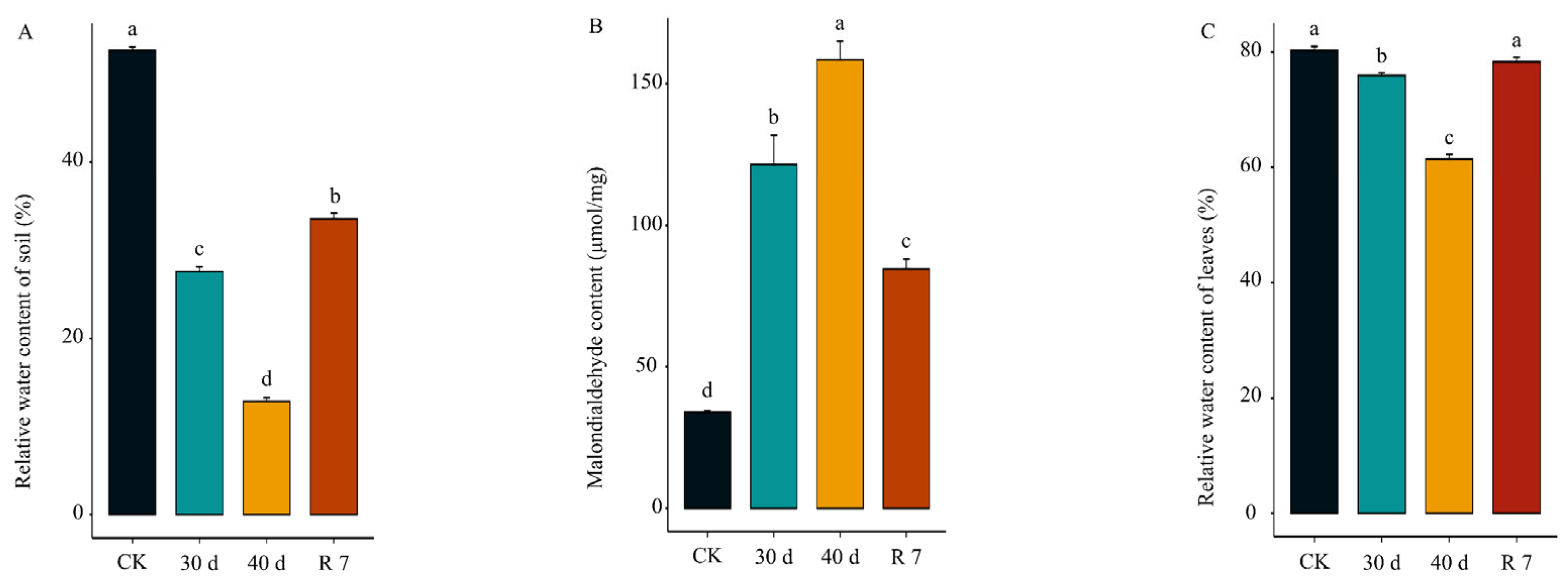
3.1. Response of H. gyantsensis to Drought Stress
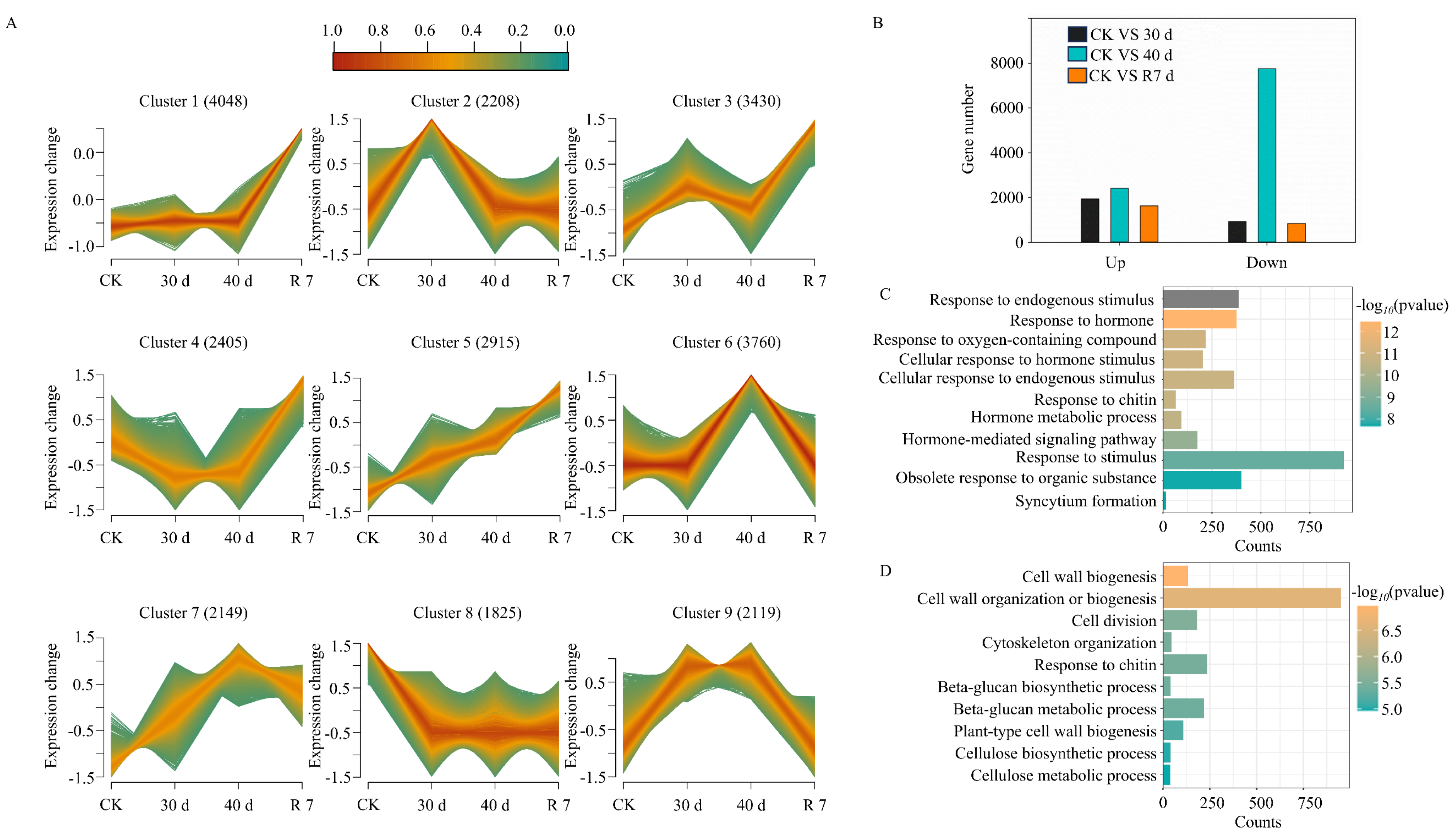
3.2. Transcriptional Changes in H. gyantsensis Under Drought Stress
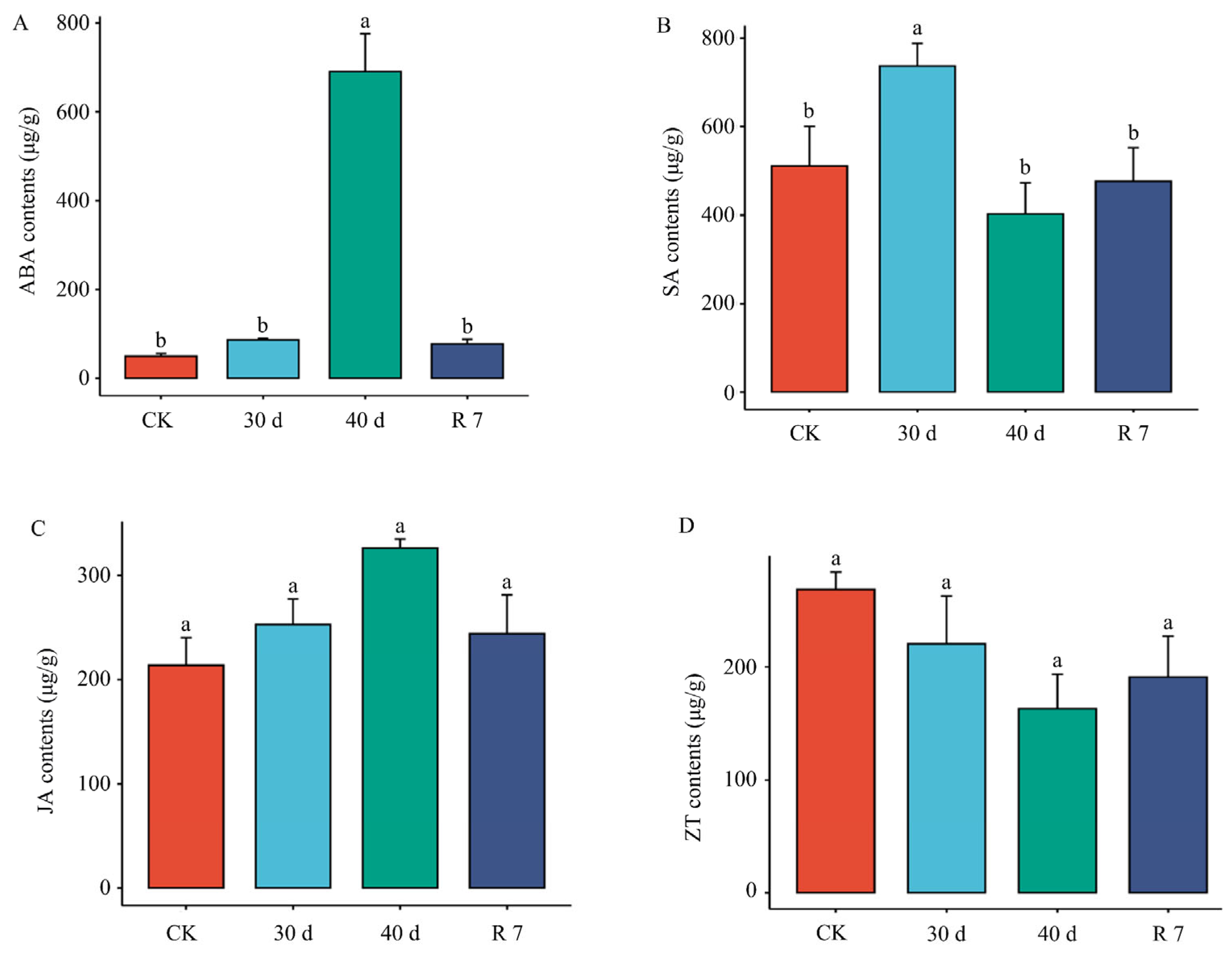
3.3. Plant Hormone Response in H. gyantsensis Under Drought Stress
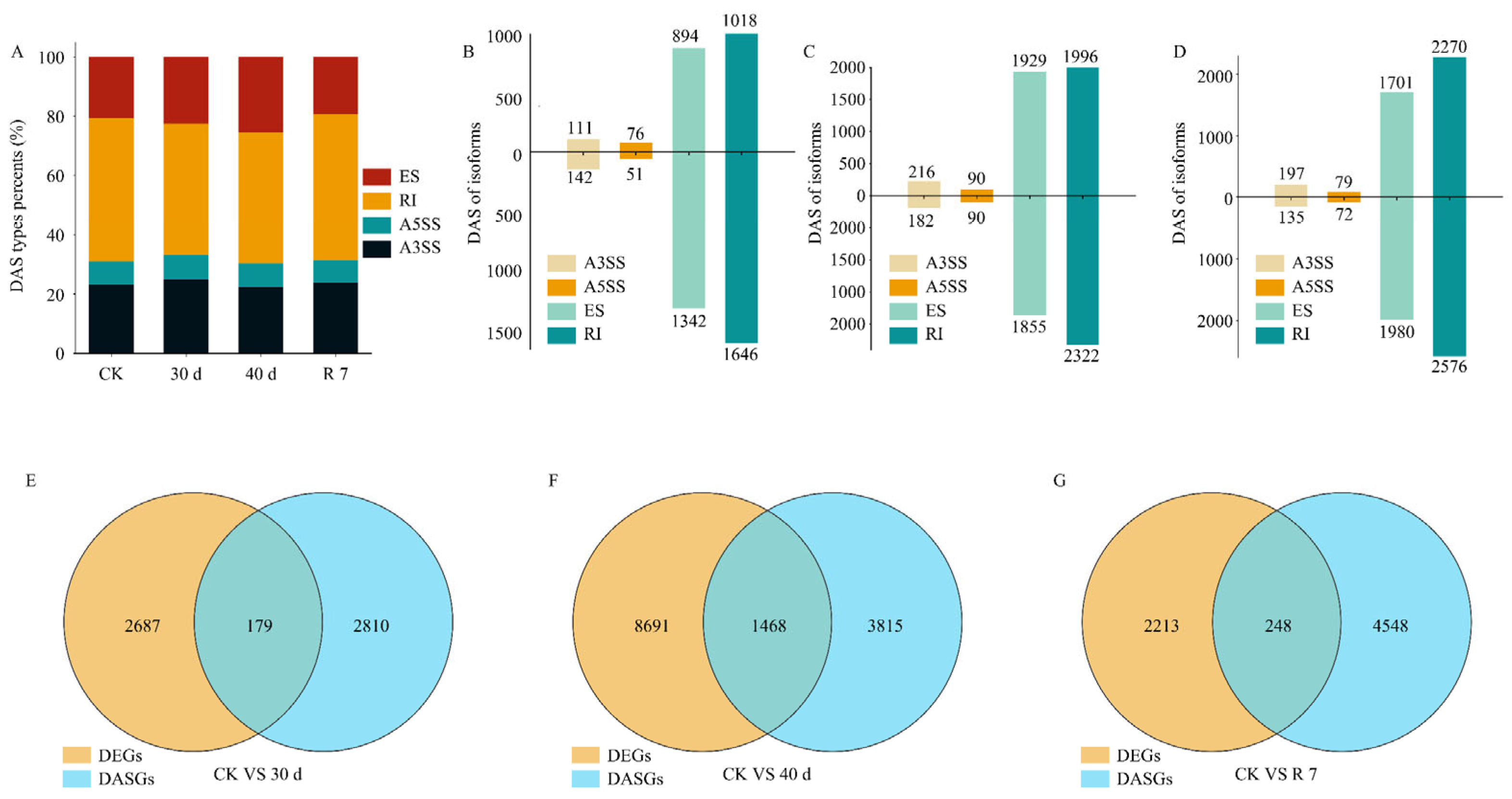
3.4. Alternative Splicing Events in Response to Drought Stress
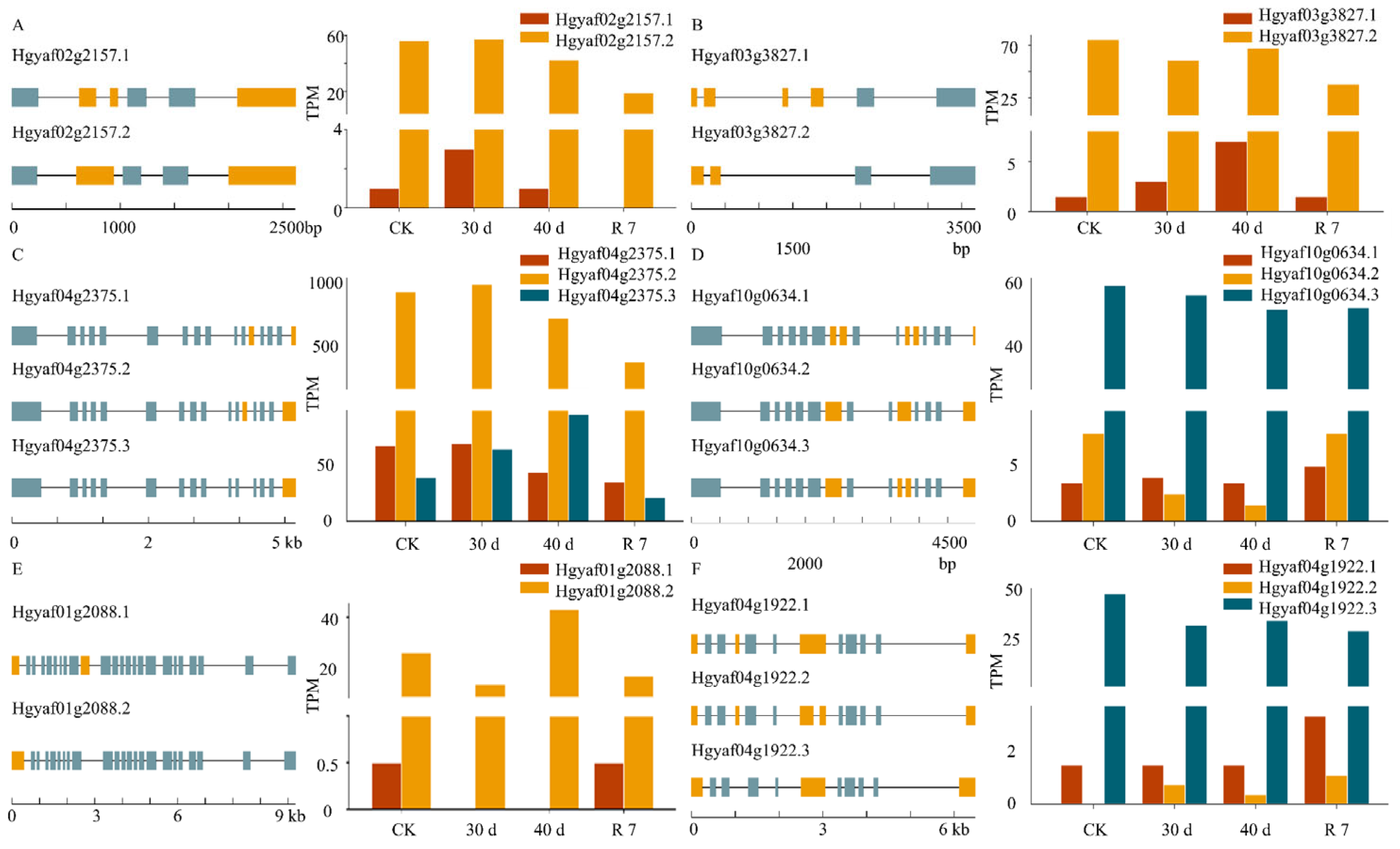
3.5. Drought Response of Alternative Splicing Events in the ABA Signaling Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, J.; Yang, B.; Yang, J.; Yu, S.; Li, X. Leaf flavonoids in Chinese sea-buckthorn (Hippophae rhamnoides subsp. sinensis Rousi) and their response to environmental gradients across northern China. ScienceAsia 2024, 50, 1–10. [Google Scholar] [CrossRef]
- Zeng, Z.; Wang, J.; Tian, Z.; Norbu, N.; Chen, Y.; Chen, J.; Zhang, W.; Qiong, L. Development of sex-specific molecular markers for early sex identification in Hippophae gyantsensis based on whole-genome resequencing. BMC Plant Biol. 2024, 24, 1187. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wu, B.; Jian, J.; Tang, Y.; Zhang, T.; Song, Z.; Zhang, W.; Qiong, L. How to survive in the world’s third poplar: Insights from the genome of the highest altitude woody plant, Hippophae tibetana (Elaeagnaceae). Front. Plant Sci. 2022, 13, 1051587. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Yang, D.; Yang, S.; Yang, X.; Chen, Z.; Yang, T.; Yang, Y.; Yang, Y. Chromosome-level genome assembly of Hippophae gyantsensis. Sci. Data 2024, 11, 126. [Google Scholar] [CrossRef]
- Zhao, A.; Xu, W.; Xu, P.; Zhang, X.; Wu, Y.; Xu, A.; Zhong, Y.; Oladipo, A.; Cao, F.; Fu, F. Establishment of Tissue Culture and Regeneration System in Hippophae gyantsensis Lian. Horticulturae 2024, 10, 460. [Google Scholar] [CrossRef]
- He, J.; Zhang, L.; Liu, Q.; Zhu, X.; Liu, X.; Feng, Q.; Luo, D.; Shi, Z. Physiological and biochemical response of typical shrubs to drought stress in the Minjiang River dry valley. Acta Ecol. Sin. 2018, 38, 2362–2371. [Google Scholar] [CrossRef]
- Kumar, S.; Sachdeva, S.; Bhat, K.V.; Vats, S. Plant Responses to Drought Stress: Physiological, Biochemical and Molecular Basis. In Biotic and Abiotic Stress Tolerance in Plants; Springer: Berlin/Heidelberg, Germany, 2018; pp. 1–25. [Google Scholar] [CrossRef]
- Zhang, T.; Gao, G.; Liu, J.; Yang, G.; Lv, Z.; Zhang, J.; He, C. Transcripts and ABA-dependent signaling in response to drought stress in Hippophae rhamnoides L. Trees Struct. Funct. 2020, 34, 1033–1045. [Google Scholar] [CrossRef]
- Rai, G.K.; Khanday, D.M.; Choudhary, S.M.; Kumar, P.; Kumari, S.; Martínez-Andújar, C.; Martínez-Melgarejo, P.A.; Rai, P.K.; Pérez-Alfocea, F. Unlocking nature’s stress buster: Abscisic acid’s crucial role in defending plants against abiotic stress. Plant Stress 2024, 11, 100359. [Google Scholar] [CrossRef]
- Xiong, Y.; Song, X.; Mehra, P.; Yu, S.; Li, Q.; Tashenmaimaiti, D.; Bennett, M.; Kong, X.; Bhosale, R.; Huang, G. ABA-auxin cascade regulates crop root angle in response to drought. Curr. Biol. 2025, 35, 542–553.e4. [Google Scholar] [CrossRef]
- Bano, A.; Singh, K.; Singh, S.P.; Sharma, P. Abscisic Acid: Metabolism, Signaling, and Crosstalk with Other Phytohormones under Heavy Metal Stress. Stresses 2023, 3, 665–686. [Google Scholar] [CrossRef]
- Hasan, M.M.; Gong, L.; Nie, Z.-F.; Li, F.-P.; Ahammed, G.J.; Fang, X.-W. ABA-induced stomatal movements in vascular plants during dehydration and rehydration. Environ. Exp. Bot. 2021, 186, 104436. [Google Scholar] [CrossRef]
- Zhao, P.; Zhao, M.; Gao, X.; Shan, Y.; Li, F.; Tian, X.; Li, Z. GhWRKY1bD improves drought tolerance by co-regulation of ABA, ROS, and proline homeostasis in cotton (Gossypium hirsutum). Ind. Crops Prod. 2024, 220, 119179. [Google Scholar] [CrossRef]
- Wang, M.; Kang, S.; Wang, Z.; Jiang, S.; Yang, Z.; Xie, Z.; Tang, H. Genome-wide analysis of the PYL-PP2C-SnRK2s family in the ABA signaling pathway of pitaya reveals its expression profiles under canker disease stress. BMC Genom. 2024, 25, 749. [Google Scholar] [CrossRef]
- Sybilska, E.; Collin, A.; Sadat Haddadi, B.; Mur, L.A.J.; Beckmann, M.; Guo, W.; Simpson, C.G.; Daszkowska-Golec, A. The cap-binding complex modulates ABA-responsive transcript splicing during germination in barley (Hordeum vulgare). Sci. Rep. 2024, 14, 18278. [Google Scholar] [CrossRef]
- Kim, N.; Lee, J.; Yeom, S.-I.; Kang, N.-J.; Kang, W.-H. The landscape of abiotic and biotic stress-responsive splice variants with deep RNA-seq datasets in hot pepper. Sci. Data 2024, 11, 381. [Google Scholar] [CrossRef]
- Albuquerque-Martins, R.; Szakonyi, D.; Rowe, J.; Jones, A.M.; Duque, P. ABA signaling prevents phosphodegradation of the SR45 splicing factor to alleviate inhibition of early seedling development in Arabidopsis. Plant Commun. 2023, 4, 100495. [Google Scholar] [CrossRef]
- Hong, Y.; Yao, J.; Shi, H.; Chen, Y.; Zhu, J.-K.; Wang, Z. The Arabidopsis spliceosomal protein SmEb modulates ABA responses by maintaining proper alternative splicing of HAB1. Stress Biol. 2021, 1, 4. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, Y.; Hou, X.; Zhang, C.; Wang, Z.; Zhang, J.; Liu, X.; Shi, X.; Duan, W.; Xiao, K. Wheat TaPYL9-involved signalling pathway impacts plant drought response through regulating distinct osmotic stress-associated physiological indices. Plant Biotechnol. J. 2025 23, 352–373. [CrossRef]
- Guo, Y.; Shang, X.; Ma, L.; Cao, Y. RNA-Binding Protein-Mediated Alternative Splicing Regulates Abiotic Stress Responses in Plants. Int. J. Mol. Sci. 2024, 25, 10548. [Google Scholar] [CrossRef]
- Khan, M.K.; Pandey, A.; Hamurcu, M.; Vyhnánek, T.; Zargar, S.M.; Kahraman, A.; Topal, A.; Gezgin, S. Exploring strigolactones for inducing abiotic stress tolerance in plants. Czech J. Genet. Plant Breed. 2024, 60, 55–69. [Google Scholar] [CrossRef]
- Mostofa, M.G.; Li, W.; Nguyen, K.H.; Fujita, M.; Tran, L.-S.P. Strigolactones in plant adaptation to abiotic stresses: An emerging avenue of plant research. Plant Cell Environ. 2018, 41, 2227–2243. [Google Scholar] [CrossRef] [PubMed]
- Lv, S.; Zhang, Y.; Li, C.; Liu, Z.; Yang, N.; Pan, L.; Wu, J.; Wang, J.; Yang, J.; Lv, Y.; et al. Strigolactone-triggered stomatal closure requires hydrogen peroxide synthesis and nitric oxide production in an abscisic acid-independent manner. New Phytol. 2017, 217, 290–304. [Google Scholar] [CrossRef] [PubMed]
- Kapulnik, Y.; Delaux, P.-M.; Resnick, N.; Mayzlish-Gati, E.; Wininger, S.; Bhattacharya, C.; Séjalon-Delmas, N.; Combier, J.-P.; Bécard, G.; Belausov, E.; et al. Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 2011, 233, 209–216. [Google Scholar] [CrossRef]
- Ha, C.V.; Leyva-González, M.A.; Osakabe, Y.; Tran, U.T.; Nishiyama, R.; Watanabe, Y.; Tanaka, M.; Seki, M.; Yamaguchi, S.; Dong, N.V.; et al. Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proc. Natl. Acad. Sci. USA 2014, 111, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Jha, A.B.; Dubey, R.S. Strigolactones: Coordination with other phytohormones and enhancement of abiotic stress responses. Environ. Exp. Bot. 2024, 223, 105782. [Google Scholar] [CrossRef]
- An, Y.; Wang, B.; Meng, Z.; Song, Y.; Wang, Y.; Wang, W.; Xu, M.; An, X. Optimization of the enzymatic hydrolysis process for sea buckthorn leaf polysaccharides: An investigation into their enhanced physicochemical properties and antioxidant activities. Chem. Biol. Technol. Agric. 2024, 11, 193. [Google Scholar] [CrossRef]
- Wang, H.; Cheng, N.; Wu, Q.; Fang, D.; Rahman, F.-U.; Hao, H.; Zhang, Y. Antioxidant activities of sea buckthorn polysaccharides and their potential application in cosmetic industry. J. Dermatol. Sci. Cosmet. Technol. 2024, 1, 100023. [Google Scholar] [CrossRef]
- Suseela, V.; Tharayil, N.; Xing, B.; Dukes, J.S. Warming alters potential enzyme activity but precipitation regulates chemical transformations in grass litter exposed to simulated climatic changes. Soil Biol. Biochem. 2014, 75, 102–112. [Google Scholar] [CrossRef]
- Sade, N.; Galkin, E.; Moshelion, M. Measuring Arabidopsis, Tomato and Barley Leaf Relative Water Content (RWC). Bio-Protoc. 2015, 5, e1451. [Google Scholar] [CrossRef]
- Kiss, T.; Karácsony, Z.; Gomba-Tóth, A.; Szabadi, K.L.; Spitzmüller, Z.; Hegyi-Kaló, J.; Cels, T.; Otto, M.; Golen, R.; Hegyi, Á.I.; et al. A modified CTAB method for the extraction of high-quality RNA from mono-and dicotyledonous plants rich in secondary metabolites. Plant Methods 2024, 20, 62. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, Y.; Liu, H.; Wu, H.; Zou, D. Simultaneous Determination of Five Plant Endogenous Hormones in Lettuce Leaves by UPLC-MS/MS. North. Hortic. 2021, 8–15. [Google Scholar] [CrossRef]
- Wang, J.; Dai, L.; Wang, A.; Lu, Y. Determination of 10 Plant Endogenous Hormones by Ultra-High-Performance Liquid Chromatography–High-Resolution Mass Spectrometry. J. Anal. Sci. 2021, 37, 81–87. [Google Scholar] [CrossRef]
- Tang, A.D.; Soulette, C.M.; van Baren, M.J.; Hart, K.; Hrabeta-Robinson, E.; Wu, C.J.; Brooks, A.N. Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. Nat. Commun. 2020, 11, 1438. [Google Scholar] [CrossRef]
- Innes, P.A.; Goebl, A.M.; Smith, C.C.R.; Rosenberger, K.; Kane, N.C. Gene expression and alternative splicing contribute to adaptive divergence of ecotypes. Heredity 2024, 132, 120–132. [Google Scholar] [CrossRef]
- Gao, G.; Lv, Z.; Zhang, G.; Li, J.; Zhang, J.; He, C. An ABA–flavonoid relationship contributes to the differences in drought resistance between different sea buckthorn subspecies. Tree Physiol. 2020, 41, 744–755. [Google Scholar] [CrossRef]
- Iuchi, S.; Kobayashi, M.; Taji, T.; Naramoto, M.; Seki, M.; Kato, T.; Tabata, S.; Kakubari, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001, 27, 325–333. [Google Scholar] [CrossRef]
- Gu, L.; Chen, P.; Yu, S. The cytochrome P450 gene GhCYP94C1 is involved in drought stress in upland cotton (Gossypium hirsutum L.). Czech J. Genet. Plant Breed. 2023, 59, 189–195. [Google Scholar] [CrossRef]
- Iuchi, S.; Kobayashi, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A Stress-Inducible Gene for 9-cis-Epoxycarotenoid Dioxygenase Involved in Abscisic Acid Biosynthesis under Water Stress in Drought-Tolerant Cowpea. Plant Physiol. 2000, 123, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Guo, H.; Xiong, H.; Xie, Y.; Gu, J.; Zhao, L.; Zhao, S.; Ding, Y.; Liu, L. Strigolactone and abscisic acid synthesis and signaling pathways are enhanced in the wheat oligo-tillering mutant ot1. Mol. Breed. 2024, 44, 12. [Google Scholar] [CrossRef] [PubMed]
- Korek, M.; Marzec, M. Strigolactones and abscisic acid interactions affect plant development and response to abiotic stresses. BMC Plant Biol. 2023, 23, 314. [Google Scholar] [CrossRef] [PubMed]
- Bu, Q.; Lv, T.; Shen, H.; Luong, P.; Wang, J.; Wang, Z.; Huang, Z.; Xiao, L.; Engineer, C.; Kim, T.H.; et al. Regulation of drought tolerance by the F-box protein MAX2 in Arabidopsis. Plant Physiol. 2014, 164, 424–439. [Google Scholar] [CrossRef]
- Díez, A.R.; Szakonyi, D.; Lozano-Juste, J.; Duque, P. Alternative splicing as a driver of natural variation in abscisic acid response. Plant J. 2024, 119, 9–27. [Google Scholar] [CrossRef]
- Lu, F.; Li, W.; Peng, Y.; Cao, Y.; Qu, J.; Sun, F.; Yang, Q.; Lu, Y.; Zhang, X.; Zheng, L.; et al. ZmPP2C26 Alternative Splicing Variants Negatively Regulate Drought Tolerance in Maize. Front. Plant Sci. 2022, 13, 851531. [Google Scholar] [CrossRef]
- Sakib, M.M.; Islam, M.S.; Bhuya, A.R.; Shuvo, M.R.K.; Abdullah-Al-Shoeb, M.; Azad, M.A.K.; Ghosh, A. Genomic identification, evolutionary analysis, and transcript profiling of protein phosphatase 2C in Solanum lycopersicum. Sci. Rep. 2024, 14, 31742. [Google Scholar] [CrossRef]
- Lu, X.; Pang, Y.; Ma, C.; Ye, F.; Liang, X.; Zhang, X.; Cao, L. Cloning of maize protein phosphatase genes ZmPP2C72 and ZmPP2C97 and study of their antistress functions. Plant Physiol. J. 2024, 60, 1588–1598. [Google Scholar] [CrossRef]
- Ye, S.; Huang, Y.; Ma, T.; Ma, X.; Li, R.; Shen, J.; Wen, J. BnaABF3 and BnaMYB44 regulate the transcription of zeaxanthin epoxidase genes in carotenoid and abscisic acid biosynthesis. Plant Physiol. 2024, 195, 2372–2388. [Google Scholar] [CrossRef]
- Dong, Y.; Du, L.; Zhang, Z.; Cheng, J.; Gao, Y.; Wang, X.; Wu, Y.; Wang, Y. Molecular cloning and functional characterization in response to saline-alkali stress of the MhZEP gene in Arabidopsis thaliana. Physiol. Mol. Biol. Plants 2024, 30, 1551–1564. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, F.; Cai, Y.; Yang, S.; Yang, Y. Alternative Splicing Events and ABA Hormone Regulation in Drought Response of Hippophae gyantsensis L. Genes 2025, 16, 350. https://doi.org/10.3390/genes16030350
Lin F, Cai Y, Yang S, Yang Y. Alternative Splicing Events and ABA Hormone Regulation in Drought Response of Hippophae gyantsensis L. Genes. 2025; 16(3):350. https://doi.org/10.3390/genes16030350
Chicago/Turabian StyleLin, Fanfan, Yifan Cai, Shihai Yang, and Yunqiang Yang. 2025. "Alternative Splicing Events and ABA Hormone Regulation in Drought Response of Hippophae gyantsensis L." Genes 16, no. 3: 350. https://doi.org/10.3390/genes16030350
APA StyleLin, F., Cai, Y., Yang, S., & Yang, Y. (2025). Alternative Splicing Events and ABA Hormone Regulation in Drought Response of Hippophae gyantsensis L. Genes, 16(3), 350. https://doi.org/10.3390/genes16030350





