Terrestrial Adaptation in Chelonoidis vicina as Revealed Based on Analysis of the Complete Mitochondrial Genome
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Complete Mitogenome Data and Species Sample
2.2. Mitogenome Analyses
2.3. Comparative Mitochondrial Genome Analyses
2.4. Phylogenetic Construction
2.5. Selection Pressure Analyses
3. Result
3.1. Mitogenome Organization and Structure
3.2. Protein-Coding Genes
3.3. RNA Genes in C. vicina Mitogenomes
3.4. Comparing Mitogenomes Among Species
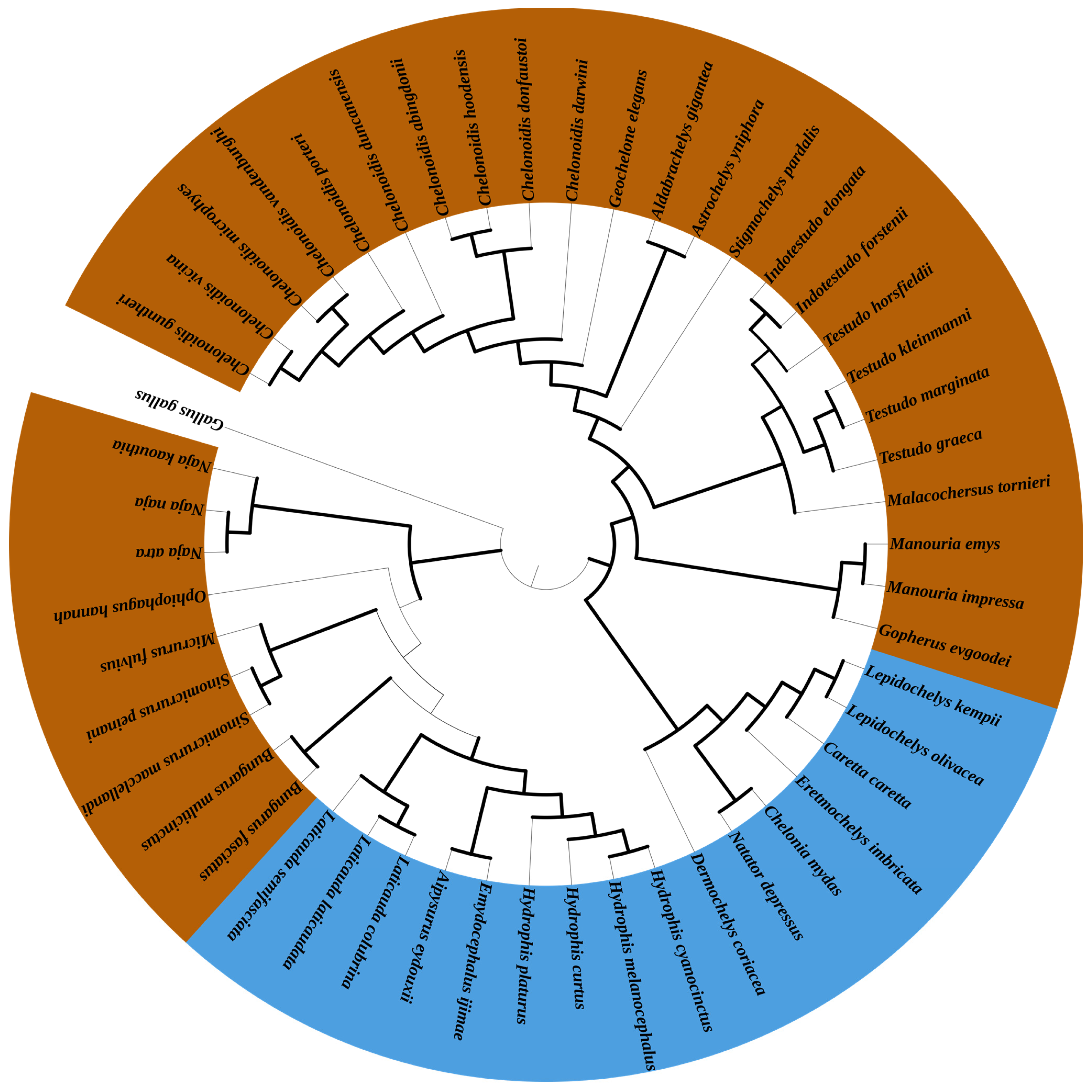
3.5. Phylogenetic Analyses
3.6. Evolutionary Analysis
4. Discussion
4.1. Mitochondrial Genome Characteristics
4.2. Phylogenetic Analysis
4.3. Evolutionary Analysis of Terrestrial Adaptability
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kruuk, H. Otters: Ecology, Behaviour and Conservation; Oxford University Press: Oxford, UK, 2006. [Google Scholar] [CrossRef]
- Timm-Davis, L.L.; DeWitt, T.J.; Marshall, C.D. Divergent Skull Morphology Supports Two Trophic Specializations in Otters (Lutrinae). PLoS ONE 2015, 10, e0143236. [Google Scholar] [CrossRef] [PubMed]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wu, P.D.; Zhang, D.Z.; Zhang, H.B.; Tang, B.P.; Liu, Q.N.; Dai, L.S. Mitochondrial genome of the yellow catfish Pelteobagrus fulvidraco and insights into Bagridae phylogenetics. Genomics 2019, 111, 1258–1265. [Google Scholar] [CrossRef] [PubMed]
- Macey, J.R.; Schulte, J.A., 2nd; Larson, A.; Papenfuss, T.J. Tandem duplication via light-strand synthesis may provide a precursor for mitochondrial genomic rearrangement. Mol. Biol. Evol. 1998, 15, 71–75. [Google Scholar] [CrossRef]
- Mascolo, C.; Ceruso, M.; Palma, G.; Anastasio, A.; Pepe, T.; Sordino, P. The complete mitochondrial genome of the Pink dentex Dentex gibbosus (Perciformes: Sparidae). Mitochondrial DNA Part B Resour. 2018, 3, 525–526. [Google Scholar] [CrossRef]
- Shinde, S.; Bhadra, U. A complex genome-microRNA interplay in human mitochondria. BioMed Res. Int. 2015, 2015, 206382. [Google Scholar] [CrossRef]
- Taanman, J.W. The mitochondrial genome: Structure, transcription, translation and replication. Biochim. Biophys. Acta 1999, 1410, 103–123. [Google Scholar] [CrossRef]
- Yang, C.; Chen, Y.; Chen, Z.; He, G.; Zhong, Z.; Xue, W. The next-generation sequencing reveals the complete mitochondrial genome of Rhinogobius formosanus (Perciformes: Gobiidae). Mitochondrial DNA Part B Resour. 2020, 5, 2673–2674. [Google Scholar] [CrossRef]
- Mindell, D.P.; Sorenson, M.D.; Dimcheff, D.E. Multiple independent origins of mitochondrial gene order in birds. Proc. Natl. Acad. Sci. USA 1998, 95, 10693–10697. [Google Scholar] [CrossRef]
- Ren, Q.; Yuan, J.; Ren, L.; Zhang, L.; Zhang, L.; Jiang, L.; Chen, D.; Kan, X.; Zhang, B. The complete mitochondrial genome of the yellow-browed bunting, Emberiza chrysophrys (Passeriformes: Emberizidae), and phylogenetic relationships within the genus Emberiza. J. Genet. 2014, 93, 699–707. [Google Scholar] [CrossRef]
- van Tuinen, M.; Sibley, C.G.; Hedges, S.B. The early history of modern birds inferred from DNA sequences of nuclear and mitochondrial ribosomal genes. Mol. Biol. Evol. 2000, 17, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Caccone, A.; Gentile, G.; Burns, C.E.; Sezzi, E.; Bergman, W.; Ruelle, M.; Saltonstall, K.; Powell, J.R. Extreme difference in rate of mitochondrial and nuclear DNA evolution in a large ectotherm, Galápagos tortoises. Mol. Phylogenetics Evol. 2004, 31, 794–798. [Google Scholar] [CrossRef] [PubMed]
- Gissi, C.; Iannelli, F.; Pesole, G. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity 2008, 101, 301–320. [Google Scholar] [CrossRef] [PubMed]
- Harrison, R.G. Animal mitochondrial DNA as a genetic marker in population and evolutionary biology. Trends Ecol. Evol. 1989, 4, 6–11. [Google Scholar] [CrossRef]
- Wang, X.; Shang, Y.; Wu, X.; Wei, Q.; Zhou, S.; Sun, G.; Mei, X.; Dong, Y.; Sha, W.; Zhang, H. Divergent evolution of mitogenomics in Cetartiodactyla niche adaptation. Org. Divers. Evol. 2023, 23, 243–259. [Google Scholar] [CrossRef]
- Wei, Q.; Zhang, H.; Wu, X.; Sha, W. The selective constraints of ecological specialization in mustelidae on mitochondrial genomes. Mammal Res. 2020, 65, 85–92. [Google Scholar] [CrossRef]
- Yang, M.; Gong, L.; Sui, J.; Li, X. The complete mitochondrial genome of Calyptogena marissinica (Heterodonta: Veneroida: Vesicomyidae): Insight into the deep-sea adaptive evolution of vesicomyids. PLoS ONE 2019, 14, e0217952. [Google Scholar] [CrossRef]
- Bjornerfeldt, S.; Webster, M.T.; Vila, C. Relaxation of selective constraint on dog mitochondrial DNA following domestication. Genome Res. 2006, 16, 990–994. [Google Scholar] [CrossRef]
- MacEachern, S.; McEwan, J.; McCulloch, A.; Mather, A.; Savin, K.; Goddard, M. Molecular evolution of the Bovini tribe (Bovidae, Bovinae): Is there evidence of rapid evolution or reduced selective constraint in Domestic cattle? BMC Genom. 2009, 10, 179. [Google Scholar] [CrossRef]
- Wang, Z.; Yonezawa, T.; Liu, B.; Ma, T.; Shen, X.; Su, J.; Guo, S.; Hasegawa, M.; Liu, J. Domestication Relaxed Selective Constraints on the Yak Mitochondrial Genome. Mol. Biol. Evol. 2011, 28, 1553–1556. [Google Scholar] [CrossRef]
- Wang, Y.; Shen, Y.; Feng, C.; Zhao, K.; Song, Z.; Zhang, Y.; Yang, L.; He, S. Mitogenomic perspectives on the origin of Tibetan loaches and their adaptation to high altitude. Sci. Rep. 2016, 6, 29690. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Lv, Y.; Wen, Z.; Bian, C.; Zhang, X.; Guo, S.; Shi, Q.; Li, D. The complete mitochondrial genome of the intertidal spider (Desis jiaxiangi) provides novel insights into the adaptive evolution of the mitogenome and the evolution of spiders. BMC Ecol. Evol. 2021, 21, 72. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Wang, X.; Liu, G.; Wu, X.; Wei, Q.; Sun, G.; Mei, X.; Dong, Y.; Sha, W.; Zhang, H. Adaptability and Evolution of Gobiidae: A Genetic Exploration. Animals 2022, 12, 1741. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, A. Comparative mitochondrial genomics within and among species of killifish. BMC Evol. Biol. 2009, 9, 11. [Google Scholar] [CrossRef]
- Jensen, E.L.; Gaughran, S.J.; Fusco, N.A.; Poulakakis, N.; Tapia, W.; Sevilla, C.; Málaga, J.; Mariani, C.; Gibbs, J.P.; Caccone, A. The Galapagos giant tortoise Chelonoidis phantasticus is not extinct. Commun. Biol. 2022, 5, 546. [Google Scholar] [CrossRef]
- Cayot, L.J.; Gibbs, J.P.; Tapia, W.; Caccone, A. Chelonoidis vicina; The IUCN Red List of Threatened Species: Cambridge, UK, 2018. [Google Scholar]
- MacFarland, C.G.; Villa, J.; Toro, B. The Galápagos giant tortoises (Geochelone elephantopus) Part I: Status of the surviving populations. Biol. Conserv. 1974, 6, 118–133. [Google Scholar] [CrossRef]
- Márquez, C.; Wiedenfeld, D.; Snell, H.; Fritts, T.; Flyway Belen, M.; MacFarland, C.; Tapia, W.; Naranjo, S. Estado actual de las poblaciones de tortugas terrestres gigantes (Geochelone spp., Chelonia: Testudinae) en las islas Galápagos. Ecol. Apl. 2004, 3, 98–111. [Google Scholar] [CrossRef][Green Version]
- Pritchard, P. The Galapagos Tortoises: Nomenclatural and Survival Status; Chelonian Research Foundation: Lunenburg, MA, USA, 1996. [Google Scholar]
- Swingland, I. Geochelone elephantopus. Galapagos giant tortoises. In The Conservation Biology of Tortoises; IUCN: Gland, Switzerland, 1989; pp. 24–28. [Google Scholar]
- Poulakakis, N.; Miller, J.M.; Jensen, E.L.; Beheregaray, L.B.; Russello, M.A.; Glaberman, S.; Boore, J.; Caccone, A. Colonization history of Galapagos giant tortoises: Insights from mitogenomes support the progression rule. J. Zool. Syst. Evol. Res. 2020, 58, 1262–1275. [Google Scholar] [CrossRef]
- Donath, A.; Juehling, F.; Al-Arab, M.; Bernhart, S.H.; Reinhardt, F.; Stadler, P.F.; Middendorf, M.; Bernt, M. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 2019, 47, 10543–10552. [Google Scholar] [CrossRef]
- Lohse, M.; Drechsel, O.; Bock, R. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Wang, N.; Tan, J.; Wang, R.; Yang, Y.; Li, G.; Guan, H.; Zheng, Y.; Shi, X.; Ye, R.; et al. Comprehensive codon usage analysis of porcine deltacoronavirus. Mol. Phylogenet. Evol. 2019, 141, 106618. [Google Scholar] [CrossRef] [PubMed]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Sun, C.H.; Liu, H.Y.; Min, X.; Lu, C.H. Mitogenome of the little owl Athene noctua and phylogenetic analysis of Strigidae. Int. J. Biol. Macromol. 2020, 151, 924–931. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Angione, C.; Costanza, J.; Carapezza, G.; Lió, P.; Nicosia, G. Analysis and design of molecular machines. Theor. Comput. Sci. 2015, 599, 102–117. [Google Scholar] [CrossRef]
- Hüttemann, M.; Helling, S.; Sanderson, T.H.; Sinkler, C.; Samavati, L.; Mahapatra, G.; Varughese, A.; Lu, G.; Liu, J.; Ramzan, R.; et al. Regulation of mitochondrial respiration and apoptosis through cell signaling: Cytochrome c oxidase and cytochrome c in ischemia/reperfusion injury and inflammation. Biochim. Biophys. Acta Bioenerg. 2012, 1817, 598–609. [Google Scholar] [CrossRef] [PubMed]
- Koch, R.E.; Buchanan, K.L.; Casagrande, S.; Crino, O.; Dowling, D.K.; Hill, G.E.; Hood, W.R.; McKenzie, M.; Mariette, M.M.; Noble, D.W.A.; et al. Integrating Mitochondrial Aerobic Metabolism into Ecology and Evolution. Trends Ecol. Evol. 2021, 36, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Huang, A.; He, Z.; Ao, L.; Ge, F.; Fan, X.; Zeng, B.; Yang, M.; Yang, D.; Ni, Q.; et al. Complete Mitogenomes of Polypedates Tree Frogs Unveil Gene Rearrangement and Concerted Evolution within Rhacophoridae. Animals 2022, 12, 2449. [Google Scholar] [CrossRef]
- Kono, N.; Tomita, M.; Arakawa, K. Accelerated Laboratory Evolution Reveals the Influence of Replication on the GC Skew in Escherichia coli. Genome Biol. Evol. 2018, 10, 3110–3117. [Google Scholar] [CrossRef]
- Ma, B.; Li, Z.; Lv, Y.; Zixuan, E.; Fang, J.; Ren, C.; Luo, P.; Hu, C. Analysis of Complete Mitochondrial Genome of Bohadschia argus (Jaeger, 1833) (Aspidochirotida, Holothuriidae). Animals 2022, 12, 1437. [Google Scholar] [CrossRef]
- Fourdrilis, S.; Martins, A.M.d.F.; Backeljau, T. Relation between mitochondrial DNA hyperdiversity, mutation rate and mitochondrial genome evolution in Melarhaphe neritoides (Gastropoda: Littorinidae) and other Caenogastropoda. Sci. Rep. 2018, 8, 17964. [Google Scholar] [CrossRef]
- Andrieux, L.O.; Arenales, D.T. Whole-genome identification of neutrally evolving pseudogenes using the evolutionary measure dN/dS. Methods Mol. Biol. 2014, 1167, 75–85. [Google Scholar] [CrossRef]
- Xia, T.; Zhang, H.; Zhang, L.; Yang, X.; Sun, G.; Chen, J.; Xu, D.; Zhao, C. Comparative and evolutionary analysis of the reptilian hedgehog gene family (Shh, Dhh, and Ihh). PeerJ 2019, 7, e7613. [Google Scholar] [CrossRef]
- Wang, X.B.; Zhou, S.Y.; Wu, X.Y.; Wei, Q.G.; Shang, Y.Q.; Sun, G.L.; Mei, X.S.; Dong, Y.H.; Sha, W.L.; Zhang, H.H. High-altitude adaptation in vertebrates as revealed by mitochondrial genome analyses. Ecol. Evol. 2021, 11, 15077–15084. [Google Scholar] [CrossRef]
- Hong, Y.H.; Huang, H.M.; Wu, L.; Storey, K.B.; Zhang, J.Y.; Zhang, Y.P.; Yu, D.N. Characterization of Two Mitogenomes of Hyla sanchiangensis (Anura: Hylidae), with Phylogenetic Relationships and Selection Pressure Analyses of Hylidae. Animals 2023, 13, 1593. [Google Scholar] [CrossRef]
- Lenaz, G.; Fato, R.; Genova, M.L.; Bergamini, C.; Bianchi, C.; Biondi, A. Mitochondrial Complex I: Structural and functional aspects. Biochim. Biophys. Acta 2006, 1757, 1406–1420. [Google Scholar] [CrossRef] [PubMed]
- Kadenbach, B. Complex IV—The regulatory center of mitochondrial oxidative phosphorylation. Mitochondrion 2021, 58, 296–302. [Google Scholar] [CrossRef] [PubMed]
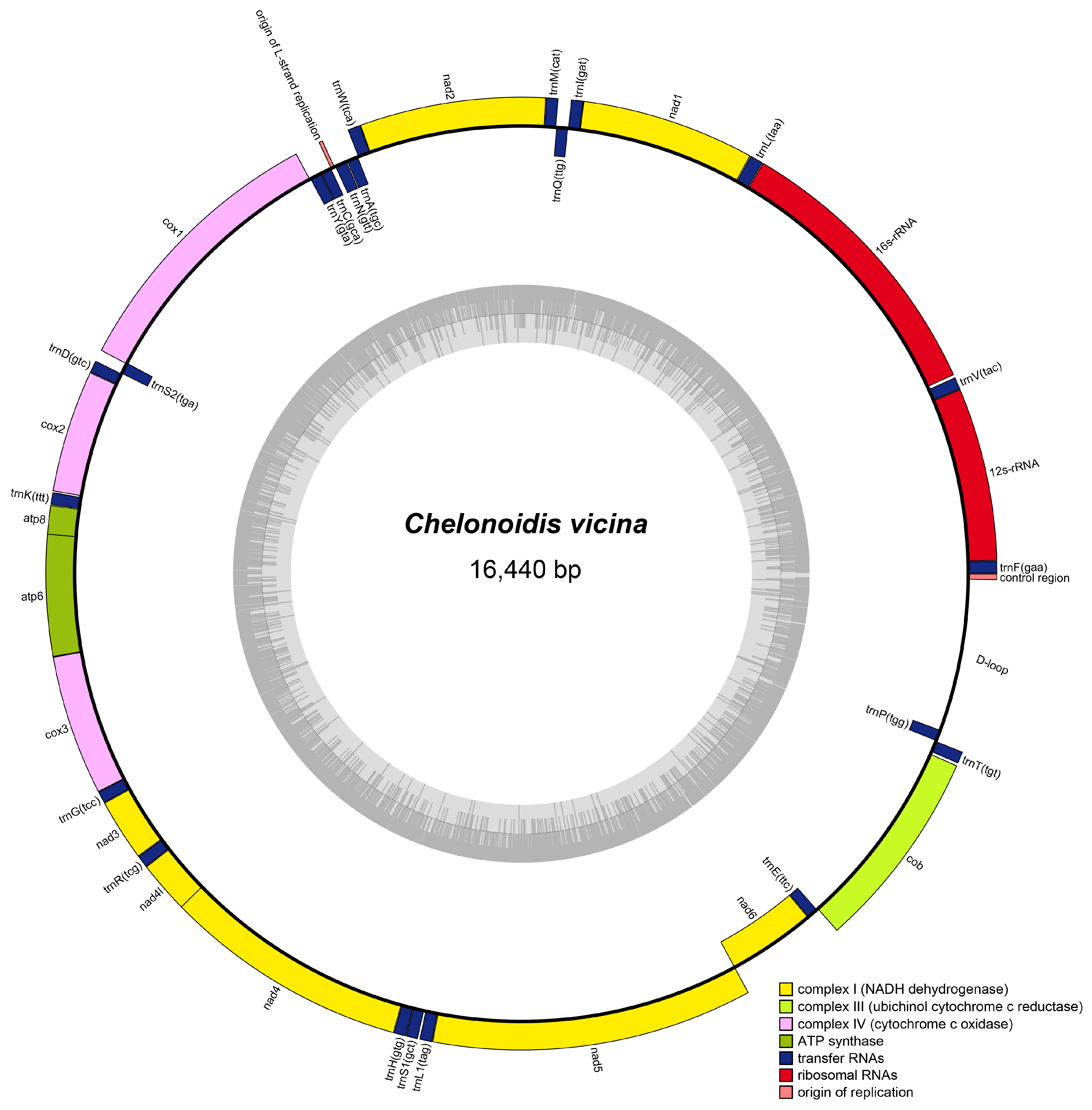
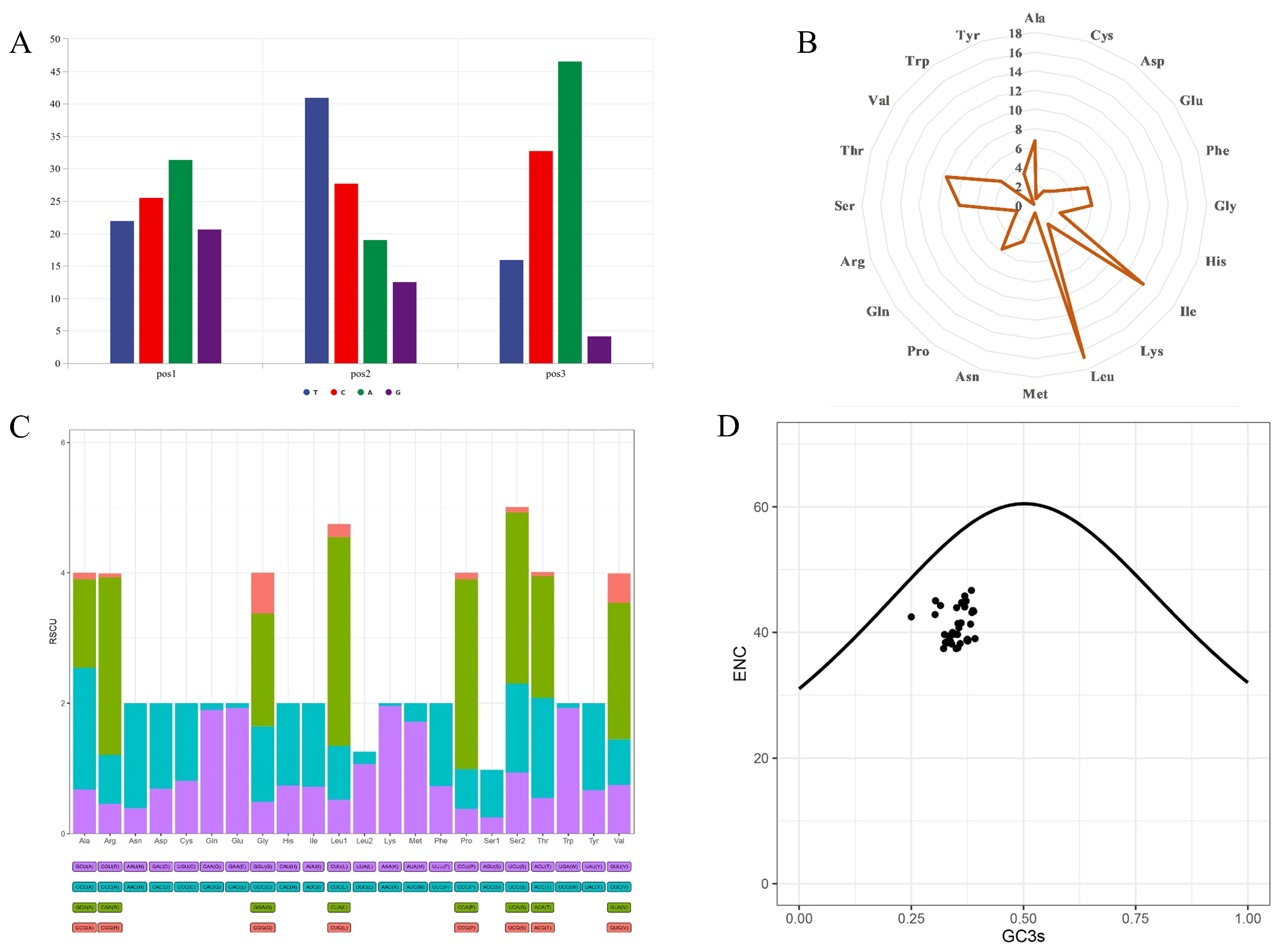

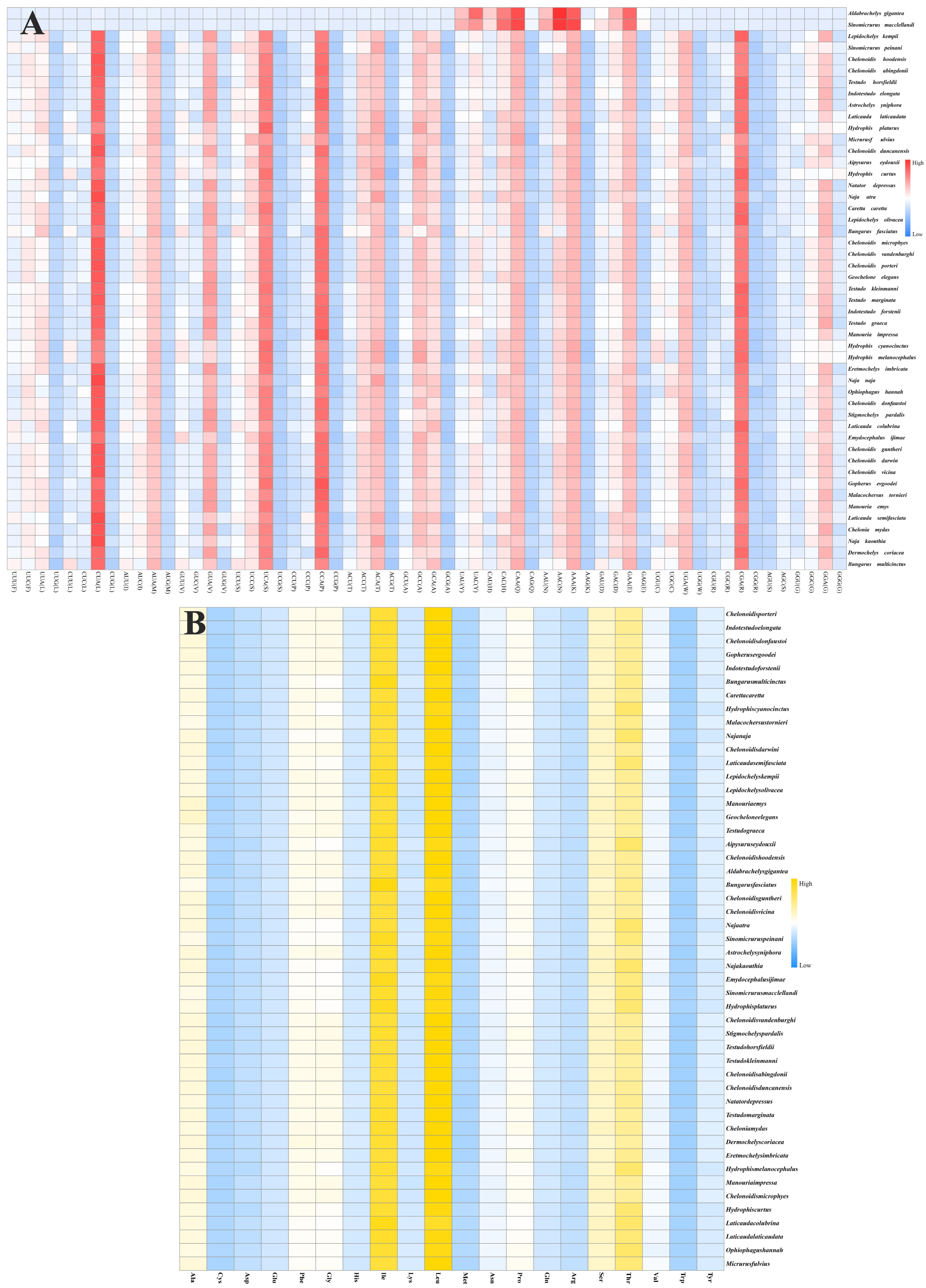
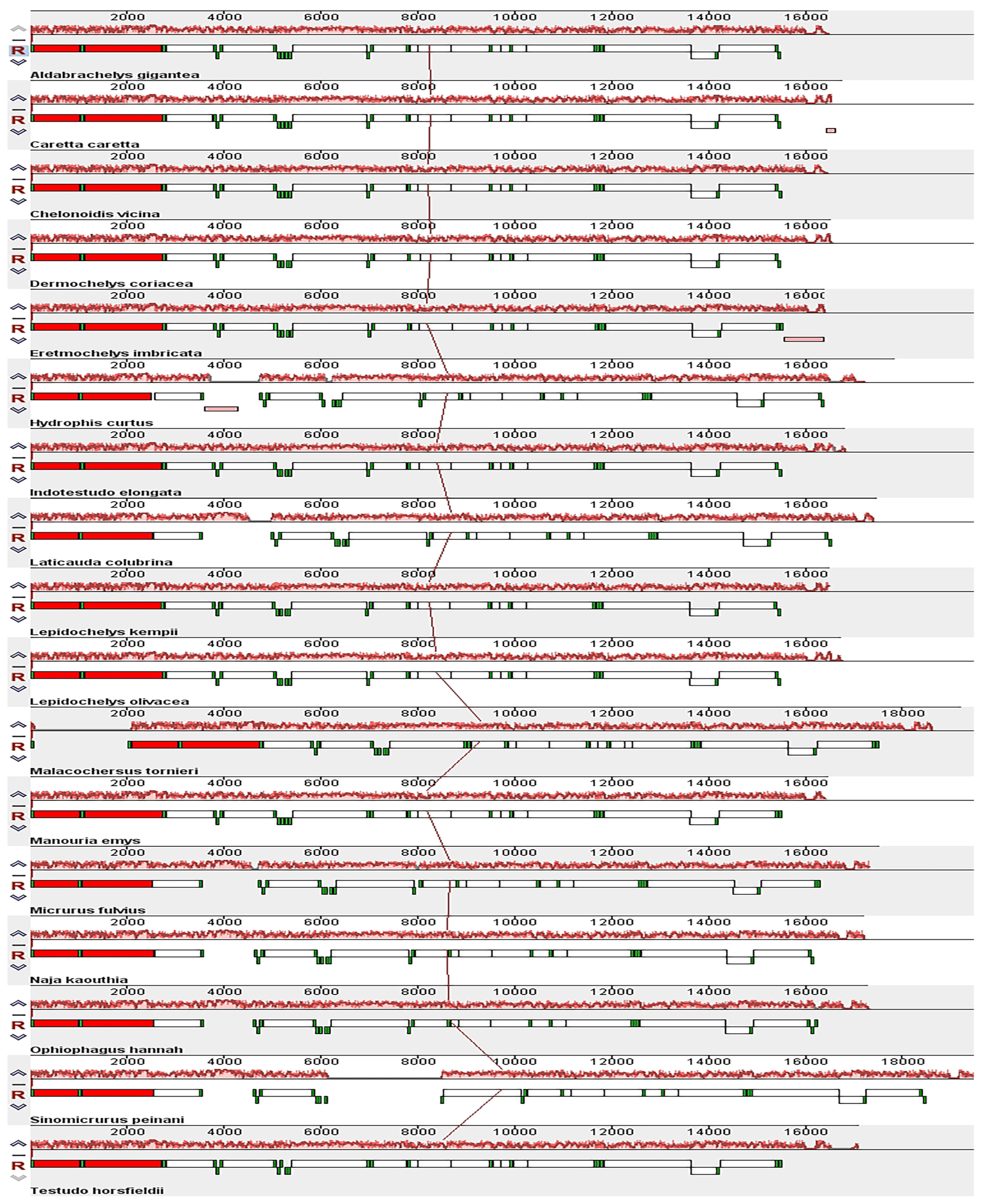
| Gene | Start | Stop | Strand | Length | Intergenic Nucleotide | Star | Stop |
|---|---|---|---|---|---|---|---|
| tRNAPhe | 1 | 70 | + | 70 | 0 | ||
| 12s rRNA | 71 | 1039 | + | 969 | 0 | ||
| tRNAVal | 1040 | 1110 | + | 71 | 14 | ||
| 16s rRNA | 1125 | 2717 | + | 1593 | 1 | ||
| tRNALeu | 2719 | 2794 | + | 76 | 1 | ||
| ND1 | 2796 | 3767 | + | 972 | −1 | ATG | TAG |
| tRNAIle | 3767 | 3836 | + | 70 | −1 | ||
| tRNAGln | 3836 | 3906 | − | 71 | −1 | ||
| tRNAMet | 3906 | 3974 | + | 69 | 0 | ||
| ND2 | 3975 | 5015 | + | 1041 | −2 | ATG | TAG |
| tRNATrp | 5014 | 5088 | + | 75 | 1 | ||
| tRNAAla | 5090 | 5158 | − | 69 | 2 | ||
| tRNAAsn | 5161 | 5234 | − | 74 | 3 | ||
| tRNACys | 5261 | 5326 | − | 66 | 26 | ||
| tRNATyr | 5327 | 5397 | − | 71 | 1 | ||
| COX1 | 5399 | 6946 | + | 1548 | 3 | GTG | AGG |
| tRNASer | 6938 | 7008 | − | 71 | 0 | ||
| tRNAAsp | 7009 | 7078 | + | 70 | 0 | ||
| COX2 | 7079 | 7765 | + | 687 | 5 | ATG | TAA |
| tRNALys | 7771 | 7840 | + | 70 | 1 | ||
| ATP8 | 7842 | 8006 | + | 165 | −10 | ATG | TAA |
| ATP6 | 7997 | 8680 | + | 684 | −1 | ATG | TAA |
| COX3 | 8680 | 9464 | + | 785 | −1 | ATG | TA |
| tRNAGly | 9464 | 9531 | + | 68 | 0 | ||
| ND3 | 9532 | 9883 | + | 180 | 0 | ATG | TA |
| tRNAArg | 9882 | 9951 | + | 70 | 1 | ||
| ND4L | 9952 | 10,248 | + | 297 | 0 | ATG | TAA |
| ND4 | 10,242 | 11,619 | + | 1378 | −7 | ATG | T |
| tRNAHis | 11,620 | 11,689 | + | 70 | 0 | ||
| tRNASer | 11,690 | 11,755 | + | 66 | 0 | ||
| tRNALeu | 11,765 | 11,836 | + | 72 | 9 | ||
| ND5 | 11,837 | 13,633 | + | 1797 | 0 | ATG | TAA |
| ND6 | 13,629 | 14,153 | − | 525 | −5 | ATG | AGG |
| tRNAGlu | 14,154 | 14,221 | − | 68 | 0 | ||
| Cytb | 14,226 | 15,357 | + | 1132 | 4 | ATG | TA |
| tRNAThr | 15,370 | 15,438 | + | 69 | 2 | ||
| tRNAPro | 15,440 | 15,508 | − | 69 | 1 |
| C. vicina | Size | A % | T % | G % | C % | AT % | GC % | AT Skew | GC Skew |
|---|---|---|---|---|---|---|---|---|---|
| mtDNA | 16,440 | 34.83 | 24.60 | 12.57 | 28.00 | 59.43 | 40.57 | 0.17 | −0.38 |
| PCGs | 11,340 | 34.02 | 24.87 | 11.34 | 29.77 | 58.89 | 41.11 | 0.16 | −0.45 |
| tRNAs | 1545 | 35.86 | 25.63 | 15.08 | 23.43 | 61.49 | 38.51 | 0.17 | −0.22 |
| rRNAs | 2562 | 38.37 | 20.07 | 16.74 | 24.82 | 58.44 | 41.56 | 0.31 | −0.19 |
| D-Loop | 932 | 34.44 | 31.97 | 11.92 | 21.67 | 66.41 | 33.59 | 0.04 | −0.29 |
| Spcies | T(U)% | A% | AT% | AT Skew | G% | C% | GC% | GC Skew |
|---|---|---|---|---|---|---|---|---|
| Bungarus fasciatus | 31.3 | 33.5 | 64.8 | 0.034 | 11.2 | 24 | 35.2 | −0.362 |
| Bungarus multicinctus | 29.9 | 32.2 | 62.1 | 0.037 | 12.1 | 25.8 | 37.9 | −0.362 |
| Micrurus fulvius | 27.7 | 30.8 | 58.5 | 0.053 | 13.0 | 28.5 | 41.5 | −0.373 |
| Naja naja | 26.2 | 32.1 | 58.3 | 0.102 | 13.2 | 28.5 | 41.7 | −0.367 |
| Naja kaouthia | 26.2 | 32.2 | 58.3 | 0.103 | 13.2 | 28.5 | 41.7 | −0.368 |
| Naja atra | 26.2 | 32.2 | 58.4 | 0.101 | 13.1 | 28.5 | 41.6 | −0.371 |
| Ophiophagus hannah | 26.1 | 32.7 | 58.8 | 0.113 | 12.1 | 29.1 | 41.2 | −0.413 |
| Sinomicrurus peinani | 29.3 | 32.4 | 61.7 | 0.051 | 11.8 | 26.5 | 38.3 | −0.383 |
| Sinomicrurus macclellandi | 27.7 | 32.0 | 59.6 | 0.072 | 12.2 | 28.2 | 40.4 | −0.397 |
| Aldabrachelys gigantea | 26.4 | 32.8 | 59.2 | 0.107 | 12.4 | 28.4 | 40.8 | −0.392 |
| Chelonoidis microphyes | 26.4 | 32.3 | 58.8 | 0.100 | 12.5 | 28.7 | 41.2 | −0.394 |
| Chelonoidis vandenburghi | 26.4 | 32.4 | 58.8 | 0.100 | 12.4 | 28.7 | 41.2 | −0.396 |
| Chelonoidis guntheri | 26.4 | 32.4 | 58.8 | 0.109 | 12.4 | 28.7 | 41.2 | −0.396 |
| Chelonoidis donfaustoi | 26.4 | 32.4 | 58.8 | 0.102 | 12.4 | 28.8 | 41.2 | −0.397 |
| Chelonoidis darwini | 26.4 | 32.5 | 58.8 | 0.104 | 12.4 | 28.8 | 41.2 | −0.399 |
| C. vicina | 26.4 | 32.4 | 58.8 | 0.108 | 12.5 | 28.7 | 41.2 | −0.395 |
| Chelonoidis porteri | 26.4 | 32.4 | 58.8 | 0.108 | 12.4 | 28.8 | 41.2 | −0.399 |
| Chelonoidis duncanensis | 26.3 | 32.5 | 58.8 | 0.104 | 12.4 | 28.8 | 41.2 | −0.399 |
| Chelonoidis hoodensis | 26.4 | 32.4 | 58.8 | 0.102 | 12.4 | 28.8 | 41.2 | −0.397 |
| Chelonoidis abingdonii | 26.4 | 32.4 | 58.8 | 0.102 | 12.4 | 28.8 | 41.2 | −0.397 |
| Gopherus evgoodei | 27.4 | 32.3 | 59.7 | 0.082 | 12.9 | 27.4 | 40.3 | −0.358 |
| Geochelone elegans | 25.6 | 32.0 | 57.6 | 0.112 | 13.2 | 29.3 | 42.4 | −0.380 |
| Malacochersus tornieri | 27.7 | 33.0 | 60.7 | 0.087 | 11.8 | 27.5 | 39.3 | −0.399 |
| Testudo kleinmanni | 27.9 | 32.3 | 60.2 | 0.073 | 12.5 | 27.2 | 39.8 | −0.370 |
| Testudo marginata | 27.8 | 32.3 | 60.1 | 0.076 | 12.7 | 27.3 | 39.9 | −0.366 |
| Testudo horsfieldii | 27.5 | 32 | 59.5 | 0.077 | 12.8 | 27.7 | 40.5 | −0.369 |
| Indotestudo forstenii | 27.8 | 32.8 | 60.7 | 0.082 | 12.1 | 27.2 | 39.3 | −0.382 |
| Stigmochelys pardalis | 27.5 | 32.5 | 60.1 | 0.084 | 12.6 | 27.4 | 39.9 | −0.371 |
| Manouria emys | 27.5 | 31.7 | 59.2 | 0.069 | 13.3 | 27.5 | 40.8 | −0.350 |
| Testudo graeca | 28.3 | 32.9 | 61.2 | 0.074 | 12.2 | 26.6 | 38.8 | −0.372 |
| Indotestudo elongata | 28.1 | 33.0 | 61.1 | 0.079 | 12.0 | 26.9 | 38.9 | −0.384 |
| Manouria impressa | 28.0 | 31.6 | 59.6 | 0.059 | 13.3 | 27.1 | 40.4 | −0.341 |
| Astrochelys yniphora | 27.5 | 32.5 | 60.1 | 0.083 | 12.5 | 27.4 | 39.9 | −0.371 |
| Laticauda semifasciata | 27.3 | 31.9 | 59.2 | 0.079 | 13.2 | 27.6 | 40.8 | −0.352 |
| Laticauda colubrina | 29.1 | 33.1 | 62.2 | 0.065 | 12.1 | 25.7 | 37.8 | −0.360 |
| Laticauda laticaudata | 28.0 | 32.1 | 60.2 | 0.068 | 12.8 | 27.1 | 39.8 | −0.358 |
| Aipysurus eydouxii | 26.8 | 31.9 | 58.7 | 0.086 | 12.7 | 28.6 | 41.3 | −0.384 |
| Emydocephalus ijimae | 28.0 | 31.9 | 59.9 | 0.066 | 12.6 | 27.6 | 40.1 | −0.373 |
| Hydrophis curtus | 28.3 | 31.0 | 59.3 | 0.046 | 13.5 | 27.2 | 40.7 | −0.337 |
| Hydrophis cyanocinctus | 28.2 | 31.4 | 59.6 | 0.054 | 13.0 | 27.3 | 40.4 | −0.354 |
| Gene | Model | 2Δlnl | p-Value | Positively Selected Sites (BEB Analysis) |
|---|---|---|---|---|
| COX2 | Model A vs. Null Mode | 0 | 1 | 5 T 0.986 *, 16 T 0.985 * |
| COX3 | 0 | 1 | 22 M 0.955 *, 175 A 0.962 * | |
| CYtb | 0 | 1 | 324 T 0.994 ** | |
| ND3 | 0 | 1 | 14 S 0.998 **, 16 L 0.975 * | |
| ND4 | 0 | 1 | 30 Y 0.975 *, 431 I 0.994** | |
| ND4L | 0 | 1 | 15 T 0.990 *,56 Q 0.962 * | |
| ND5 | 0 | 1 | 535 S 0.970 * | |
| ND6 | 0 | 1 | 130 G 0.977 * |
| Gene | Model Compared | |2ΔlnL| | p-Value | ωAR | ωTR |
|---|---|---|---|---|---|
| ATP6 | M2 vs. M0 | 0.635 | 0.426 | 0.095 | 0.085 |
| ATP8 | 0.049 | 0.825 | 0.209 | 0.198 | |
| COX1 | 0.518 | 0.472 | 0.017 | 0.015 | |
| COX2 | 6.322 | 0.012 * | 0.024 | 0.039 | |
| COX3 | 0.021 | 0.885 | 0.033 | 0.034 | |
| Cytb | 0.087 | 0.768 | 0.044 | 0.046 | |
| ND1 | 8.989 | 0.003 ** | 0.049 | 0.035 | |
| ND2 | 0.133 | 0.715 | 0.067 | 0.064 | |
| ND3 | 10.256 | 0.002 ** | 0.060 | 0.112 | |
| ND4 | 3.462 | 0.063 | 0.061 | 0.050 | |
| ND4L | 0.017 | 0.898 | 0.057 | 0.055 | |
| ND5 | 0.609 | 0.435 | 0.071 | 0.066 | |
| ND6 | 2.074 | 0.150 | 0.090 | 0.116 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Wang, X.; Wu, X.; Shang, Y.; Wei, Q.; Cai, H.; Sha, W.; Qi, Y.; Liu, S.; Zhang, H. Terrestrial Adaptation in Chelonoidis vicina as Revealed Based on Analysis of the Complete Mitochondrial Genome. Genes 2025, 16, 173. https://doi.org/10.3390/genes16020173
Chen Y, Wang X, Wu X, Shang Y, Wei Q, Cai H, Sha W, Qi Y, Liu S, Zhang H. Terrestrial Adaptation in Chelonoidis vicina as Revealed Based on Analysis of the Complete Mitochondrial Genome. Genes. 2025; 16(2):173. https://doi.org/10.3390/genes16020173
Chicago/Turabian StyleChen, Yao, Xibao Wang, Xiaoyang Wu, Yongquan Shang, Qinguo Wei, Haotian Cai, Weilai Sha, Yan Qi, Shuli Liu, and Honghai Zhang. 2025. "Terrestrial Adaptation in Chelonoidis vicina as Revealed Based on Analysis of the Complete Mitochondrial Genome" Genes 16, no. 2: 173. https://doi.org/10.3390/genes16020173
APA StyleChen, Y., Wang, X., Wu, X., Shang, Y., Wei, Q., Cai, H., Sha, W., Qi, Y., Liu, S., & Zhang, H. (2025). Terrestrial Adaptation in Chelonoidis vicina as Revealed Based on Analysis of the Complete Mitochondrial Genome. Genes, 16(2), 173. https://doi.org/10.3390/genes16020173





