A Comparative Analysis and Phylogenetic Relationship of the Chloroplast Genome Sequences of Illicium verum and Illicium difengpi
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Sequencing and Genome Assembly
2.3. Gene Annotation
2.4. Comparison and Differential Analysis of Cp Genomes
3. Results
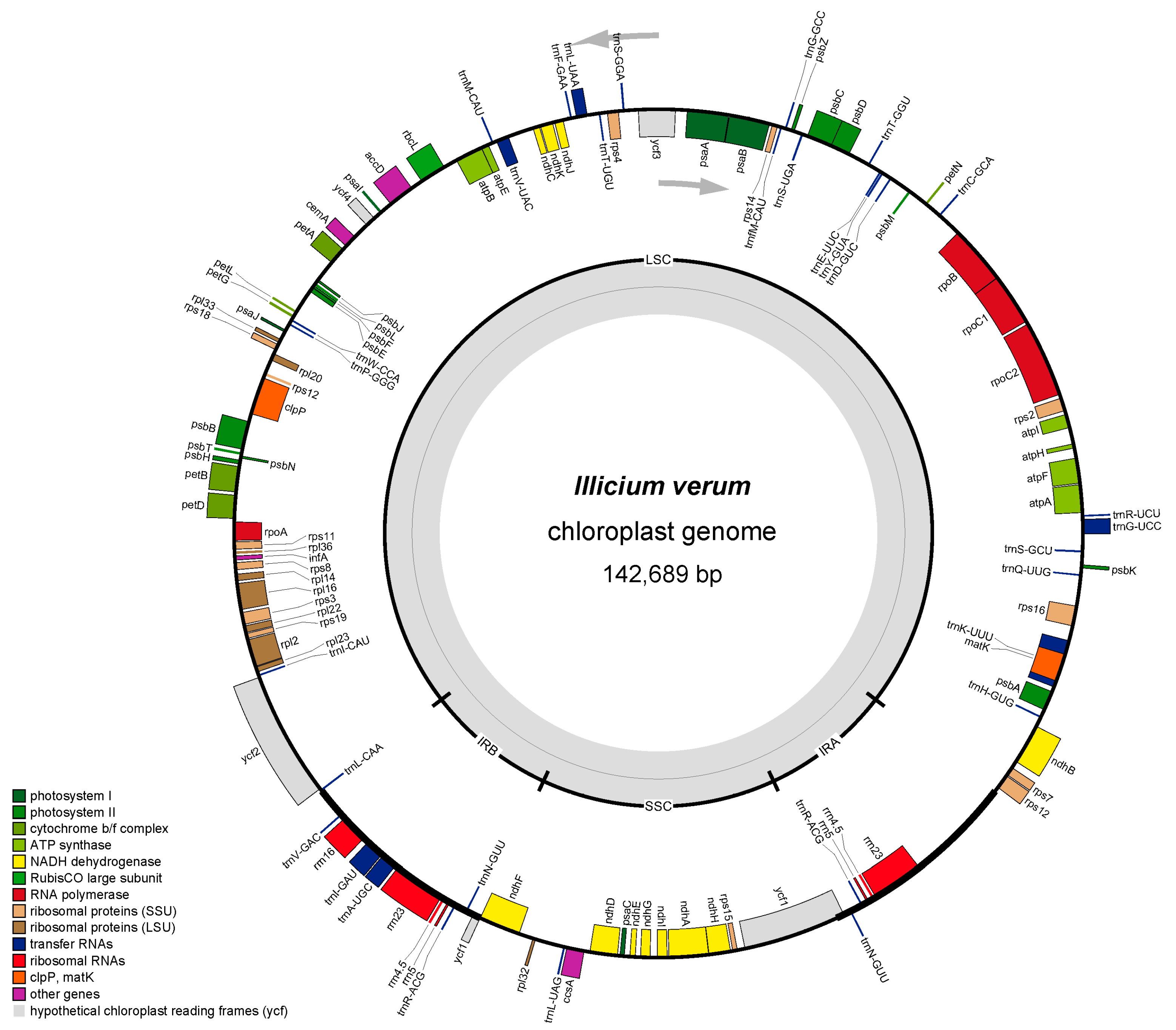
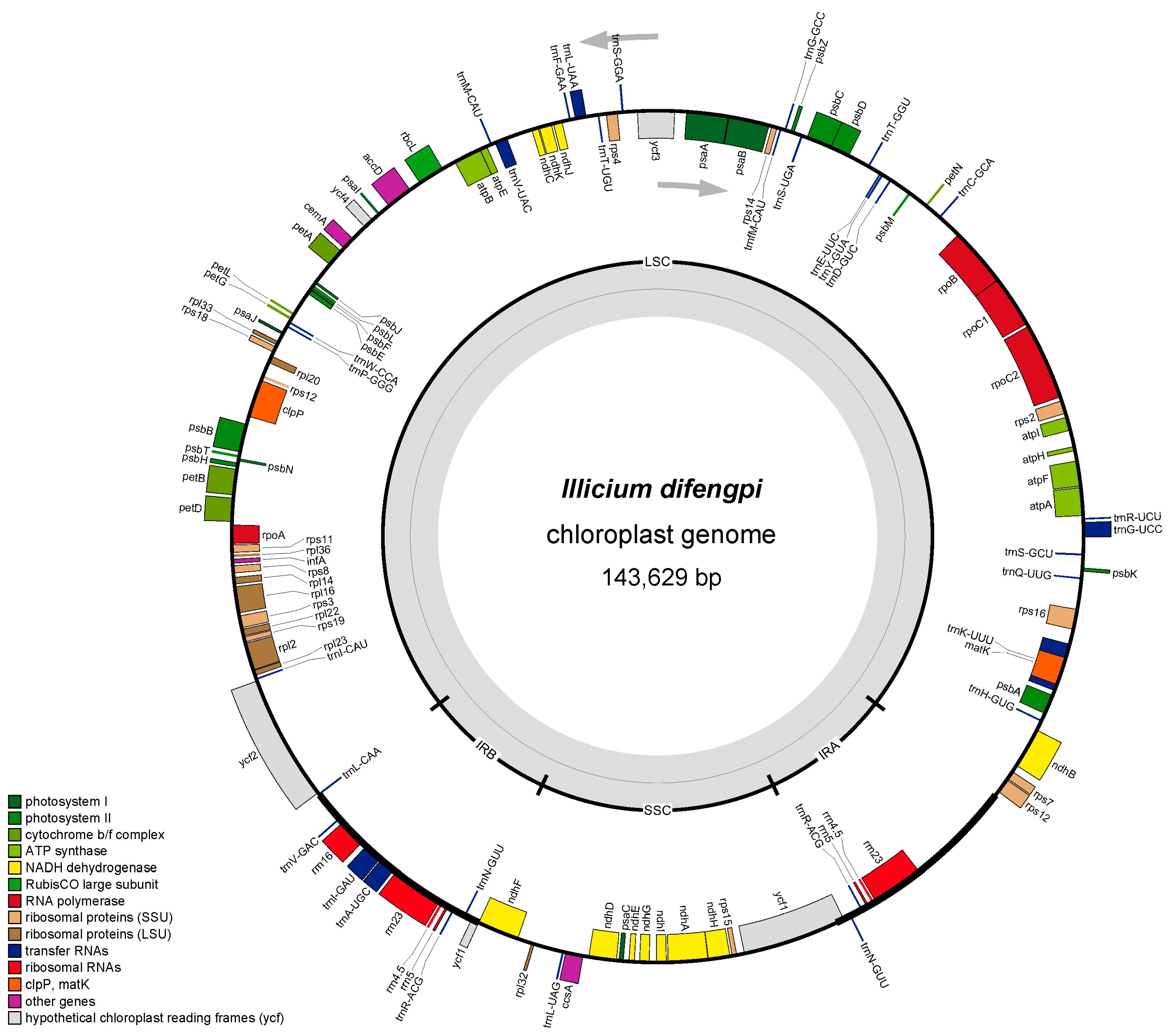
3.1. Chloroplast Genome Structure and Composition
3.2. Codon Bias Analysis
3.3. Simple Sequence Repeats (SSRs) and Long Repeats
3.4. IR Expansion and Contraction
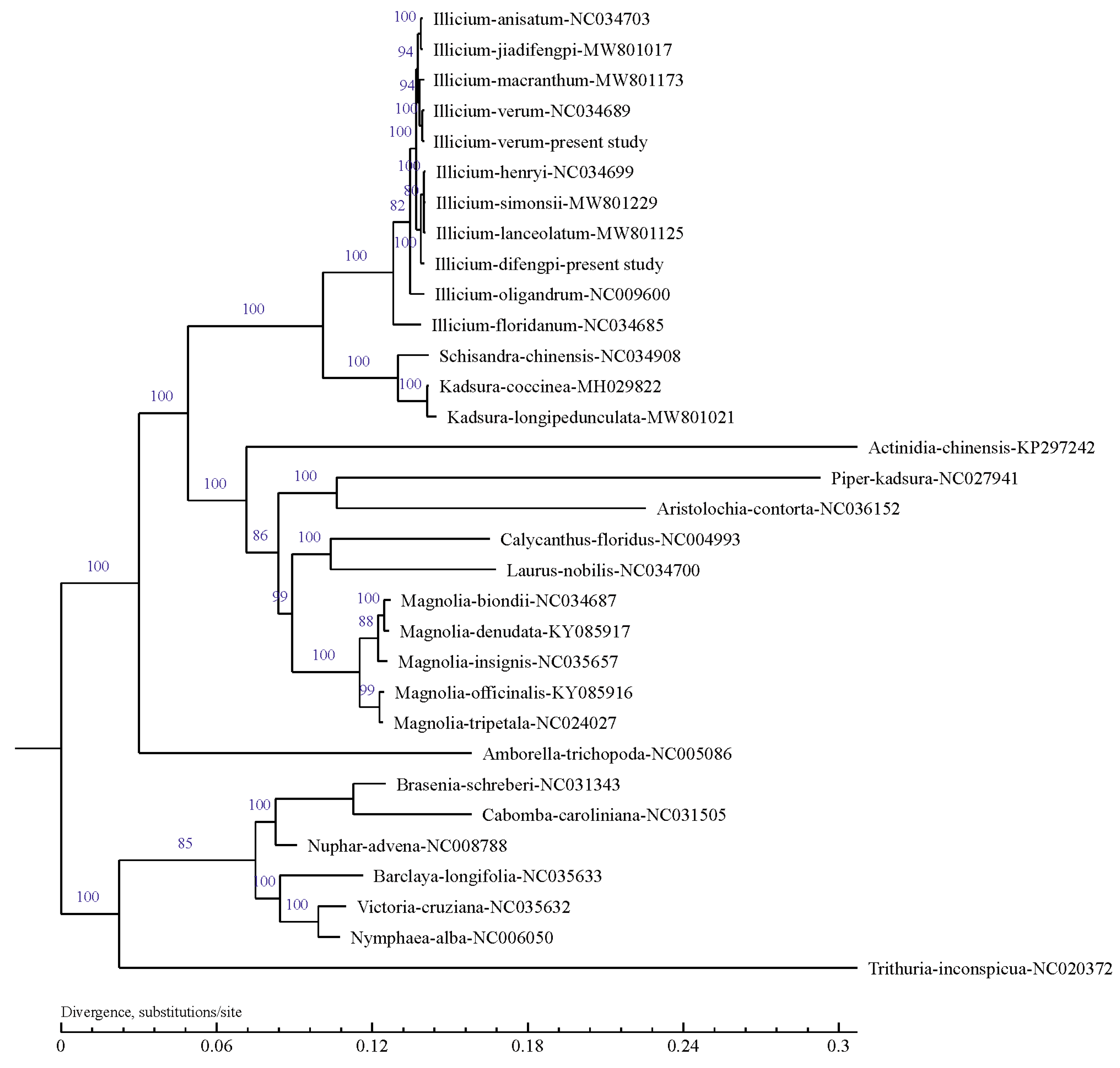
3.5. Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| cp | Chloroplast |
| IRs | Inverted repeats |
| LSC | Large-single-copy |
| SSC | Small-single-copy |
| tRNA | Transfer RNA |
| rRNA | Ribosome RNA |
| Nr | Non-redundant |
| COGs | Clusters of Orthologous Groups |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| GO | Gene Ontology |
| RSCU | Relative synonymous codon usage |
| MISA | MIcroSAtellite |
| ML | Maximum likelihood |
| SSRs | Simple sequence repeats |
| ML BSs | ML bootstrap support values |
References
- Bremer, B.; Bremer, K.; Chase, M.; Reveal, J.L.; Soltis, P.; Stevens, P.; Anderberg, A.; Fay, M.; Goldblatt, P.; Judd, W.; et al. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot. J. Linn. Soc. 2003, 141, 399–436. [Google Scholar]
- Iii, A.; Bremer, K. Angiosperm Phylogeny Group III (APG III). An update of The Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Botanical Journal of the Linnean Society. Bot. J. Linn. Soc. 2009, 161, 105–121. [Google Scholar]
- Zou, Q.; Huang, Y.; Zhang, W.; Lu, C.; Yuan, J. A Comprehensive Review of the Pharmacology, Chemistry, Traditional Uses and Quality Control of Star Anise (Illicium verum Hook. F.): An Aromatic Medicinal Plant. Molecules 2023, 28, 7378. [Google Scholar] [CrossRef]
- Shahrajabian, M.; Sun, W.; Cheng, Q. Chinese star anise (Illicium verum) and pyrethrum (Chrysanthemum cinerariifolium) as natural alternatives for organic farming and health care- A review. Aust. J. Crop Sci. 2020, 14, 517–523. [Google Scholar] [CrossRef]
- Wu, C.; Liang, H.; Qi, B.; Liu, B.; Wang, M.; Tang, H.; Li, D.; Fahad, S. Research progress on Illicium difengpi (Illiciaceae): A review. Horticulturae 2021, 8, 19. [Google Scholar] [CrossRef]
- Zhu, L.; Luo, Y.; Xiao, J.; Hao, E.; Wei, J.; Zhao, J.; Yao, C.; Wang, Y.; Luo, H. Star anise (Illicium verum Hook. f.): Dual therapeutic and nutritional potential in food and medicine. Acupunct. Herb. Med. 2024, 4, 563–587. [Google Scholar] [CrossRef]
- Ning, D.S.; Fu, Y.X.; Peng, L.Y.; Tang, H.; Li, L.C.; Wu, X.D.; Huang, Y.S.; Pan, Z.H. Phytochemical constituents of the pericarps of Illicium difengpi and their anti-inflammatory activity. Nat. Prod. Res. 2020, 34, 1756–1762. [Google Scholar] [CrossRef]
- Pan, Z.-H.; Cheng, L.; Ning, D.-S.; Peng, L.-Y.; Fu, Y.-X.; Li, L.-C. Difengpienols A and B, two new sesqui-neolignans with anti-inflammatory activity from the bark of Illicium difengpi. Phytochem. Lett. 2019, 30, 210–214. [Google Scholar] [CrossRef]
- Huang, J.; Liu, H.; Yang, C.; Ye, J.; Xue, Y. Morphological identification of fruits from 16 illicium species. Chin. Tradit. Herb. Drugs 2000, 31, 54–58. [Google Scholar]
- Wen, S. Comparison study of volatile oil content and toxicity test between Illicium verum and its adulterating species. China J. Chin. Mater. Medica 1990, 15, 8–9. [Google Scholar]
- Mower, J.P.; Vickrey, T.L. Chapter Nine—Structural Diversity Among Plastid Genomes of Land Plants. In Advances in Botanical Research; Chaw, S.-M., Jansen, R.K., Eds.; Academic Press: Cambridge, MA, USA, 2018; Volume 85, pp. 263–292. [Google Scholar]
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Lai, Y.; Li, Z.; Zhai, S.; Dai, Y.; Tao, J.; Wang, Q.; Xu, Z.; Jiang, M.; Yu, L.; et al. The complete chloroplast genome of Illicium verum and comparative analysis with related species from Magnoliaceae and Illiciaceae. Front. Genet. 2024, 15, 1452680. [Google Scholar] [CrossRef]
- Park, J.; Kim, Y.; Xi, H. The complete chloroplast genome of aniseed tree, Illicium anisatum L. (Schisandraceae). Mitochondrial DNA Part B 2019, 4, 1023–1024. [Google Scholar] [CrossRef]
- Zhou, F.; Liu, Y.; Xiong, S.; Huang, Y. The complete chloroplast genome of Illicium simonsii Maxim. (Illiciaceae), a species with important medicinal properties. Mitochondrial DNA Part B 2024, 9, 678–682. [Google Scholar] [CrossRef] [PubMed]
- Hansen, D.R.; Dastidar, S.G.; Cai, Z.; Penaflor, C.; Kuehl, J.V.; Boore, J.L.; Jansen, R.K. Phylogenetic and evolutionary implications of complete chloroplast genome sequences of four early-diverging angiosperms: Buxus (Buxaceae), Chloranthus (Chloranthaceae), Dioscorea (Dioscoreaceae), and Illicium (Schisandraceae). Mol. Phylogenetics Evol. 2007, 45, 547–563. [Google Scholar] [CrossRef]
- MeiZi, L.; Hui, Y.; Kun, L.; Pei, M.; Wenbin, Z.; Ping, L. Authentication of Illicium verum using a DNA barcode psbA-trnH. J. Med. Plants Res. 2012, 6, 3156–3161. [Google Scholar]
- Zhou, Q.-R.; Ma, Y.-Y.; Lv, H.-Q.; Lu, Z.-C.; Wang, L.-S.; Liang, J.-S.; Li, J.-J. Chloroplast mini-barcodes combined with high resolution melting analysis to identify herbal medicine Difengpi (Illicium difengpi). Heliyon 2024, 10, e38700. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zheng, Y. Dynamic evolution and phylogenomic analysis of the chloroplast genome in Schisandraceae. Sci. Rep. 2018, 8, 9285. [Google Scholar] [CrossRef]
- Morris, A.B.; Bell, C.D.; Clayton, J.W.; Judd, W.S.; Soltis, D.E.; Soltis, P.S. Phylogeny and Divergence time estimation in Illicium with implications for New World biogeography. Syst. Bot. 2007, 32, 236–249. [Google Scholar] [CrossRef]
- White, D.A.; Leonard, B.T. The pollination of Illicium parviflorum (Illiciaceae). J. Elisha Mitchell Sci. Soc. 1985, 101, 15–18. [Google Scholar]
- Roberts, M.L.; Robert, R.H. Ballistic seed dispersal in Illicium (Illiciaceae). Plant Syst. Evol. 1983, 143, 227–232. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, M.; Dong, X.; Lin, R.; Fan, J.; Chen, Z. Evaluation of four commonly used DNA barcoding Loci for chinese medicinal plants of the family schisandraceae. PLoS ONE 2015, 10, e0125574. [Google Scholar] [CrossRef] [PubMed]

| Category | Gene Groups | Gene Names |
|---|---|---|
| Photosynthesis | Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Subunits of NADH dehydrogenase | ndhA a, ndhB a, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Subunits of cytochrome b/f complex | petA, petB a, petD a, petG, petL, petN | |
| Subunits of ATP synthase | atpA, atpB, atpE, atpF a, atpH, atpI | |
| Large subunit of Rubisco | rbcL | |
| Self-replication | Large subunits of ribosome | rpl14, rpl16 a, rpl2 a, rpl20, rpl22, rpl23, rpl32, rpl33, rpl36 |
| Small subunits of ribosome | rps11, rps12 a, rps14, rps15, rps16 a, rps18, rps19, rps2, rps3, rps4, rps7, rps8 | |
| DNA-dependent RNA polymerase | rpoA, rpoB, rpoC1 a, rpoC2 | |
| Ribosomal RNAs | rrn16 (×2), rrn23 (×2), rrn4.5 (×2), rrn5 (×2) | |
| Transfer RNAs | trnA-UGC (×2) a, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC a, trnH-GUG, trnI-CAU, trnI-GAU (×2) a, trnK-UUU a, trnL-CAA, trnL-UAA a, trnL-UAG, trnM-CAU, trnN-GUU (×2), trnP-GGG, trnQ-UUG, trnR-ACG, trnR-ACG, trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC (×2), trnV-UAC a, trnW-CCA, trnY-GUA, trnfM-CAU | |
| Other genes | Maturase | matK |
| Protease | clpP b | |
| Envelope membrane protein | cemA | |
| Acetyl-CoA carboxylase | accD | |
| C-type cytochrome synthesis gene | ccsA | |
| Translation initiation factor | infA | |
| Protochlorophyllide reductase subunit | -- | |
| Genes of unknown | Proteins of unknown function | ycf1 (×2), ycf2, ycf3 b, ycf4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, S.; Wu, X.; Peng, F.; Zhang, K.; Sooranna, S.R.; Tan, G. A Comparative Analysis and Phylogenetic Relationship of the Chloroplast Genome Sequences of Illicium verum and Illicium difengpi. Genes 2025, 16, 321. https://doi.org/10.3390/genes16030321
Guo S, Wu X, Peng F, Zhang K, Sooranna SR, Tan G. A Comparative Analysis and Phylogenetic Relationship of the Chloroplast Genome Sequences of Illicium verum and Illicium difengpi. Genes. 2025; 16(3):321. https://doi.org/10.3390/genes16030321
Chicago/Turabian StyleGuo, Suqin, Xiqun Wu, Feng Peng, Kun Zhang, Suren Rao Sooranna, and Guiyu Tan. 2025. "A Comparative Analysis and Phylogenetic Relationship of the Chloroplast Genome Sequences of Illicium verum and Illicium difengpi" Genes 16, no. 3: 321. https://doi.org/10.3390/genes16030321
APA StyleGuo, S., Wu, X., Peng, F., Zhang, K., Sooranna, S. R., & Tan, G. (2025). A Comparative Analysis and Phylogenetic Relationship of the Chloroplast Genome Sequences of Illicium verum and Illicium difengpi. Genes, 16(3), 321. https://doi.org/10.3390/genes16030321







