Genetic Insight into Expression-Defined Melanoma Subtypes and Network Mechanisms: An in Silico Study
Abstract
1. Introduction
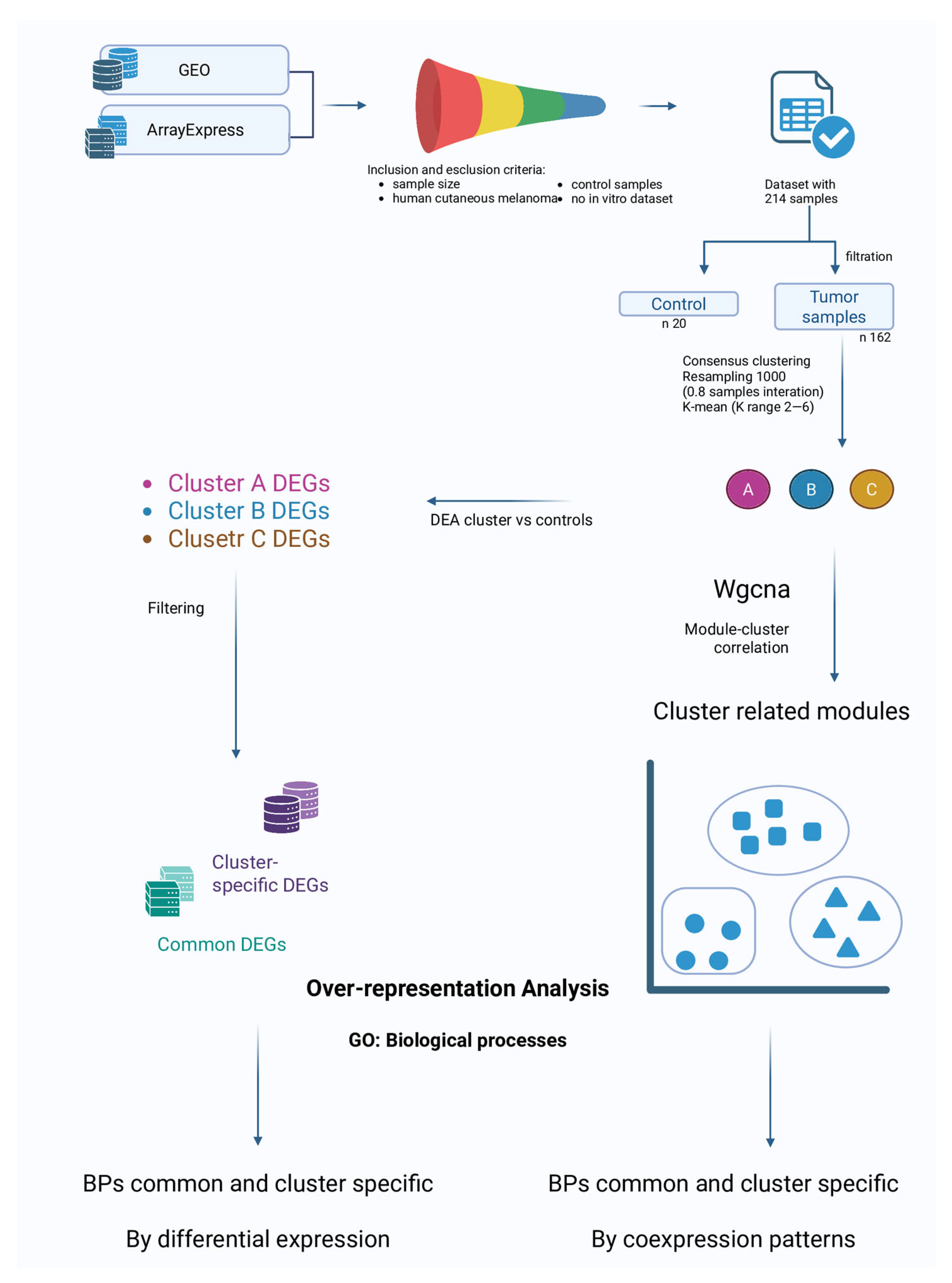
2. Materials and Methods
2.1. Dataset Search and Selection
2.2. Probes and Genes Annotation
2.3. Clustering of Melanoma Samples
2.4. Clusters vs. Control: Differential Expression and Over-Representation Analysis
2.5. In-Between Clusters Characteristics: Weighted-Gene Co-Expression Network Analysis
3. Results
3.1. Dataset Selection
3.2. Clustering
3.3. Differential Expression Analysis (DEA)
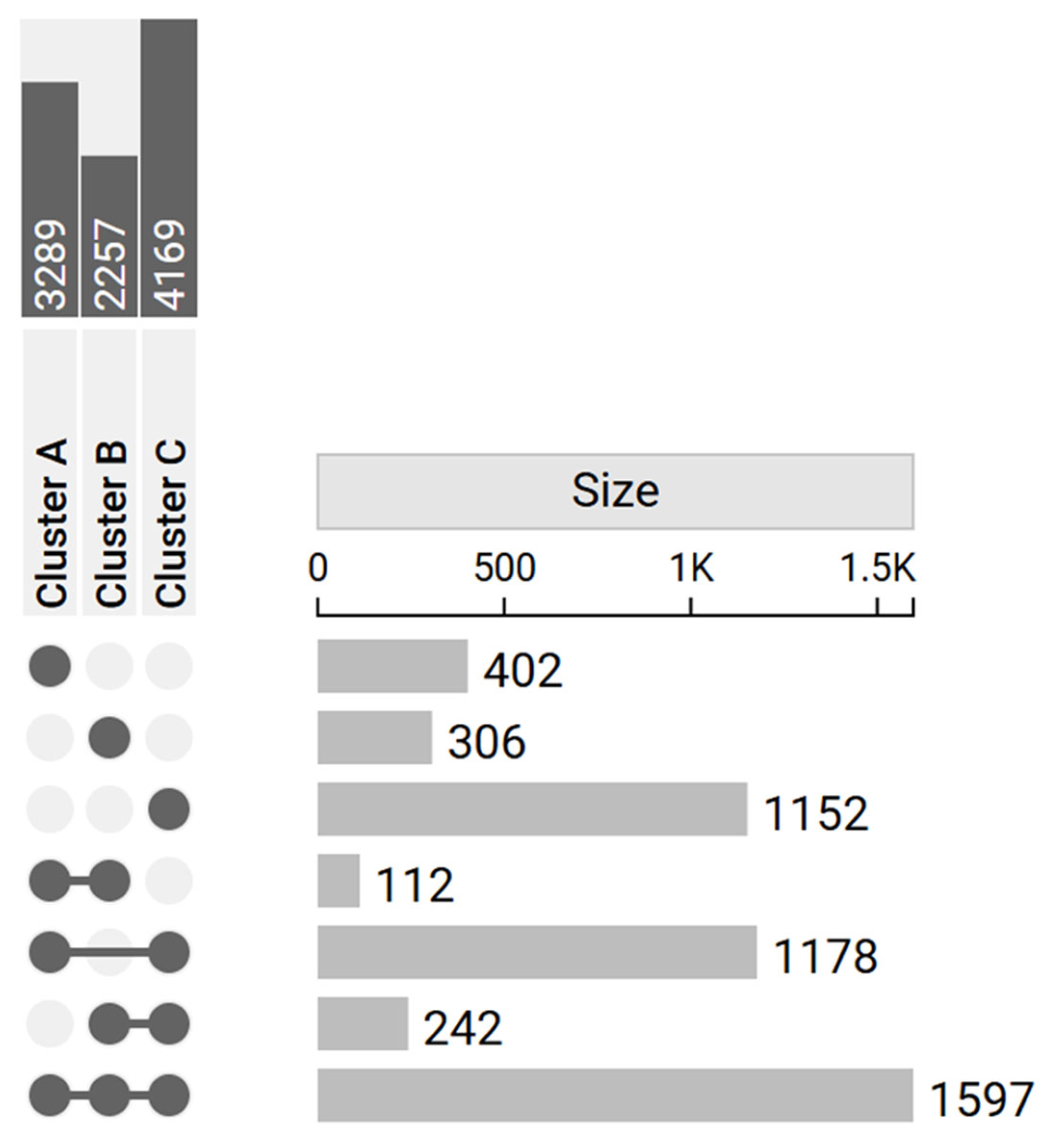
3.4. DEGs Comparison
3.5. Over-Representation Analysis (ORA)
3.6. Weighted-Gene Co-Expression Network Analysis (WGCNA) and Module Correlation with the Clusters
4. Discussion
4.1. Biological Processes Involved in Differentiation
4.2. Proliferative and Pro-Survival Biological Processes
4.3. Phenotypic Switching and Invasive Phenotype
Hub Genes in Melanoma Proliferation/Invasive Pathways
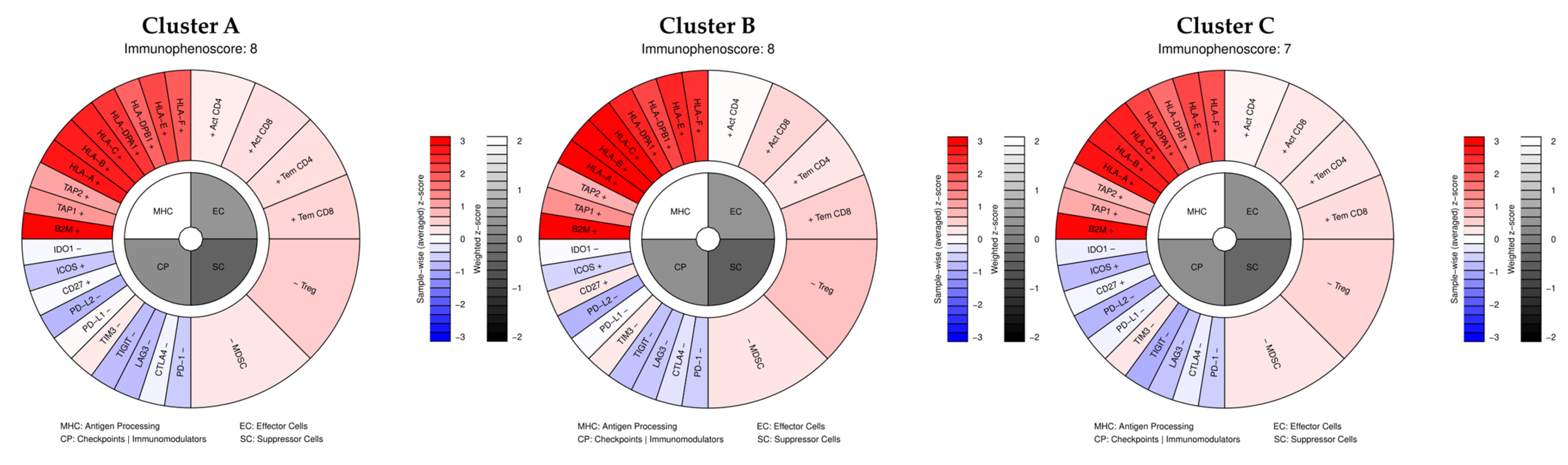
4.4. Immune and Inflammatory Processes
4.5. General Alteration of Basic Cellular Functions
4.6. Lipid Metabolism: Association and Roles Within Melanoma Progression
4.7. Limits
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CM | Cutaneous Melanoma |
| RR | Relative Risk |
| GEO | Gene Expression Omnibus |
| UMAP | Uniform Manifold Approximation and Projection |
| PCA | Principal Component Analysis |
| t-SNE | t-Distributed Stochastic Neighbor Embedding |
| CDF | Cumulative Distribution Function |
| DEA | Differential Expression Analysis |
| DEGs | Differentially Expressed Genes |
| FDR | False Discovery Rate |
| GO | Gene Ontology |
| WGCNA | Weighted-Gene Co-Expression Network Analysis |
| RP | Rank Product |
| ORA | Over-Representation Analysis |
| BP | Biological Process |
| GO:BP | Gene Ontology: Biological Processes |
| TCA | Tricarboxylic Acid |
| OXPHOS | Oxidative Phosphorylation |
| PFS | Progression-Free Survival |
| TME | Tumor Microenvironment |
| TAFs | Tumor-Associated Fibroblasts |
| ECM | Extracellular Matrix |
| Bregs | Regulatory B Cells |
| NLR | Neutrophil-To-Lymphocyte Ratios |
| TAMs | Tumor-Associated Macrophages |
| ICIs | Immune Checkpoint Inhibitors |
| NMD | Nonsense-Mediated Decay |
| RBP | RNA-Binding Protein |
| ALA | Alpha-Linolenic Acid |
| SCD | Stearoyl-Coa Desaturase |
| FADS2 | Fatty Acid Desaturase-2 |
| EMT | Epithelial–Mesenchymal Transition |
| MUFAs | Monounsaturated Fatty Acids |
| FADS1 | Fatty Acid Desaturase 1 |
| FADS3 | Fatty Acid Desaturase 3 |
| OS | Overall Survival |
| DFS | Disease-Free Survival |
| FAO | Fatty Acid Oxidation |
References
- Saginala, K.; Barsouk, A.; Aluru, J.S.; Rawla, P.; Barsouk, A. Epidemiology of Melanoma. Med. Sci. 2021, 9, 63. [Google Scholar] [CrossRef]
- Soura, E.; Eliades, P.J.; Shannon, K.; Stratigos, A.J.; Tsao, H. Hereditary melanoma: Update on syndromes and management: Genetics of familial atypical multiple mole melanoma syndrome. J. Am. Acad. Dermatol. 2016, 74, 395–407. [Google Scholar] [CrossRef]
- Ali, Z.; Yousaf, N.; Larkin, J. Melanoma epidemiology, biology and prognosis. EJC Suppl. 2013, 11, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Gershenwald, J.E.; Scolyer, R.A.; Hess, K.R.; Sondak, V.K.; Long, G.V.; Ross, M.I.; Lazar, A.J.; Faries, M.B.; Kirkwood, J.M.; McArthur, G.A.; et al. Melanoma staging: Evidence-based changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J. Clin. 2017, 67, 472–492. [Google Scholar] [CrossRef] [PubMed]
- Breslow, A. Thickness, cross-sectional areas and depth of invasion in the prognosis of cutaneous melanoma. Ann. Surg. 1970, 172, 902–908. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro Moura Brasil Arnaut, J.; Dos Santos Guimaraes, I.; Evangelista Dos Santos, A.C.; de Moraes Lino da Silva, F.; Machado, J.R.; de Melo, A.C. Molecular landscape of Hereditary Melanoma. Crit. Rev. Oncol. Hematol. 2021, 164, 103425. [Google Scholar] [CrossRef]
- Cancer Genome Atlas, N. Genomic Classification of Cutaneous Melanoma. Cell 2015, 161, 1681–1696. [Google Scholar] [CrossRef]
- Savoia, P.; Fava, P.; Casoni, F.; Cremona, O. Targeting the ERK Signaling Pathway in Melanoma. Int. J. Mol. Sci. 2019, 20, 1483. [Google Scholar] [CrossRef]
- Chiba, K.; Lorbeer, F.K.; Shain, A.H.; McSwiggen, D.T.; Schruf, E.; Oh, A.; Ryu, J.; Darzacq, X.; Bastian, B.C.; Hockemeyer, D. Mutations in the promoter of the telomerase gene TERT contribute to tumorigenesis by a two-step mechanism. Science 2017, 357, 1416–1420. [Google Scholar] [CrossRef]
- Haugh, A.M.; Osorio, R.C.; Francois, R.A.; Tawil, M.E.; Tsai, K.K.; Tetzlaff, M.; Daud, A.; Vasudevan, H.N. Targeted DNA Sequencing of Cutaneous Melanoma Identifies Prognostic and Predictive Alterations. Cancers 2024, 16, 1347. [Google Scholar] [CrossRef]
- Valdez-Salazar, F.; Jimenez-Del Rio, L.A.; Padilla-Gutierrez, J.R.; Valle, Y.; Munoz-Valle, J.F.; Valdes-Alvarado, E. Advances in Melanoma: From Genetic Insights to Therapeutic Innovations. Biomedicines 2024, 12, 1851. [Google Scholar] [CrossRef] [PubMed]
- Najem, A.; Soumoy, L.; Sabbah, M.; Krayem, M.; Awada, A.; Journe, F.; Ghanem, G.E. Understanding Molecular Mechanisms of Phenotype Switching and Crosstalk with TME to Reveal New Vulnerabilities of Melanoma. Cells 2022, 11, 1157. [Google Scholar] [CrossRef] [PubMed]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Eddy, K.; Chen, S. Overcoming Immune Evasion in Melanoma. Int. J. Mol. Sci. 2020, 21, 8984. [Google Scholar] [CrossRef]
- Restifo, N.P.; Smyth, M.J.; Snyder, A. Acquired resistance to immunotherapy and future challenges. Nat. Rev. Cancer 2016, 16, 121–126. [Google Scholar] [CrossRef]
- Munn, D.H.; Mellor, A.L. IDO in the Tumor Microenvironment: Inflammation, Counter-Regulation, and Tolerance. Trends Immunol. 2016, 37, 193–207. [Google Scholar] [CrossRef]
- Sadrkhanloo, M.; Entezari, M.; Orouei, S.; Ghollasi, M.; Fathi, N.; Rezaei, S.; Hejazi, E.S.; Kakavand, A.; Saebfar, H.; Hashemi, M.; et al. STAT3-EMT axis in tumors: Modulation of cancer metastasis, stemness and therapy response. Pharmacol. Res. 2022, 182, 106311. [Google Scholar] [CrossRef]
- Read, J.; Wadt, K.A.; Hayward, N.K. Melanoma genetics. J. Med. Genet. 2016, 53, 1–14. [Google Scholar] [CrossRef]
- Rossi, M.; Pellegrini, C.; Cardelli, L.; Ciciarelli, V.; Di Nardo, L.; Fargnoli, M.C. Familial Melanoma: Diagnostic and Management Implications. Dermatol. Pract. Concept. 2019, 9, 10–16. [Google Scholar] [CrossRef]
- Grzywa, T.M.; Paskal, W.; Wlodarski, P.K. Intratumor and Intertumor Heterogeneity in Melanoma. Transl. Oncol. 2017, 10, 956–975. [Google Scholar] [CrossRef]
- Krepler, C.; Sproesser, K.; Brafford, P.; Beqiri, M.; Garman, B.; Xiao, M.; Shannan, B.; Watters, A.; Perego, M.; Zhang, G.; et al. A Comprehensive Patient-Derived Xenograft Collection Representing the Heterogeneity of Melanoma. Cell Rep. 2017, 21, 1953–1967. [Google Scholar] [CrossRef]
- Ng, M.F.; Simmons, J.L.; Boyle, G.M. Heterogeneity in Melanoma. Cancers 2022, 14, 3030. [Google Scholar] [CrossRef] [PubMed]
- Reuben, A.; Spencer, C.N.; Prieto, P.A.; Gopalakrishnan, V.; Reddy, S.M.; Miller, J.P.; Mao, X.; De Macedo, M.P.; Chen, J.; Song, X.; et al. Genomic and immune heterogeneity are associated with differential responses to therapy in melanoma. NPJ Genom. Med. 2017, 2, 10. [Google Scholar] [CrossRef] [PubMed]
- Smalley, I.; Kim, E.; Li, J.; Spence, P.; Wyatt, C.J.; Eroglu, Z.; Sondak, V.K.; Messina, J.L.; Babacan, N.A.; Maria-Engler, S.S.; et al. Leveraging transcriptional dynamics to improve BRAF inhibitor responses in melanoma. EBioMedicine 2019, 48, 178–190. [Google Scholar] [CrossRef] [PubMed]
- Pillai, M.; Jolly, M.K. Systems-level network modeling deciphers the master regulators of phenotypic plasticity and heterogeneity in melanoma. iScience 2021, 24, 103111. [Google Scholar] [CrossRef]
- Motwani, J.; Eccles, M.R. Genetic and Genomic Pathways of Melanoma Development, Invasion and Metastasis. Genes 2021, 12, 1543. [Google Scholar] [CrossRef]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H., 2nd; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G.; et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef]
- Parkinson, H.; Kapushesky, M.; Shojatalab, M.; Abeygunawardena, N.; Coulson, R.; Farne, A.; Holloway, E.; Kolesnykov, N.; Lilja, P.; Lukk, M.; et al. ArrayExpress--a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 2007, 35, D747–D750. [Google Scholar] [CrossRef]
- Miller, J.A.; Cai, C.; Langfelder, P.; Geschwind, D.H.; Kurian, S.M.; Salomon, D.R.; Horvath, S. Strategies for aggregating gene expression data: The collapseRows R function. BMC Bioinform. 2011, 12, 322. [Google Scholar] [CrossRef]
- Xin, J.; Mark, A.; Afrasiabi, C.; Tsueng, G.; Juchler, M.; Gopal, N.; Stupp, G.S.; Putman, T.E.; Ainscough, B.J.; Griffith, O.L.; et al. High-performance web services for querying gene and variant annotation. Genome Biol. 2016, 17, 91. [Google Scholar] [CrossRef] [PubMed]
- Monti, S.; Tamayo, P.; Mesirov, J.; Golub, T. Consensus Clustering: A Resampling-Based Method for Class Discovery and Visualization of Gene Expression Microarray Data. Mach. Learn. 2003, 52, 91–118. [Google Scholar] [CrossRef]
- Melville, L.M.J.H.J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2020, arXiv:1802.03426. [Google Scholar] [CrossRef]
- Pedregosa, F.V.G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; Vanderplas, J.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- The pandas development team. Pandas-Dev/Pandas: Pandas (v2.3.2); Zenodo: Geneva, Switzerland, 2025. [Google Scholar]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef]
- Visentin, L.; Scarpellino, G.; Chinigo, G.; Munaron, L.; Ruffinatti, F.A. BioTEA: Containerized Methods of Analysis for Microarray-Based Transcriptomics Data. Biology 2022, 11, 1346. [Google Scholar] [CrossRef]
- Del Carratore, F.; Jankevics, A.; Eisinga, R.; Heskes, T.; Hong, F.; Breitling, R. RankProd 2.0: A refactored bioconductor package for detecting differentially expressed features in molecular profiling datasets. Bioinformatics 2017, 33, 2774–2775. [Google Scholar] [CrossRef]
- Fang, Z.; Liu, X.; Peltz, G. GSEApy: A comprehensive package for performing gene set enrichment analysis in Python. Bioinformatics 2023, 39, btac757. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Rezaie, N.; Reese, F.; Mortazavi, A. PyWGCNA: A Python package for weighted gene co-expression network analysis. Bioinformatics 2023, 39, btad415. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Waskom, M.L. seaborn: Statistical data visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Wang, J.X.; Fukunaga-Kalabis, M.; Herlyn, M. Crosstalk in skin: Melanocytes, keratinocytes, stem cells, and melanoma. J. Cell Commun. Signal. 2016, 10, 191–196. [Google Scholar] [CrossRef]
- Han, W.; Hu, C.; Fan, Z.J.; Shen, G.L. Transcript levels of keratin 1/5/6/14/15/16/17 as potential prognostic indicators in melanoma patients. Sci. Rep. 2021, 11, 1023. [Google Scholar] [CrossRef] [PubMed]
- Marrapodi, R.; Bellei, B. The Keratinocyte in the Picture Cutaneous Melanoma Microenvironment. Cancers 2024, 16, 913. [Google Scholar] [CrossRef] [PubMed]
- Anastas, J.N.; Kulikauskas, R.M.; Tamir, T.; Rizos, H.; Long, G.V.; von Euw, E.M.; Yang, P.T.; Chen, H.W.; Haydu, L.; Toroni, R.A.; et al. WNT5A enhances resistance of melanoma cells to targeted BRAF inhibitors. J. Clin. Investig. 2014, 124, 2877–2890. [Google Scholar] [CrossRef] [PubMed]
- Weeraratna, A.T.; Jiang, Y.; Hostetter, G.; Rosenblatt, K.; Duray, P.; Bittner, M.; Trent, J.M. Wnt5a signaling directly affects cell motility and invasion of metastatic melanoma. Cancer Cell 2002, 1, 279–288. [Google Scholar] [CrossRef]
- Miao, B.; Ji, Z.; Tan, L.; Taylor, M.; Zhang, J.; Choi, H.G.; Frederick, D.T.; Kumar, R.; Wargo, J.A.; Flaherty, K.T.; et al. EPHA2 is a mediator of vemurafenib resistance and a novel therapeutic target in melanoma. Cancer Discov. 2015, 5, 274–287. [Google Scholar] [CrossRef]
- Paraiso, K.H.; Das Thakur, M.; Fang, B.; Koomen, J.M.; Fedorenko, I.V.; John, J.K.; Tsao, H.; Flaherty, K.T.; Sondak, V.K.; Messina, J.L.; et al. Ligand-independent EPHA2 signaling drives the adoption of a targeted therapy-mediated metastatic melanoma phenotype. Cancer Discov. 2015, 5, 264–273. [Google Scholar] [CrossRef]
- Wang, J.; Huang, S.K.; Marzese, D.M.; Hsu, S.C.; Kawas, N.P.; Chong, K.K.; Long, G.V.; Menzies, A.M.; Scolyer, R.A.; Izraely, S.; et al. Epigenetic changes of EGFR have an important role in BRAF inhibitor-resistant cutaneous melanomas. J. Investig. Dermatol. 2015, 135, 532–541. [Google Scholar] [CrossRef]
- Boone, B.; Jacobs, K.; Ferdinande, L.; Taildeman, J.; Lambert, J.; Peeters, M.; Bracke, M.; Pauwels, P.; Brochez, L. EGFR in melanoma: Clinical significance and potential therapeutic target. J. Cutan. Pathol. 2011, 38, 492–502. [Google Scholar] [CrossRef]
- Green, K.J.; Pokorny, J.; Jarrell, B. Dangerous liaisons: Loss of keratinocyte control over melanocytes in melanomagenesis. Bioessays 2024, 46, e2400135. [Google Scholar] [CrossRef] [PubMed]
- Jobe, N.P.; Asberg, L.; Andersson, T. Reduced WNT5A signaling in melanoma cells favors an amoeboid mode of invasion. Mol. Oncol. 2021, 15, 1835–1848. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, P.; Yadav, V.; Toftdahl, M.; Andersson, T. WNT5A-Induced Activation of the Protein Kinase C Substrate MARCKS Is Required for Melanoma Cell Invasion. Cancers 2020, 12, 346. [Google Scholar] [CrossRef] [PubMed]
- Jenei, V.; Sherwood, V.; Howlin, J.; Linnskog, R.; Safholm, A.; Axelsson, L.; Andersson, T. A t-butyloxycarbonyl-modified Wnt5a-derived hexapeptide functions as a potent antagonist of Wnt5a-dependent melanoma cell invasion. Proc. Natl. Acad. Sci. USA 2009, 106, 19473–19478. [Google Scholar] [CrossRef]
- Cheng, G.; Hardy, M.; Zielonka, J.; Weh, K.; Zielonka, M.; Boyle, K.A.; Abu Eid, M.; McAllister, D.; Bennett, B.; Kresty, L.A.; et al. Mitochondria-targeted magnolol inhibits OXPHOS, proliferation, and tumor growth via modulation of energetics and autophagy in melanoma cells. Cancer Treat. Res. Commun. 2020, 25, 100210. [Google Scholar] [CrossRef]
- Vashisht Gopal, Y.N.; Gammon, S.; Prasad, R.; Knighton, B.; Pisaneschi, F.; Roszik, J.; Feng, N.; Johnson, S.; Pramanik, S.; Sudderth, J.; et al. A Novel Mitochondrial Inhibitor Blocks MAPK Pathway and Overcomes MAPK Inhibitor Resistance in Melanoma. Clin. Cancer Res. 2019, 25, 6429–6442. [Google Scholar] [CrossRef]
- Kauffmann, A.; Rosselli, F.; Lazar, V.; Winnepenninckx, V.; Mansuet-Lupo, A.; Dessen, P.; van den Oord, J.J.; Spatz, A.; Sarasin, A. High expression of DNA repair pathways is associated with metastasis in melanoma patients. Oncogene 2008, 27, 565–573. [Google Scholar] [CrossRef]
- Winnepenninckx, V.; Lazar, V.; Michiels, S.; Dessen, P.; Stas, M.; Alonso, S.R.; Avril, M.F.; Ortiz Romero, P.L.; Robert, T.; Balacescu, O.; et al. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J. Natl. Cancer Inst. 2006, 98, 472–482. [Google Scholar] [CrossRef]
- Zhao, S.; Wen, S.; Liu, H.; Zhou, Z.; Liu, Y.; Zhong, J.; Xie, J. High Expression of TIMELESS Predicts Poor Prognosis: A Potential Therapeutic Target for Skin Cutaneous Melanoma. Front. Surg. 2022, 9, 917776. [Google Scholar] [CrossRef]
- Patel, J.A.; Kim, H. The TIMELESS effort for timely DNA replication and protection. Cell Mol. Life Sci. 2023, 80, 84. [Google Scholar] [CrossRef]
- Makino, E.; Frohlich, L.M.; Sinnberg, T.; Kosnopfel, C.; Sauer, B.; Garbe, C.; Schittek, B. Targeting Rad51 as a strategy for the treatment of melanoma cells resistant to MAPK pathway inhibition. Cell Death Dis. 2020, 11, 581. [Google Scholar] [CrossRef]
- Gonzalez-Acosta, D.; Blanco-Romero, E.; Ubieto-Capella, P.; Mutreja, K.; Miguez, S.; Llanos, S.; Garcia, F.; Munoz, J.; Blanco, L.; Lopes, M.; et al. PrimPol-mediated repriming facilitates replication traverse of DNA interstrand crosslinks. EMBO J. 2021, 40, e106355. [Google Scholar] [CrossRef] [PubMed]
- Guilliam, T.A.; Doherty, A.J. PrimPol-Prime Time to Reprime. Genes 2017, 8, 20. [Google Scholar] [CrossRef] [PubMed]
- Zamani, G.Y.; Khan, R.; Karim, N.; Ahmed, Z.M.; Naeem, M. Identification of Frameshift Variants in POLH Gene Causing Xeroderma Pigmentosum in Two Consanguineous Pakistani Families. Genes 2022, 13, 543. [Google Scholar] [CrossRef] [PubMed]
- Di Lucca, J.; Guedj, M.; Lacapere, J.J.; Fargnoli, M.C.; Bourillon, A.; Dieude, P.; Dupin, N.; Wolkenstein, P.; Aegerter, P.; Saiag, P.; et al. Variants of the xeroderma pigmentosum variant gene (POLH) are associated with melanoma risk. Eur. J. Cancer 2009, 45, 3228–3236. [Google Scholar] [CrossRef]
- Szadai, L.; Guedes, J.S.; Woldmar, N.; de Almeida, N.P.; Janosi, A.J.; Rajeh, A.; Kovacs, F.; Kriston, A.; Migh, E.; Wan, G.; et al. Mitochondrial and immune response dysregulation in melanoma recurrence. Clin. Transl. Med. 2023, 13, e1495. [Google Scholar] [CrossRef]
- Hughes, T.P.; Kerry, J.T.; Baird, A.H.; Connolly, S.R.; Dietzel, A.; Eakin, C.M.; Heron, S.F.; Hoey, A.S.; Hoogenboom, M.O.; Liu, G.; et al. Global warming transforms coral reef assemblages. Nature 2018, 556, 492–496. [Google Scholar] [CrossRef]
- Peres, J.; Prince, S. The T-box transcription factor, TBX3, is sufficient to promote melanoma formation and invasion. Mol. Cancer 2013, 12, 117. [Google Scholar] [CrossRef]
- Slipicevic, A.; Jorgensen, K.; Skrede, M.; Rosnes, A.K.; Troen, G.; Davidson, B.; Florenes, V.A. The fatty acid binding protein 7 (FABP7) is involved in proliferation and invasion of melanoma cells. BMC Cancer 2008, 8, 276. [Google Scholar] [CrossRef]
- Loria, R.; Laquintana, V.; Scalera, S.; Fraioli, R.; Caprara, V.; Falcone, I.; Bazzichetto, C.; Di Martile, M.; Rosano, L.; Del Bufalo, D.; et al. SEMA6A/RhoA/YAP axis mediates tumor-stroma interactions and prevents response to dual BRAF/MEK inhibition in BRAF-mutant melanoma. J. Exp. Clin. Cancer Res. 2022, 41, 148. [Google Scholar] [CrossRef]
- D’Aguanno, S.; Valentini, E.; Tupone, M.G.; Desideri, M.; Di Martile, M.; Spagnuolo, M.; Buglioni, S.; Ercolani, C.; Falcone, I.; De Dominici, M.; et al. Semaphorin 5A drives melanoma progression: Role of Bcl-2, miR-204 and c-Myb. J. Exp. Clin. Cancer Res. 2018, 37, 278. [Google Scholar] [CrossRef] [PubMed]
- Deborde, S.; Wong, R.J. How Schwann cells facilitate cancer progression in nerves. Cell Mol. Life Sci. 2017, 74, 4405–4420. [Google Scholar] [CrossRef] [PubMed]
- Deborde, S.; Omelchenko, T.; Lyubchik, A.; Zhou, Y.; He, S.; McNamara, W.F.; Chernichenko, N.; Lee, S.Y.; Barajas, F.; Chen, C.H.; et al. Schwann cells induce cancer cell dispersion and invasion. J. Clin. Investig. 2016, 126, 1538–1554. [Google Scholar] [CrossRef] [PubMed]
- Kruglov, O.; Vats, K.; Soman, V.; Tyurin, V.A.; Tyurina, Y.Y.; Wang, J.; Williams, L.; Zhang, J.; Donahue Carey, C.; Jaklitsch, E.; et al. Melanoma-associated repair-like Schwann cells suppress anti-tumor T-cells via 12/15-LOX/COX2-associated eicosanoid production. Oncoimmunology 2023, 12, 2192098. [Google Scholar] [CrossRef]
- Slutsky, S.G.; Kamaraju, A.K.; Levy, A.M.; Chebath, J.; Revel, M. Activation of myelin genes during transdifferentiation from melanoma to glial cell phenotype. J. Biol. Chem. 2003, 278, 8960–8968. [Google Scholar] [CrossRef]
- Garraway, L.A.; Widlund, H.R.; Rubin, M.A.; Getz, G.; Berger, A.J.; Ramaswamy, S.; Beroukhim, R.; Milner, D.A.; Granter, S.R.; Du, J.; et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 2005, 436, 117–122. [Google Scholar] [CrossRef]
- Moller, K.; Sigurbjornsdottir, S.; Arnthorsson, A.O.; Pogenberg, V.; Dilshat, R.; Fock, V.; Brynjolfsdottir, S.H.; Bindesboll, C.; Bessadottir, M.; Ogmundsdottir, H.M.; et al. MITF has a central role in regulating starvation-induced autophagy in melanoma. Sci. Rep. 2019, 9, 1055. [Google Scholar] [CrossRef]
- Carotenuto, P.; Romano, A.; Barbato, A.; Quadrano, P.; Brillante, S.; Volpe, M.; Ferrante, L.; Tammaro, R.; Morleo, M.; De Cegli, R.; et al. Targeting the MITF/APAF-1 axis as salvage therapy for MAPK inhibitors in resistant melanoma. Cell Rep. 2022, 41, 111601. [Google Scholar] [CrossRef]
- Thurber, A.E.; Douglas, G.; Sturm, E.C.; Zabierowski, S.E.; Smit, D.J.; Ramakrishnan, S.N.; Hacker, E.; Leonard, J.H.; Herlyn, M.; Sturm, R.A. Inverse expression states of the BRN2 and MITF transcription factors in melanoma spheres and tumour xenografts regulate the NOTCH pathway. Oncogene 2011, 30, 3036–3048. [Google Scholar] [CrossRef]
- Fane, M.E.; Chhabra, Y.; Hollingsworth, D.E.J.; Simmons, J.L.; Spoerri, L.; Oh, T.G.; Chauhan, J.; Chin, T.; Harris, L.; Harvey, T.J.; et al. NFIB Mediates BRN2 Driven Melanoma Cell Migration and Invasion Through Regulation of EZH2 and MITF. EBioMedicine 2017, 16, 63–75. [Google Scholar] [CrossRef]
- Muller, J.; Krijgsman, O.; Tsoi, J.; Robert, L.; Hugo, W.; Song, C.; Kong, X.; Possik, P.A.; Cornelissen-Steijger, P.D.; Geukes Foppen, M.H.; et al. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat. Commun. 2014, 5, 5712. [Google Scholar] [CrossRef]
- Simmons, J.L.; Neuendorf, H.M.; Boyle, G.M. BRN2 and MITF together impact AXL expression in melanoma. Exp. Dermatol. 2022, 31, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Strnadova, K.; Pfeiferova, L.; Prikryl, P.; Dvorankova, B.; Vlcak, E.; Frydlova, J.; Vokurka, M.; Novotny, J.; Sachova, J.; Hradilova, M.; et al. Exosomes produced by melanoma cells significantly influence the biological properties of normal and cancer-associated fibroblasts. Histochem. Cell Biol. 2022, 157, 153–172. [Google Scholar] [CrossRef] [PubMed]
- Remsik, J.; Tong, X.; Kunes, R.Z.; Li, M.J.; Estrera, R.; Snyder, J.; Thomson, C.; Osman, A.M.; Chabot, K.; Sener, U.T.; et al. Interferon-gamma orchestrates leptomeningeal anti-tumour response. Nature 2025, 643, 1087–1096. [Google Scholar] [CrossRef] [PubMed]
- Wellbrock, C.; Weisser, C.; Hassel, J.C.; Fischer, P.; Becker, J.; Vetter, C.S.; Behrmann, I.; Kortylewski, M.; Heinrich, P.C.; Schartl, M. STAT5 contributes to interferon resistance of melanoma cells. Curr. Biol. 2005, 15, 1629–1639. [Google Scholar] [CrossRef]
- Galdiero, M.R.; Varricchi, G.; Loffredo, S.; Mantovani, A.; Marone, G. Roles of neutrophils in cancer growth and progression. J. Leukoc. Biol. 2018, 103, 457–464. [Google Scholar] [CrossRef]
- Zhong, Q.; Hao, H.; Li, S.; Ning, Y.; Li, H.; Hu, X.; McMasters, K.M.; Yan, J.; Ding, C. B cell c-Maf signaling promotes tumor progression in animal models of pancreatic cancer and melanoma. J. Immunother. Cancer 2024, 12, e009861. [Google Scholar] [CrossRef]
- Prevost-Blondel, A.; Richard, Y. Interleukin 4-Induced Gene 1 as an Emerging Regulator of B-Cell Biology and its Role in Cutaneous Melanoma. Crit. Rev. Immunol. 2019, 39, 39–57. [Google Scholar] [CrossRef]
- Li, H.; van der Leun, A.M.; Yofe, I.; Lubling, Y.; Gelbard-Solodkin, D.; van Akkooi, A.C.J.; van den Braber, M.; Rozeman, E.A.; Haanen, J.; Blank, C.U.; et al. Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell 2019, 176, 775–789.e718. [Google Scholar] [CrossRef]
- Mallardo, D.; Fordellone, M.; White, A.; Ottaviano, M.; Sparano, F.; Bailey, M.; Facchini, A.B.; Ong, S.; Maiolino, P.; Caraco, C.; et al. CD39 and LDHA affects the prognostic role of NLR in metastatic melanoma patients treated with immunotherapy. J. Transl. Med. 2023, 21, 610. [Google Scholar] [CrossRef]
- Mollinedo, F. Neutrophil Degranulation, Plasticity, and Cancer Metastasis. Trends Immunol. 2019, 40, 228–242. [Google Scholar] [CrossRef]
- Lee-Rueckert, M.; Lappalainen, J.; Kovanen, P.T.; Escola-Gil, J.C. Lipid-Laden Macrophages and Inflammation in Atherosclerosis and Cancer: An Integrative View. Front. Cardiovasc. Med. 2022, 9, 777822. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Huang, M.; Zhang, L.; Huang, Q.; Wang, Y.; Liang, Y. Inflammatory response signature score model for predicting immunotherapy response and pan-cancer prognosis. Comput. Struct. Biotechnol. J. 2024, 23, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Yang, C.; Luo, R. Induction of CCL2 by siMAML1 through upregulation of TweakR in melanoma cells. Biochem. Biophys. Res. Commun. 2008, 372, 629–633. [Google Scholar] [CrossRef] [PubMed]
- Gorbatenko, A.; Olesen, C.W.; Loebl, N.; Sigurdsson, H.H.; Bianchi, C.; Pedraz-Cuesta, E.; Christiansen, J.; Pedersen, S.F. Oncogenic p95HER2 regulates Na+-HCO3- cotransporter NBCn1 mRNA stability in breast cancer cells via 3′UTR-dependent processes. Biochem. J. 2016, 473, 4027–4044. [Google Scholar] [CrossRef]
- Xing, H.; Jiang, X.; Yang, C.; Tan, B.; Hu, J.; Zhang, M. High expression of RPL27A predicts poor prognosis in patients with hepatocellular carcinoma. World J. Surg. Oncol. 2023, 21, 209. [Google Scholar] [CrossRef]
- Rapino, F.; Delaunay, S.; Rambow, F.; Zhou, Z.; Tharun, L.; De Tullio, P.; Sin, O.; Shostak, K.; Schmitz, S.; Piepers, J.; et al. Codon-specific translation reprogramming promotes resistance to targeted therapy. Nature 2018, 558, 605–609. [Google Scholar] [CrossRef]
- Wurth, L.; Papasaikas, P.; Olmeda, D.; Bley, N.; Calvo, G.T.; Guerrero, S.; Cerezo-Wallis, D.; Martinez-Useros, J.; Garcia-Fernandez, M.; Huttelmaier, S.; et al. UNR/CSDE1 Drives a Post-transcriptional Program to Promote Melanoma Invasion and Metastasis. Cancer Cell 2016, 30, 694–707. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, C.; Suo, C.; Zhao, R.; Jin, L.; Zhang, T.; Chen, X. Metabolic dysfunction-associated fatty liver disease and the risk of 24 specific cancers. Metabolism 2022, 127, 154955. [Google Scholar] [CrossRef]
- Chen, Z.; Gong, Y.; Chen, F.; Lee, H.J.; Qian, J.; Zhao, J.; Zhang, W.; Li, Y.; Zhou, Y.; Xu, Q.; et al. Orchestrated desaturation reprogramming from stearoyl-CoA desaturase to fatty acid desaturase 2 in cancer epithelial-mesenchymal transition and metastasis. Cancer Commun. 2025, 45, 245–280. [Google Scholar] [CrossRef]
- Lee, H.J.; Chen, Z.; Collard, M.; Chen, F.; Chen, J.G.; Wu, M.; Alani, R.M.; Cheng, J.X. Multimodal Metabolic Imaging Reveals Pigment Reduction and Lipid Accumulation in Metastatic Melanoma. BME Front. 2021, 2021, 9860123. [Google Scholar] [CrossRef] [PubMed]
- Koundouros, N.; Poulogiannis, G. Reprogramming of fatty acid metabolism in cancer. Br. J. Cancer 2020, 122, 4–22. [Google Scholar] [CrossRef] [PubMed]
- Kothapalli, K.S.D.; Park, H.G.; Kothapalli, N.S.L.; Brenna, J.T. FADS2 function at the major cancer hotspot 11q13 locus alters fatty acid metabolism in cancer. Prog. Lipid Res. 2023, 92, 101242. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Pang, S.; Sun, B.; Zhang, M.; Jiao, X.; Lai, L.; Qian, Y.; Yang, N.; Yang, W. ELOVLs Predict Distinct Prognosis Value and Immunotherapy Efficacy in Patients with Hepatocellular Carcinoma. Front. Oncol. 2022, 12, 884066. [Google Scholar] [CrossRef]
- Zhang, J.; He, Y.; Liang, S.; Liao, X.; Li, T.; Qiao, Z.; Chang, C.; Jia, H.; Chen, X. Non-invasive, opsin-free mid-infrared modulation activates cortical neurons and accelerates associative learning. Nat. Commun. 2021, 12, 2730. [Google Scholar] [CrossRef]
- Sella, R.; Chou, L.; Schuster, A.K.; Gali, H.E.; Weinreb, R.N.; Afshari, N.A. Accuracy of IOL power calculations in the very elderly. Eye 2020, 34, 1848–1855. [Google Scholar] [CrossRef]




| Tumor Samples | Control Samples | Upregulated DEGs | Downregulated DEGs | Total DEGs | |
|---|---|---|---|---|---|
| N° | N° | ||||
| Cluster A | 51 | 20 | 1606 | 1683 | 3289 |
| Cluster B | 52 | 20 | 860 | 1397 | 2257 |
| Cluster C | 59 | 20 | 2035 | 2134 | 4169 |
| Term | Overlap | p-Value | Adjusted p-Value | Odds | Combined Score |
|---|---|---|---|---|---|
| Ratio | |||||
| Cluster A-specific | |||||
| - | - | - | - | - | - |
| Cluster B-specific | |||||
| Positive regulation of T-cell activation (GO:0050870) | 13/111 | 1.53 × 10−8 | 2.02 × 10−5 | 8.87 × 1000 | 1.60 × 102 |
| Lymphocyte differentiation (GO:0030098) | 9/76 | 2.13 × 10−8 | 2.02 × 10−5 | 1.13 × 101 | 1.99 × 102 |
| Antigen receptor-mediated signaling pathway (GO:0050851) | 12/114 | 1.82 × 10−7 | 1.16 × 10−4 | 7.84 × 1000 | 1.22 × 102 |
| B cell-mediated immunity (GO:0019724) | 6/20 | 3.95 × 10−7 | 1.88 × 10−4 | 2.81 × 101 | 4.15 × 102 |
| Regulation of Interleukin-12 Production (GO:0032655) | 8/50 | 8.42 × 10−7 | 2.89 × 10−4 | 1.26 × 101 | 1.76 × 102 |
| Cluster C-specific | |||||
| Regulation of intracellular signal transduction (GO:1902531) | 38/302 | 5× 10−6 | 0.0169708 | 2.40123 | 29.29741 |
| Common | |||||
| Epidermis development (GO:0008544) | 38/86 | 1.07 × 10−19 | 4.09 × 10−16 | 9.32 × 1000 | 4.07 × 102 |
| Intermediate filament organization (GO:0045109) | 32/72 | 6.77 × 10−17 | 1.29 × 10−13 | 9.39 × 1000 | 3.49 × 102 |
| Supramolecular fiber organization (GO:0097435) | 66/339 | 9.05 × 10−12 | 1.15 × 10−8 | 2.86 × 1000 | 7.28 × 101 |
| Long-chain fatty acid metabolic process (GO:0001676) | 26/85 | 1.16 × 10−9 | 1.02 × 10−6 | 5.15 × 1000 | 1.06 × 102 |
| Very long-chain fatty acid metabolic process (GO:0000038) | 15/30 | 1.57 × 10−9 | 1.02 × 10−6 | 1.16 × 101 | 2.36 × 102 |
| Hub Gene/Axis | Linked Pathway(s) | Mechanistic Role (Literature) | Evidence from Our Dataset | Functional Hypothesis | References |
|---|---|---|---|---|---|
| MITF | MAPK, PI3K/AKT, EMT | Lineage survival oncogene; rheostat model; suppressed by Notch/BRN2 → invasive switch | Higher than AXL across clusters (MITF-high/AXL-low); Cluster B with reduced MITF suggests transitional state | Restoring MITF activity could resensitize tumors to MAPKi | [78] |
| AXL | EMT, PI3K/AKT, drug resistance | Marker of MITF-low phenotype; promotes invasion and MAPKi resistance | Lower than MITF in all clusters; no AXL-driven subgroup detected | AXL inhibition may counteract EMT-like resistance | [83] |
| BRN2 (POU3F2) | MAPK, EMT, Notch cross-talk | Antagonist of MITF; cooperates in AXL regulation; drives invasion | Weakly expressed; not a cluster driver | Targeting BRN2 may restore MITF expression and reduce invasion | [84] |
| NGFR (p75NTR) | EMT, stemness, immune evasion | Marker of neural crest-like MITF-low state; supports plasticity and immune escape | Not strongly represented in our dataset | Targeting NGFR-positive subpopulations could limit relapse and immune evasion | [83] |
| MITF/APAF-1 axis | Apoptosis, MAPK inhibitor resistance | MITF represses APAF-1 → impaired apoptosome and resistance | Not directly clustered, but consistent with MITF dominance | Pharmacologic inhibition of MITF or APAF-1 reactivation (quinacrine, MBZ) may restore apoptosis and sensitize tumors | [79,80] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Speranza, D.; Marafioti, M.; Musarra, M.; Cianci, V.; Mondello, C.; Astorino, M.F.; Santarpia, M.; Irrera, N.; Vaccaro, M.; Silvestris, N.; et al. Genetic Insight into Expression-Defined Melanoma Subtypes and Network Mechanisms: An in Silico Study. Genes 2025, 16, 1428. https://doi.org/10.3390/genes16121428
Speranza D, Marafioti M, Musarra M, Cianci V, Mondello C, Astorino MF, Santarpia M, Irrera N, Vaccaro M, Silvestris N, et al. Genetic Insight into Expression-Defined Melanoma Subtypes and Network Mechanisms: An in Silico Study. Genes. 2025; 16(12):1428. https://doi.org/10.3390/genes16121428
Chicago/Turabian StyleSperanza, Desirèe, Mariapia Marafioti, Martina Musarra, Vincenzo Cianci, Cristina Mondello, Maria Francesca Astorino, Mariacarmela Santarpia, Natasha Irrera, Mario Vaccaro, Nicola Silvestris, and et al. 2025. "Genetic Insight into Expression-Defined Melanoma Subtypes and Network Mechanisms: An in Silico Study" Genes 16, no. 12: 1428. https://doi.org/10.3390/genes16121428
APA StyleSperanza, D., Marafioti, M., Musarra, M., Cianci, V., Mondello, C., Astorino, M. F., Santarpia, M., Irrera, N., Vaccaro, M., Silvestris, N., Crisafulli, C., Calabrò, M., & Briuglia, S. (2025). Genetic Insight into Expression-Defined Melanoma Subtypes and Network Mechanisms: An in Silico Study. Genes, 16(12), 1428. https://doi.org/10.3390/genes16121428








