Epigenetic and Transcriptomic Alterations of Protein Aggregation-Linked Genes in Suicide: A Pilot Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Brain Region Selection
2.3. DNA Methylation Reanalysis and Gene Selection
2.4. RNA Isolation and Gene Expression
2.5. Statistical Analysis
3. Results
3.1. DNA Methylation Reanalysis in the Hippocampus
3.2. Gene Expression
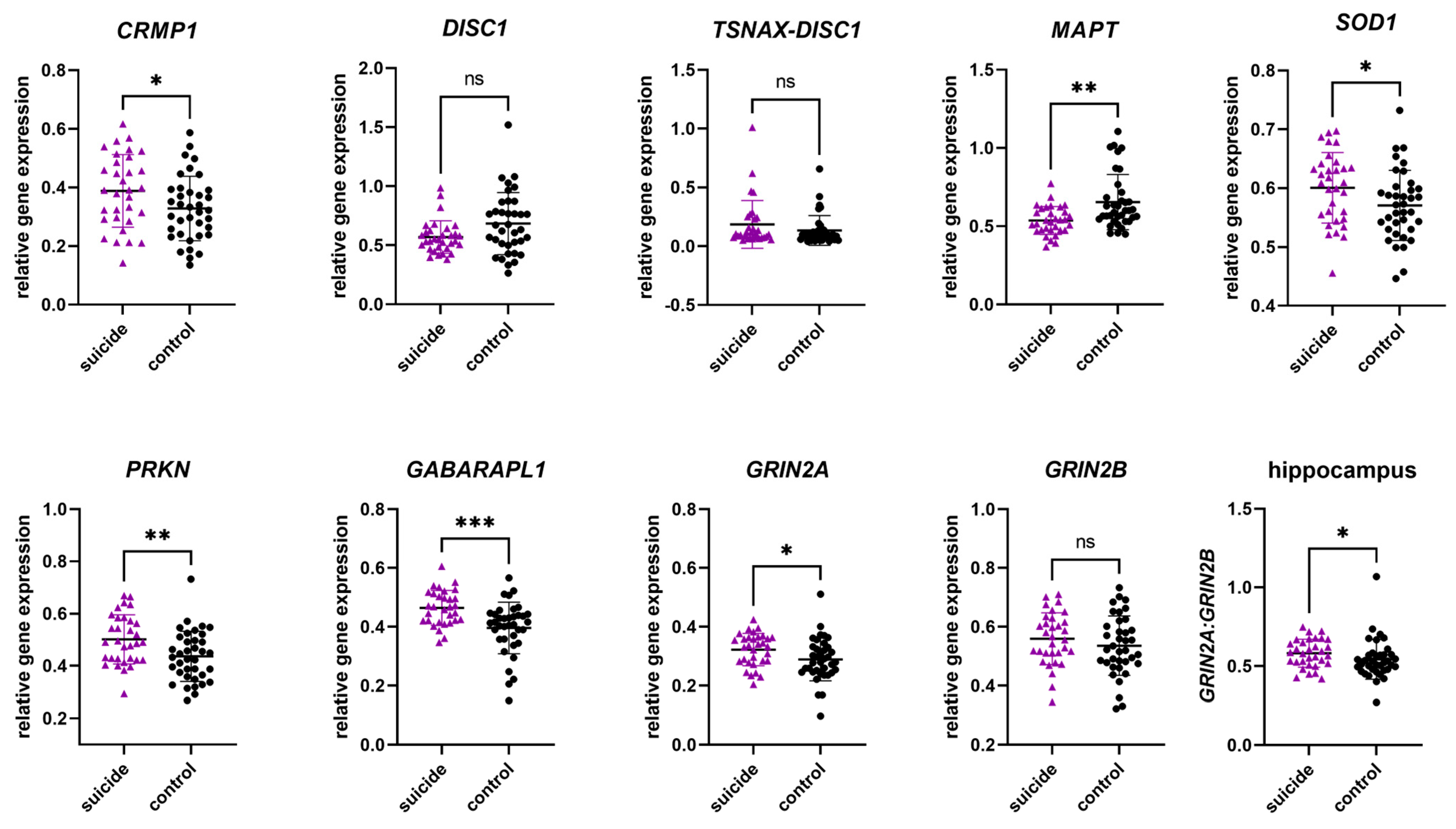
3.2.1. Gene Expression in the Hippocampus
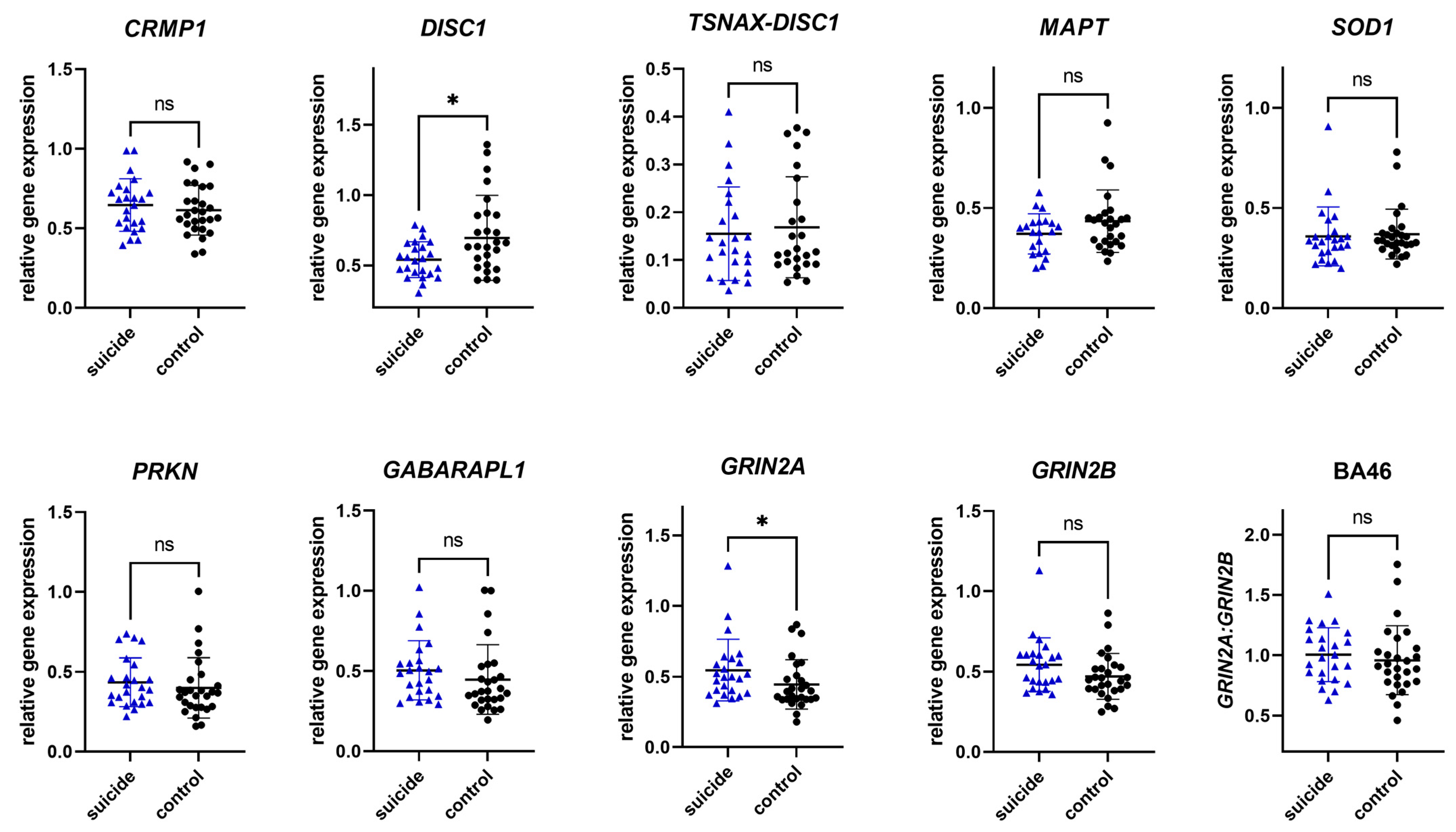
3.2.2. Gene Expression in Brodmann Area 46
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s disease |
| ALS | amyotrophic lateral sclerosis |
| DISC1 | disrupted-in-schizophrenia 1 |
| DMC | differentially methylated cytosine |
| DTNBP1 | dystrobrevin-binding protein 1 |
| FTD | frontotemporal dementia |
| GWAS | genome-wide association studies |
| HPA | hypothalamic–pituitary–adrenal |
| MDD | major depressive disorder |
| mDNA | DNA methylation |
| NMDAR | N-methyl-D-aspartate receptor |
| NPAS3 | neuronal PAS domain protein 3 |
| PD | Parkinson’s disease |
| PMI | Post-mortem interval |
| TRIOBP | TRIO and F-actin binding protein |
References
- WHO. Suicide Worldwide in 2019: Global Health Estimates; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- Abou Chahla, M.N.; Khalil, M.I.; Comai, S.; Brundin, L.; Erhardt, S.; Guillemin, G.J. Biological Factors Underpinning Suicidal Behaviour: An Update. Brain Sci. 2023, 13, 505. [Google Scholar] [CrossRef]
- Turecki, G.; Brent, D.A.; Gunnell, D.; O’Connor, R.C.; Oquendo, M.A.; Pirkis, J.; Stanley, B.H. Suicide and suicide risk. Nat. Rev. Dis. Primers 2019, 5, 74. [Google Scholar] [CrossRef]
- Chiu, C.-C.; Liu, H.-C.; Li, W.-H.; Tsai, S.-Y.; Chen, C.-C.; Kuo, C.-J. Incidence, risk and protective factors for suicide mortality among patients with major depressive disorder. Asian J. Psychiatry 2023, 80, 103399. [Google Scholar] [CrossRef]
- Oquendo, M.A.; Porras-Segovia, A. Barriers for the Research, Prevention, and Treatment of Suicidal Behavior. Curr. Top. Behav. Neurosci. 2020, 46, 25–40. [Google Scholar] [CrossRef]
- Costanza, A.; D’Orta, I.; Perroud, N.; Burkhardt, S.; Malafosse, A.; Mangin, P.; La Harpe, R. Neurobiology of suicide: Do biomarkers exist? Int. J. Leg. Med. 2014, 128, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Heath, A.C.; Bucholz, K.K.; Nelson, E.C.; Glowinski, A.L.; Goldberg, J.; Lyons, M.J.; Tsuang, M.T.; Jacob, T.; True, M.R.; et al. A twin study of genetic and environmental influences on suicidality in men. Psychol. Med. 2002, 32, 11–24. [Google Scholar] [CrossRef]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; de Vries, J.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-wide association studies. Nat. Rev. Methods Primers 2021, 1, 59. [Google Scholar] [CrossRef]
- DiBlasi, E.; Kang, J.; Docherty, A.R. Genetic contributions to suicidal thoughts and behaviors. Psychol. Med. 2021, 51, 2148–2155. [Google Scholar] [CrossRef]
- Turecki, G. The molecular bases of the suicidal brain. Nat. Rev. Neurosci. 2014, 15, 802–816. [Google Scholar] [CrossRef] [PubMed]
- Kouter, K.; Videtic Paska, A. ‘Omics’ of suicidal behaviour: A path to personalised psychiatry. World J. Psychiatry 2021, 11, 774–790. [Google Scholar] [CrossRef]
- Lutz, P.E.; Mechawar, N.; Turecki, G. Neuropathology of suicide: Recent findings and future directions. Mol. Psychiatry 2017, 22, 1395–1412. [Google Scholar] [CrossRef]
- van Heeringen, K.; Mann, J.J. The neurobiology of suicide. Lancet Psychiatry 2014, 1, 63–72. [Google Scholar] [CrossRef]
- Roy, B.; Dwivedi, Y. Understanding epigenetic architecture of suicide neurobiology: A critical perspective. Neurosci. Biobehav. Rev. 2017, 72, 10–27. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, G.; Heard, E. Advances in epigenetics link genetics to the environment and disease. Nature 2019, 571, 489–499. [Google Scholar] [CrossRef]
- Zhang, T.Y.; Labonté, B.; Wen, X.L.; Turecki, G.; Meaney, M.J. Epigenetic Mechanisms for the Early Environmental Regulation of Hippocampal Glucocorticoid Receptor Gene Expression in Rodents and Humans. Neuropsychopharmacology 2013, 38, 111–123. [Google Scholar] [CrossRef]
- Brivio, P.; Sbrini, G.; Tarantini, L.; Parravicini, C.; Gruca, P.; Lason, M.; Litwa, E.; Favero, C.; Riva, M.A.; Eberini, I.; et al. Stress Modifies the Expression of Glucocorticoid-Responsive Genes by Acting at Epigenetic Levels in the Rat Prefrontal Cortex: Modulatory Activity of Lurasidone. Int. J. Mol. Sci. 2021, 22, 6197. [Google Scholar] [CrossRef]
- Labonte, B.; Yerko, V.; Gross, J.; Mechawar, N.; Meaney, M.J.; Szyf, M.; Turecki, G. Differential Glucocorticoid Receptor Exon 1B, 1C, and 1H Expression and Methylation in Suicide Completers with a History of Childhood Abuse. Biol. Psychiatry 2012, 72, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Policicchio, S.; Washer, S.; Viana, J.; Iatrou, A.; Burrage, J.; Hannon, E.; Turecki, G.; Kaminsky, Z.; Mill, J.; Dempster, E.L.; et al. Genome-wide DNA methylation meta-analysis in the brains of suicide completers. Transl. Psychiatry 2020, 10, 69. [Google Scholar] [CrossRef]
- Kimbrel, N.A.; Garrett, M.E.; Evans, M.K.; Mellows, C.; Dennis, M.F.; Hair, L.P.; Hauser, M.A.; Ashley-Koch, A.E.; Beckham, J.C. Large epigenome-wide association study identifies multiple novel differentially methylated CpG sites associated with suicidal thoughts and behaviors in veterans. Front. Psychiatry 2023, 14, 1145375. [Google Scholar] [CrossRef] [PubMed]
- Kouter, K.; Zupanc, T.; Videtič Paska, A. Genome-wide DNA methylation in suicide victims revealing impact on gene expression. J. Affect. Disord. 2019, 253, 419–425. [Google Scholar] [CrossRef]
- Bradshaw, N.J.; Korth, C. Protein misassembly and aggregation as potential convergence points for non-genetic causes of chronic mental illness. Mol. Psychiatry 2019, 24, 936–951. [Google Scholar] [CrossRef]
- Hui, K.K.; Endo, R.; Sawa, A.; Tanaka, M. A Perspective on the Potential Involvement of Impaired Proteostasis in Neuropsychiatric Disorders. Biol. Psychiatry 2022, 91, 335–345. [Google Scholar] [CrossRef]
- Ochneva, A.; Zorkina, Y.; Abramova, O.; Pavlova, O.; Ushakova, V.; Morozova, A.; Zubkov, E.; Pavlov, K.; Gurina, O.; Chekhonin, V. Protein Misfolding and Aggregation in the Brain: Common Pathogenetic Pathways in Neurodegenerative and Mental Disorders. Int. J. Mol. Sci. 2022, 23, 14498. [Google Scholar] [CrossRef]
- Wu, J.; Wu, J.N.; Chen, T.; Cai, J.; Ren, R. Protein Aggregation and its Affecting Mechanisms in Neurodegenerative Diseases. Neurochem. Int. 2024, 180, 105880. [Google Scholar] [CrossRef] [PubMed]
- Trojsi, F.; Christidi, F.; Migliaccio, R.; Santamaría-García, H.; Santangelo, G. Behavioural and Cognitive Changes in Neurodegenerative Diseases and Brain Injury. Behav. Neurol. 2018, 2018, 4935915. [Google Scholar] [CrossRef] [PubMed]
- van der Linde, R.M.; Dening, T.; Stephan, B.C.; Prina, A.M.; Evans, E.; Brayne, C. Longitudinal course of behavioural and psychological symptoms of dementia: Systematic review. Br. J. Psychiatry J. Ment. Sci. 2016, 209, 366–377. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J. The Role of Neuropsychiatric Symptoms in Research Diagnostic Criteria for Neurodegenerative Diseases. Am. J. Geriatr. Psychiatry Off. J. Am. Assoc. Geriatr. Psychiatry 2021, 29, 375–383. [Google Scholar] [CrossRef]
- Nucifora, L.G.; MacDonald, M.L.; Lee, B.J.; Peters, M.E.; Norris, A.L.; Orsburn, B.C.; Yang, K.; Gleason, K.; Margolis, R.L.; Pevsner, J.; et al. Increased Protein Insolubility in Brains from a Subset of Patients With Schizophrenia. Am. J. Psychiatry 2019, 176, 730–743. [Google Scholar] [CrossRef]
- Samardžija, B.; Juković, M.; Zaharija, B.; Renner, É.; Palkovits, M.; Bradshaw, N.J. Co-Aggregation and Parallel Aggregation of Specific Proteins in Major Mental Illness. Cells 2023, 12, 1848. [Google Scholar] [CrossRef]
- Nucifora, L.G.; Ishizuka, K.; El Demerdash, N.; Lee, B.J.; Imai, M.T.; Ayala-Grosso, C.; Yenokyan, G.; Cascella, N.G.; Lin, S.; Schretlen, D.J.; et al. Protein aggregation identified in olfactory neuronal cells is associated with cognitive impairments in a subset of living schizophrenia patients. Mol. Psychiatry 2025, 30, 3673–3685. [Google Scholar] [CrossRef]
- Zaharija, B.; Bradshaw, N.J. Aggregation of Disrupted in Schizophrenia 1 arises from a central region of the protein. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2024, 130, 110923. [Google Scholar] [CrossRef]
- Korth, C. DISCopathies: Brain disorders related to DISC1 dysfunction. Rev. Neurosci. 2009, 20, 321–330. [Google Scholar] [CrossRef]
- Martínez-Magaña, J.J.; Genis-Mendoza, A.D.; Villatoro Velázquez, J.A.; Bustos-Gamiño, M.; Juárez-Rojop, I.E.; Tovilla-Zarate, C.A.; Sarmiento, E.; Saucedo, E.; Rodríguez-Mayoral, O.; Fleiz-Bautista, C.; et al. Genome-wide association study of psychiatric and substance use comorbidity in Mexican individuals. Sci. Rep. 2021, 11, 6771. [Google Scholar] [CrossRef] [PubMed]
- Saez, E.; Erkoreka, L.; Moreno-Calle, T.; Berjano, B.; Gonzalez-Pinto, A.; Basterreche, N.; Arrue, A. Genetic variables of the glutamatergic system associated with treatment-resistant depression: A review of the literature. World J. Psychiatry 2022, 12, 884–896. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef]
- Mohd Murshid, N.; Aminullah Lubis, F.; Makpol, S. Epigenetic Changes and Its Intervention in Age-Related Neurodegenerative Diseases. Cell. Mol. Neurobiol. 2022, 42, 577–595. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Du, P.; Zhao, Z. Impacts of DNA methylation on Tau protein related genes in the brains of patients with Alzheimer’s disease. Neurosci. Lett. 2021, 763, 136196. [Google Scholar] [CrossRef]
- Ottis, P.; Bader, V.; Trossbach, S.V.; Kretzschmar, H.; Michel, M.; Leliveld, S.R.; Korth, C. Convergence of two independent mental disease genes on the protein level: Recruitment of dysbindin to cell-invasive disrupted-in-schizophrenia 1 aggresomes. Biol. Psychiatry 2011, 70, 604–610. [Google Scholar] [CrossRef]
- Navarro, D.; Marín-Mayor, M.; Gasparyan, A.; García-Gutiérrez, M.S.; Rubio, G.; Manzanares, J. Molecular Changes Associated with Suicide. Int J Mol Sci 2023, 24, 16726. [Google Scholar] [CrossRef]
- Morgane, P.J.; Galler, J.R.; Mokler, D.J. A review of systems and networks of the limbic forebrain/limbic midbrain. Prog. Neurobiol. 2005, 75, 143–160. [Google Scholar] [CrossRef]
- Knierim, J.J. The hippocampus. Curr. Biol. 2015, 25, R1116–R1121. [Google Scholar] [CrossRef]
- Molnár, E. Long-term potentiation in cultured hippocampal neurons. Semin. Cell Dev. Biol. 2011, 22, 506–513. [Google Scholar] [CrossRef] [PubMed]
- Chan, R.C.; Shum, D.; Toulopoulou, T.; Chen, E.Y. Assessment of executive functions: Review of instruments and identification of critical issues. Arch. Clin. Neuropsychol. 2008, 23, 201–216. [Google Scholar] [CrossRef]
- Greene, J.D.; Sommerville, R.B.; Nystrom, L.E.; Darley, J.M.; Cohen, J.D. An fMRI investigation of emotional engagement in moral judgment. Science 2001, 293, 2105–2108. [Google Scholar] [CrossRef]
- Snow, P.J. The Structural and Functional Organization of Cognition. Front. Hum. Neurosci. 2016, 10, 501. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Posit Team. RStudio: Integrated Development Environment for R; Posit Software, PBC: Boston, MA, USA, 2025; Available online: http://www.posit.co/ (accessed on 5 January 2025).
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, Research0034.1. [Google Scholar] [CrossRef]
- Yu-Kemp, H.C.; Kemp, J.P., Jr.; Brieher, W.M. CRMP-1 enhances EVL-mediated actin elongation to build lamellipodia and the actin cortex. J. Cell Biol. 2017, 216, 2463–2479. [Google Scholar] [CrossRef]
- Bader, V.; Tomppo, L.; Trossbach, S.V.; Bradshaw, N.J.; Prikulis, I.; Leliveld, S.R.; Lin, C.Y.; Ishizuka, K.; Sawa, A.; Ramos, A.; et al. Proteomic, genomic and translational approaches identify CRMP1 for a role in schizophrenia and its underlying traits. Hum. Mol. Genet. 2012, 21, 4406–4418. [Google Scholar] [CrossRef]
- Knuesel, I. Reelin-mediated signaling in neuropsychiatric and neurodegenerative diseases. Prog. Neurobiol. 2010, 91, 257–274. [Google Scholar] [CrossRef]
- Soares, D.C.; Carlyle, B.C.; Bradshaw, N.J.; Porteous, D.J. DISC1: Structure, Function, and Therapeutic Potential for Major Mental Illness. ACS Chem. Neurosci. 2011, 2, 609–632. [Google Scholar] [CrossRef] [PubMed]
- Dahoun, T.; Trossbach, S.V.; Brandon, N.J.; Korth, C.; Howes, O.D. The impact of Disrupted-in-Schizophrenia 1 (DISC1) on the dopaminergic system: A systematic review. Transl. Psychiatry 2017, 7, e1015. [Google Scholar] [CrossRef] [PubMed]
- McCartney, D.L.; Walker, R.M.; Morris, S.W.; Anderson, S.M.; Duff, B.J.; Marioni, R.E.; Millar, J.K.; McCarthy, S.E.; Ryan, N.M.; Lawrie, S.M.; et al. Altered DNA methylation associated with a translocation linked to major mental illness. npj Schizophr. 2018, 4, 5. [Google Scholar] [CrossRef]
- Schosser, A.; Gaysina, D.; Cohen-Woods, S.; Chow, P.C.; Martucci, L.; Craddock, N.; Farmer, A.; Korszun, A.; Gunasinghe, C.; Gray, J.; et al. Association of DISC1 and TSNAX genes and affective disorders in the depression case-control (DeCC) and bipolar affective case-control (BACCS) studies. Mol. Psychiatry 2010, 15, 844–849. [Google Scholar] [CrossRef]
- Ruiz-Gabarre, D.; Carnero-Espejo, A.; Ávila, J.; García-Escudero, V. What’s in a Gene? The Outstanding Diversity of MAPT. Cells 2022, 11, 840. [Google Scholar] [CrossRef]
- Caillet-Boudin, M.L.; Buée, L.; Sergeant, N.; Lefebvre, B. Regulation of human MAPT gene expression. Mol. Neurodegener. 2015, 10, 28. [Google Scholar] [CrossRef]
- Tsang, C.K.; Liu, Y.; Thomas, J.; Zhang, Y.; Zheng, X.F.S. Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat. Commun. 2014, 5, 3446. [Google Scholar] [CrossRef] [PubMed]
- Gagliardi, S.; Cova, E.; Davin, A.; Guareschi, S.; Abel, K.; Alvisi, E.; Laforenza, U.; Ghidoni, R.; Cashman, J.R.; Ceroni, M.; et al. SOD1 mRNA expression in sporadic amyotrophic lateral sclerosis. Neurobiol. Dis. 2010, 39, 198–203. [Google Scholar] [CrossRef]
- Djordjevic, V. Superoxide Dismutase in Psychiatric Diseases. In Reactive Oxygen Species; Ahmad, R., Ed.; IntechOpen: London, UK, 2022. [Google Scholar]
- Hattori, N.; Mizuno, Y. Twenty years since the discovery of the parkin gene. J. Neural Transm. 2017, 124, 1037–1054. [Google Scholar] [CrossRef]
- Chan, N.C.; Salazar, A.M.; Pham, A.H.; Sweredoski, M.J.; Kolawa, N.J.; Graham, R.L.; Hess, S.; Chan, D.C. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum. Mol. Genet. 2011, 20, 1726–1737. [Google Scholar] [CrossRef]
- Srivastava, A.; Tang, M.X.; Mejia-Santana, H.; Rosado, L.; Louis, E.D.; Caccappolo, E.; Comella, C.; Colcher, A.; Siderowf, A.; Jennings, D.; et al. The relation between depression and parkin genotype: The CORE-PD study. Park. Relat. Disord. 2011, 17, 740–744. [Google Scholar] [CrossRef]
- Wang, C.; Ko, H.S.; Thomas, B.; Tsang, F.; Chew, K.C.; Tay, S.P.; Ho, M.W.; Lim, T.M.; Soong, T.W.; Pletnikova, O.; et al. Stress-induced alterations in parkin solubility promote parkin aggregation and compromise parkin’s protective function. Hum. Mol. Genet. 2005, 14, 3885–3897. [Google Scholar] [CrossRef] [PubMed]
- Wingo, T.S.; Liu, Y.; Gerasimov, E.S.; Vattathil, S.M.; Wynne, M.E.; Liu, J.; Lori, A.; Faundez, V.; Bennett, D.A.; Seyfried, N.T.; et al. Shared mechanisms across the major psychiatric and neurodegenerative diseases. Nat. Commun. 2022, 13, 4314. [Google Scholar] [CrossRef] [PubMed]
- Moreira, P.I.; Smith, M.A.; Zhu, X.; Nunomura, A.; Castellani, R.J.; Perry, G. Oxidative stress and neurodegeneration. Ann. N. Y. Acad. Sci. 2005, 1043, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Le Grand, J.N.; Chakrama, F.Z.; Seguin-Py, S.; Fraichard, A.; Delage-Mourroux, R.; Jouvenot, M.; Boyer-Guittaut, M. GABARAPL1 (GEC1): Original or copycat? Autophagy 2011, 7, 1098–1107. [Google Scholar] [CrossRef]
- Ye, J.; Zou, G.; Zhu, R.; Kong, C.; Miao, C.; Zhang, M.; Li, J.; Xiong, W.; Wang, C. Structural basis of GABARAP-mediated GABAA receptor trafficking and functions on GABAergic synaptic transmission. Nat. Commun. 2021, 12, 297. [Google Scholar] [CrossRef]
- Simunovic, F.; Yi, M.; Wang, Y.; Macey, L.; Brown, L.T.; Krichevsky, A.M.; Andersen, S.L.; Stephens, R.M.; Benes, F.M.; Sonntag, K.C. Gene expression profiling of substantia nigra dopamine neurons: Further insights into Parkinson’s disease pathology. Brain 2009, 132, 1795–1809. [Google Scholar] [CrossRef]
- Schür, R.R.; Draisma, L.W.; Wijnen, J.P.; Boks, M.P.; Koevoets, M.G.; Joëls, M.; Klomp, D.W.; Kahn, R.S.; Vinkers, C.H. Brain GABA levels across psychiatric disorders: A systematic literature review and meta-analysis of (1) H-MRS studies. Hum. Brain Mapp. 2016, 37, 3337–3352. [Google Scholar] [CrossRef] [PubMed]
- Hansen, K.B.; Wollmuth, L.P.; Bowie, D.; Furukawa, H.; Menniti, F.S.; Sobolevsky, A.I.; Swanson, G.T.; Swanger, S.A.; Greger, I.H.; Nakagawa, T.; et al. Structure, Function, and Pharmacology of Glutamate Receptor Ion Channels. Pharmacol. Rev. 2021, 73, 298–487. [Google Scholar] [CrossRef]
- Juszczyk, G.; Mikulska, J.; Kasperek, K.; Pietrzak, D.; Mrozek, W.; Herbet, M. Chronic Stress and Oxidative Stress as Common Factors of the Pathogenesis of Depression and Alzheimer’s Disease: The Role of Antioxidants in Prevention and Treatment. Antioxidants 2021, 10, 1439. [Google Scholar] [CrossRef]
- Shepard, N.; Baez-Nieto, D.; Iqbal, S.; Kurganov, E.; Budnik, N.; Campbell, A.J.; Pan, J.Q.; Sheng, M.; Farsi, Z. Differential functional consequences of GRIN2A mutations associated with schizophrenia and neurodevelopmental disorders. Sci. Rep. 2024, 14, 2798. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.J.; Brown, A.M.; Purves-Tyson, T.D.; Huang, X.F.; Shannon Weickert, C.; Newell, K.A. GRIN2B gene expression is increased in the anterior cingulate cortex in major depression. J. Psychiatr. Res. 2023, 160, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Rahman, T.; Purves-Tyson, T.; Geddes, A.E.; Huang, X.F.; Newell, K.A.; Weickert, C.S. N-Methyl-d-Aspartate receptor and inflammation in dorsolateral prefrontal cortex in schizophrenia. Schizophr. Res. 2022, 240, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zhang, H.; Huang, S.; Yin, L.; Wang, F.; Luo, P.; Huang, H. Epigenetic regulation in cardiovascular disease: Mechanisms and advances in clinical trials. Signal Transduct. Target. Ther. 2022, 7, 200. [Google Scholar] [CrossRef]
- Smith, C.; Millar, T. Chapter 2—Brain donation procedures in the Sudden Death Brain Bank in Edinburgh. In Handbook of Clinical Neurology; Huitinga, I., Webster, M.J., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; Volume 150, pp. 17–27. [Google Scholar]
| Subjects | Age (Years) | PMI (Hours) |
|---|---|---|
| Individuals who died by suicide (n = 32) | 41.09 ± 12.63 | 28.83 ± 14.28 |
| Control group subjects (n = 37) | 56.51 ± 6.83 | 26.66 ± 18.35 |
| p-value | <0.0001 | 0.5904 |
| Function | Gene | Assay ID |
|---|---|---|
| reference gene | GAPDH | Hs02758991_g1 |
| reference gene | BECN1 | Hs01007018_m1 |
| reference gene | DCTN2 | Hs00197379_m1 |
| candidate gene | CRMP1 | Hs00609716_m1 |
| candidate gene | DISC1 | Hs07287401_m1 |
| candidate gene | TSNAX–DISC1 | Hs03826399_s1 |
| candidate gene | MAPT | Hs00902194_m1 |
| candidate gene | SOD1 | Hs00533490_m1 |
| candidate gene | PRKN | Hs01038323_m1 |
| candidate gene | GABARAPL1 | Hs00740588_mH |
| candidate gene | GRIN2A | Hs00168219_m1 |
| candidate gene | GRIN2B | Hs01002012_m1 |
| Gene | Position (hg19) | Gene Location | mDNA % Difference | q-Value |
|---|---|---|---|---|
| CRMP1 | chr4:5865655 | intron | −16.20 | 2.25 × 10−3 |
| chr4:5867801 | intron | −29.38 | 1.97 × 10−11 | |
| chr4:5872659 | intron | −12.38 | 1.58 × 10−4 | |
| chr4:5892328 | promoter | −7.81 | 6.30 × 10−4 | |
| chr4:5893333 | 1 to 5 kb | 15.64 | 2.03 × 10−3 | |
| chr4:5893366 | 1 to 5 kb | −30.80 | 1.40 × 10−10 | |
| chr4:5894303 | 1 to 5 kb | 5.64 | 2.31 × 10−5 | |
| chr4:5899566 | 1 to 5 kb | −14.31 | 4.63 × 10−3 | |
| DISC1 | chr1:231826172 | intron | −18.96 | 2.62 × 10−3 |
| chr1:232073343 | intron | −20.61 | 2.91 × 10−5 | |
| chr1:232073364 | intron | −24.51 | 1.46 × 10−7 | |
| chr1:232073393 | intron | −32.39 | 4.30 × 10−5 | |
| TSNAX–DISC1 | chr1:231663797 | promoter | −9.17 | 2.81 × 10−3 |
| chr1:231826172 | intron | −18.96 | 2.62 × 10−3 | |
| chr1:232073343 | intron | −20.61 | 2.91 × 10−5 | |
| chr1:232073364 | intron | −24.51 | 1.46 × 10−7 | |
| chr1:232073393 | intron | −32.39 | 4.30 × 10−5 | |
| MAPT | chr17:43974616 | intron | −10.79 | 1.32 × 10−3 |
| chr17:43974622 | intron | −15.80 | 2.94 × 10−6 | |
| chr17:43974635 | intron | −11.40 | 5.34 × 10−4 | |
| chr17:44101476 | exon | −18.77 | 2.25 × 10−4 | |
| chr17:44101486 | exon | −25.83 | 3.78 × 10−8 | |
| chr17:44101491 | exon | −22.69 | 3.19 × 10−6 | |
| SOD1 | chr21:33031523 | promoter | −24.27 | 6.00 × 10−8 |
| chr21:33031539 | promoter | −30.97 | 2.15 × 10−12 | |
| PRKN | chr1:89166724 | intron | −9.70 | 4.27 × 10−3 |
| GABARAPL1 | chr12:10368728 | intron | −26.63 | 7.32 × 10−5 |
| GRIN2A | chr16:9970416 | intron | −21.76 | 1.13 × 10−6 |
| chr16:10001979 | intron | −32.04 | 1.36 × 10−5 | |
| chr16:10070665 | intron | −12.92 | 4.80 × 10−3 | |
| chr16:10277015 | promoter | −5.26 | 8.29 × 10−4 | |
| GRIN2B | chr12:13989920 | intron | −16.79 | 9.57 × 10−3 |
| chr12:13994346 | intron | −7.21 | 2.49 × 10−3 | |
| chr12:14108596 | intron | −54.37 | 3.20 × 10−18 |
| Gene | p-Value | Mann–Whitney or Student T-Test Statistics |
|---|---|---|
| CRMP1 | 0.0369 | t = 2.129, df = 67, 95% CI [0.003–0.028] |
| DISC1 | 0.0642 | U = 425, mean S = 0.5419, mean C = 0.6515 |
| TSNAX–DISC1 | 0.2376 | U = 493, mean S = 0.1031, mean C = 0.09710 |
| MAPT | 0.0020 | U = 339, mean S = 0.5323, mean C = 0.6006 |
| SOD1 | 0.0425 | t = 2.069, df = 67, % CI [−0.05864–0.001046] |
| PRKN | 0.0061 | t = 2.834, df = 67, % CI [−0.1108–0.01921] |
| GABARAPL1 | 0.0005 | U = 296, mean S = 0.4679, mean C = 0.4127 |
| GRIN2A | 0.0368 | t = 2.131, df = 67, % CI [−0.06504–0.002128] |
| GRIN2B | 0.3067 | t = 1.030, df = 67, 95% CI [−0.06969–0.02225] |
| GRIN2A/GRIN2B | 0.0321 | U = 414, mean S = 0.5827, mean C = 0.5220 |
| Gene | p-Value | Mann–Whitney or Student T-Test Statistics |
|---|---|---|
| CRMP1 | 0.4662 | t = 0.7344, df = 49, % CI [−0.1233–0.05729] |
| DISC1 | 0.0241 | t = 2.328, df = 49, % CI [0.02117–0.2882] |
| TSNAX–DISC1 | 0.7738 | U = 285, mean S = 0.1286, mean C = 0.1173 |
| MAPT | 0.1691 | U = 208, mean S = 0.3823, mean C = 0.4231 |
| SOD1 | 0.4942 | U = 287, mean S = 0.3343, mean C = 0.3374 |
| PRKN | 0.2482 | U = 262, mean S = 0.3947, mean C = 0.3682 |
| GABARAPL1 | 0.0990 | U = 236, mean S = 0.4927, mean C = 0.3725 |
| GRIN2A | 0.0232 | U = 204, mean S = 0.4979, mean C = 0.3946 |
| GRIN2B | 0.0914 | U = 234, mean S = 0.5405, mean C = 0.4495 |
| GRIN2A/GRIN2B | 0.3436 | U = 273, mean S = 0.9923, mean C = 0.9107 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bedene, T.; Šmon, J.; Videtič Paska, A.; Zupanc, T.; Kouter, K. Epigenetic and Transcriptomic Alterations of Protein Aggregation-Linked Genes in Suicide: A Pilot Study. Genes 2025, 16, 1467. https://doi.org/10.3390/genes16121467
Bedene T, Šmon J, Videtič Paska A, Zupanc T, Kouter K. Epigenetic and Transcriptomic Alterations of Protein Aggregation-Linked Genes in Suicide: A Pilot Study. Genes. 2025; 16(12):1467. https://doi.org/10.3390/genes16121467
Chicago/Turabian StyleBedene, Taja, Julija Šmon, Alja Videtič Paska, Tomaž Zupanc, and Katarina Kouter. 2025. "Epigenetic and Transcriptomic Alterations of Protein Aggregation-Linked Genes in Suicide: A Pilot Study" Genes 16, no. 12: 1467. https://doi.org/10.3390/genes16121467
APA StyleBedene, T., Šmon, J., Videtič Paska, A., Zupanc, T., & Kouter, K. (2025). Epigenetic and Transcriptomic Alterations of Protein Aggregation-Linked Genes in Suicide: A Pilot Study. Genes, 16(12), 1467. https://doi.org/10.3390/genes16121467






