Abstract
Background: Diabetic nephropathy (DN) is a leading cause of end-stage renal disease, arising from complex interactions between metabolic, hemodynamic, and genetic factors. Among candidate genes, vascular endothelial growth factor A (VEGFA) has been extensively investigated due to its role in endothelial homeostasis and microvascular complications of diabetes. The present study aimed to examine the association of VEGFA polymorphisms with DN in a Greek population and to perform a comprehensive meta-analysis of available evidence. Methods: A case–control study was conducted, including 197 patients with type 2 diabetes mellitus (T2DM) and DN, 155 diabetic patients without nephropathy, and 246 healthy controls. Ten tagging single-nucleotide polymorphisms (SNPs) across VEGFA were genotyped. Statistical analyses employed the generalized odds ratio (ORG). To contextualize these findings, a meta-analysis of 13 eligible studies was performed, encompassing 7520 cases, 6951 diabetic controls, and 1718 healthy controls. Results: Of the tested variants in the present case–control study, only rs833070 was significantly associated with DN across all comparisons. Nine VEGFA variants were evaluated in meta-analysis, with rs2146323 showing a protective effect (allelic OR = 0.85; 95% CI: 0.76–0.95), while other variants yielded non-significant associations. Conclusions: Overall, the data suggest that VEGFA polymorphisms, particularly rs833070 and rs2146323, contribute to genetic susceptibility to DN, although population-specific differences and heterogeneity across studies remain substantial. Future research in large, ethnically diverse cohorts with functional analyses is warranted to clarify causal mechanisms and enable the integration of VEGFA genetic variation into risk stratification and personalized therapeutic strategies.
1. Introduction
Diabetic nephropathy (DN) is a progressive microvascular complication associated with both type 1 and type 2 diabetes and continues to be the primary cause of end-stage renal disease (ESRD) globally, affecting approximately 30–40% of individuals with diabetes [1]. It is clinically defined by persistent micro/macroalbuminuria, a sustained decline in estimated glomerular filtration rate (eGFR), and often hypertension, confirmed on at least two occasions over 3–6 months [2]. Pathophysiologically, the disease is driven by hyperglycemia-induced metabolic dysregulation (e.g., advanced glycation end-products), hemodynamic changes (glomerular hyperfiltration, RAAS activation), and inflammatory and profibrotic cytokine signaling, resulting in glomerular basement membrane thickening, mesangial expansion, podocyte injury, and eventual fibrosis [3,4]. Major risk factors include poor glycemic control, uncontrolled hypertension, dyslipidemia, obesity, smoking, family history, and certain ethnic predispositions [5]. Early intervention—comprising strict glucose and blood pressure control, renin-angiotensin system blockade, SGLT2 inhibitors, GLP-1 receptor agonists, and lifestyle modification—can delay progression and improve renal and cardiovascular outcomes [6,7].
Although glycemic and hemodynamic disturbances play a central role, the genetic contribution to DN is undeniable yet remains poorly understood [8]. Multiple genetic loci have been associated with the disease’s pathogenesis; however, each locus exerts only a modest effect size [9]. Different methodological approaches have been implemented. Among them, linkage studies [10,11] and genetic association studies [12,13] are the most common study designs, as well as meta-analyses of these studies [14,15,16].
Vascular endothelial growth factor A (VEGFA) is a crucial regulator of endothelial cell proliferation, differentiation, and survival, and is essential for both physiological and pathological angiogenesis [17,18]. In the healthy kidney, podocytes secrete VEGFA to support the integrity and function of glomerular and peritubular endothelial cells, helping maintain the filtration barrier [19,20]. However, in diabetes, local VEGFA expression becomes dysregulated, as rodent models of diabetic nephropathy show elevated renal VEGFA, and pharmacological blockade of VEGF signaling in these models reduces albuminuria and glomerular injury [21,22]. More specifically, diabetes is associated with increased levels of glomerular VEGFA expression [23]. However, human data present a nuanced picture, as some studies find higher circulating VEGF associated with worse albuminuria and glycemic control, while others find no or even inverse relationships, suggesting that timing, isoform, receptor context, and disease stage critically shape VEGF’s impact in diabetic nephropathy [24].
Several genetic association studies (GAS), as well as many meta-analyses, have implicated certain VEGFA variants in the pathophysiology of DN, but the results are inconsistent. For instance, the promoter variant rs833061 (−460 T/C) was associated with DN in a meta-analysis of two studies (n = 543) containing only type 1 diabetes mellitus patients of European origin [25]. A broader meta-analysis covering multiple VEGFA variants also identified rs833061 as significantly associated with DN risk across diverse populations [26]. In a large case–control study among Han Chinese type 2 diabetic patients, VEGFA rs2010963 and rs69947 variants were linked to increased DN susceptibility [27]. These findings support the hypothesis that common functional VEGFA polymorphisms modulate VEGF expression and microvascular vulnerability in diabetes, although effects vary by ancestry, diabetes type, and study design—highlighting the need for larger, ethnically diverse cohorts and deeper functional validation. Another variant across VEGFA that has been examined in meta-analyses is rs2146323 [14,16,28].
Given the pivotal role of VEGFA in angiogenesis and glomerular endothelial function, genetic variations within this gene are strong candidates for influencing susceptibility to diabetic nephropathy. However, findings from previous studies remain inconsistent, likely due to differences in study design, sample size, ethnicity, and diabetes type. To address these gaps, we conducted a case–control association study in a well-characterized Greek cohort, genotyping ten tagging SNPs across the VEGFA gene. In parallel, we performed a systematic review and meta-analysis of all available genetic data on VEGFA variants and DN, thereby providing the most comprehensive evaluation to date. It is noteworthy to mention that the odds ratio generalized (ORG) will be used for the analysis of genotypic data in the context of a genetic model-free approach. This combined approach aims to clarify the contribution of VEGFA genetic variation to DN risk and progression, with the ultimate goal of improving understanding of disease pathogenesis and guiding future precision medicine strategies.
2. Materials and Methods
2.1. Association Study
2.1.1. Subjects
Comprehensive information on the study design and demographic characteristics of the participants has been reported previously [29]. Briefly, the study included 197 patients with DN, 155 diabetics (T2DM) but without microvascular complications, and 246 healthy controls. The participants were evaluated at the Ophthalmology and Nephrology clinics of the University Hospital of Larissa, Greece.
DN was defined as persistent macroalbuminuria, with urinary albumin excretion > 300 mg/24 h (>200 μg/min), irrespective of serum creatinine levels. The study protocol was approved by the Ethics Committee of the University of Thessaly, and written informed consent was obtained from all participants prior to enrollment.
2.1.2. Genotyping
Genotyping was performed as has been reported previously [29]. A total of 10 tag SNPs across VEGFA were retrieved (rs3025053, rs3025040, rs10434, rs25648, rs3024994, rs3025035, rs2146323, rs3024997, rs2010963, rs833070). The selection of tagging SNPs was performed using an r2 threshold of ≥0.8 and a minor allele frequency (MAF) greater than 0.05. To ensure accuracy, at least 10% of the samples were randomly selected for repeat genotyping as an internal quality control. All genotyping was conducted by staff blinded to the participants’ clinical status.
2.1.3. Data Analysis
The generalized odds ratio (ORG) was used for the assessment of the association between genotype distribution and disease risk [30,31]. In healthy controls, genotype distributions were also tested for deviation from Hardy–Weinberg equilibrium (HWE). The ORG was calculated with the ORGGASMA software (http://biomath.med.uth.gr; accessed 30 August 2025) [30,31]. All statistical analyses were performed using SPSS version 29.0 for Windows (SPSS 29.0 Inc., Chicago, IL, USA).
2.2. Meta-Analysis
All studies published up to August 2025 were identified through a comprehensive PubMed search. The following search terms were applied: [‘diabetic nephropathy’ AND ‘association’ AND (‘VEGFA’ OR ‘vascular endothelial growth factor’)]. In addition, relevant records were retrieved from the Genome-Wide Association Studies (GWAS) Catalog (https://www.ebi.ac.uk/gwas/; accessed 30 August 2025). Case reports, editorials, and review articles were excluded. The search was restricted to English-language publications.
Eligible studies for inclusion in the meta-analysis were case–control designs that investigated VEGFA gene polymorphisms in patients with DN compared with either diabetic controls without DN or healthy controls. The inclusion criteria for DN cases and both control groups were consistent with those applied in the present association study. Genome linkage scans were excluded, as they represent a different study design.
From each eligible study, the following data were extracted: first author, year of publication, ethnicity, type of diabetes, and phenotype. For both cases and controls, we recorded sample size and selection criteria. Genotypic information was extracted as complete genotype counts or allele frequencies.
For meta-analysis, a minimum of two studies per genetic variant was required. Pooled ORs were calculated with the DerSimonian and Laird random-effects model [32]. Between-study heterogeneity was assessed using Cochran’s Q test [33] and quantified with the I2 statistic [34].
3. Results
3.1. Association Analysis
The study population included 197 cases with DN (DM+DN), 155 diabetics without DN (DN-DN), and 246 healthy controls (HC). All participants were Caucasians of Greek origin. Demographic data and clinical profile are summarized in Table 1, as have also been described elsewhere [35]. Among the 197 DN cases, 11 had progressed to end-stage renal disease (ESRD).

Table 1.
Clinical profiles of the study cohort.
The genotype distributions of the ten variants (rs3025053, rs3025040, rs10434, rs25648, rs3024994, rs3025035, rs2146323, rs3024997, rs2010963, rs833070) and the respective ORG are shown in Table 2, Table 3 and Table 4. Only rs833070 was found statistically significant [ORG = 1.26, 95% CI (1.01, 1.59)] in healthy controls versus diseased controls versus cases and in diseased controls versus cases [ORG = 1.46, 95% CI (1.01, 2.12)], as well as in healthy controls versus cases [ORG = 1.43, 95% CI (1.03, 1.99)].

Table 2.
Association between VEGFA variants and T2DM-nephropathy in healthy cases versus diseased controls versus cases.

Table 3.
Association between VEGFA variants and T2DM-nephropathy in diseased controls versus cases.

Table 4.
Association between VEGFA variants and T2DM-nephropathy in healthy controls versus cases.
3.2. Meta-Analysis
The literature search yielded 312 PubMed records that met the inclusion criteria. When a study reported data from different populations, each population was treated as a separate dataset. A flowchart summarizing the selection process and reasons for exclusion is provided in Figure 1, while the characteristics of the included studies are detailed in Table 5.
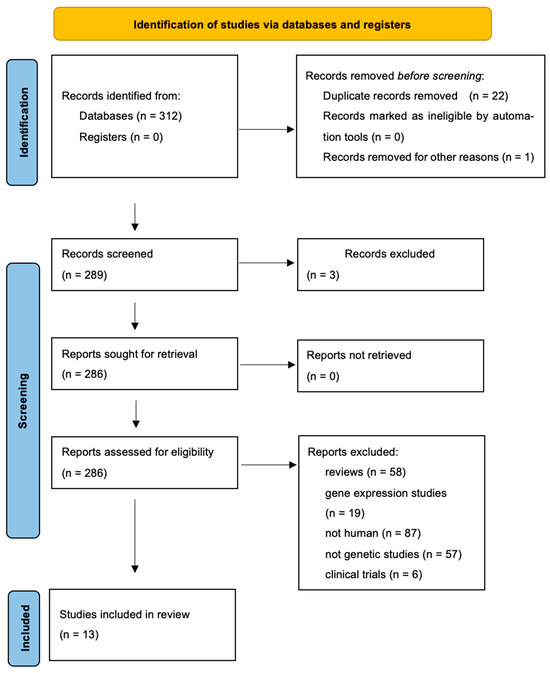
Figure 1.
Flowchart showing how studies were selected for meta-analysis.

Table 5.
Characteristics of studies included in meta-analysis.
In meta-analysis, nine variants (rs2010963, rs699947, rs833061, rs35569394, rs6921438, rs10738760, rs2146323, rs3024997, rs3025000) were included. The studies were published between 2003 and 2024. The forest plot is presented in Figure 2. Meta-analysis results are presented in Table 6 and Table 7.
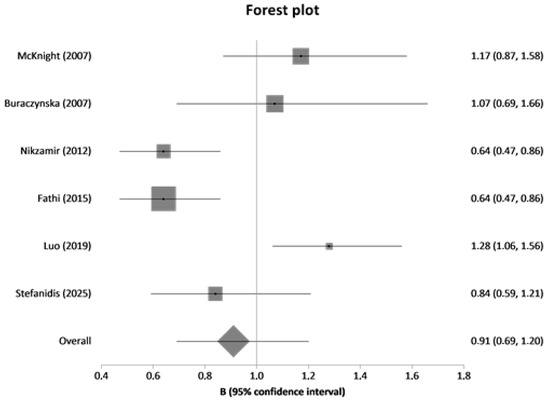
Figure 2.
Forest plot of rs2010963 between diseased controls (diabetics without DN) versus cases (diabetics with DN).

Table 6.
Results from meta-analyses based on genotype counts.

Table 7.
Results from meta-analyses based on allele counts.
4. Discussion
While hyperglycemia and hypertension are recognized as crucial pathogenic factors, growing evidence highlights the multifactorial nature of DN, with genetic predisposition playing a significant role in susceptibility and progression [47]. Among the candidate genes, VEGFA has attracted considerable attention due to its established involvement in diabetic microvascular complications [20,48,49]. Genetic variations in VEGFA are believed to influence the risk of developing DN, although findings vary among studies and populations [14]. To provide a comprehensive assessment of genetic variation in the VEGFA gene, we selected ten tag SNPs for genotyping in a Greek-origin cohort and conducted a systematic review and meta-analysis incorporating all available genetic data on variants of this gene, providing the most comprehensive overview assessing the contribution of VEGFA variants in DN risk and progression.
Out of ten variants, only rs833070 was revealed statistically significant in all analyses in the context of the present GAS. From these variants, available data from other studies were only for rs2146323, rs3024997, and rs2010963. In agreement with our findings, Trégouet et al. did not find statistically significant differences regarding rs2146323 and rs3024997 [46]. Findings from the Ensembl genome browser suggest that rs833070, which is an intron variant, is positioned within a predicted regulatory element enriched in CTCF and DNase I binding sites, which may influence VEGF expression levels [50]. This observation was further supported by a second bioinformatics analysis using FastSNP [51,52].
Regarding rs2010963, the results of previous studies are conflicting. In Iranian T2DM patients, both the GG genotype and G allele of the VEGF +405 G/C polymorphism were significantly more frequent in patients with microalbuminuria (indicative of DN) times [39]. More specifically, the GG genotype was an independent predictor, increasing the risk of microalbuminuria by 2.22 times [39]. Another Iranian study confirmed the GG genotype as an independent predictor for albuminuria (p = 0.014, OR = 1.771), also noting that glomerular filtration rate (GFR) varied significantly by genotype [36]. For Han Chinese individuals, the GC/CC genotypes at rs2010963 were also significantly associated with an increased risk of T2DN (OR = 1.15), and wild-type VEGFA protein levels were higher than mutant types [27]. In contrast, a Polish study and an Irish study found no significant association between this polymorphism and DN [38]. Similarly, a separate study failed to find an association between this variant and diabetic nephropathy [37]. It has been found that the C allele of rs2010963 was associated with increased VEGFA expression, suggesting that rs2010963 functions as an eQTL for the VEGFA gene and can affect the gene expression of VEGFA in multiple normal human tissues [53].
The findings of a study indicate that the I/D polymorphism in the VEGF gene promoter is not linked to DN in West Indian patients with type 2 diabetes, but it may contribute to the development of non-diabetic nephropathy [42]. Another study in the Indian population with type 2 diabetes (T2DM), the DD genotype and D allele of the 18 bp I/D polymorphism at −2549 position were significantly associated with DN, conferring a 4.2-fold increased risk for the DD genotype and a 2.2-fold increased risk for the D allele [43]. The D allele is hypothesized to be associated with increased transcription of VEGFA. Similarly, a study on Type 1 diabetes (T1DM) patients in the UK observed a significantly increased frequency of the D/D genotype (40.2% vs. 22.7%) and the D allele in patients with nephropathy compared to uncomplicated diabetic patients [44]. Functional studies indicated that the construct containing the 18 bp deletion had a 1.95-fold increase in transcriptional activity. Conversely, a Polish study in T2DM patients found no significant association of the DD genotype or D allele with DN. However, this study did observe a significant association between the DD genotype and diabetic retinopathy and suggested an inadequate sample size for the DN analysis [38].
In addition, despite their influence on circulating VEGF levels, Bonnefond et al. reported no association between SNPs rs6921438 and rs10738760 and the risk of type 2 diabetes, diabetic nephropathy, or retinopathy [44]. However, this study reported an association between the G-allele of rs6921438 (linked to higher circulating VEGF levels) and increased T2DM risk and higher HbA1c in the French population, an observation not replicated in the Danish cohort [44]. However, in Slovenian patients with type 2 diabetes, the G allele of rs6921438-VEGF was associated with a reduced risk of developing diabetic nephropathy [45]. More specifically, individuals with the G allele (GG+AG genotypes) had a 0.66-fold lower risk of DN.
Regarding VEGF-1499 C/T polymorphism (rs833061), an Irish study on T1DM patients found a positive association between the VEGF-1499T allele and DN, which was replicated in an independent population. Carriage of the T allele was associated with a 2.24-fold increased risk of developing DN [37].
This study has several strengths, including the combined use of a case–control association study and a comprehensive meta-analysis, which together provide robust insights into the contribution of VEGFA variants to diabetic nephropathy (DN). The use of a well-characterized Greek cohort, systematic selection of tag SNPs, and rigorous statistical methods further enhance the reliability of the findings.
Nonetheless, some limitations should be acknowledged. The relatively modest sample size may have reduced statistical power for detecting small effect sizes, and the restriction to a single ethnic group limits generalizability. In addition, the lack of multiple-testing correction entails the risk significant association of rs833070 is a false-positive result. Regarding the meta-analysis, heterogeneity across studies in ethnicity, diabetes type, and DN diagnostic criteria complicates interpretation. Moreover, the lack of functional validation prevents mechanistic conclusions, and the cross-sectional design of the association study precludes causal inferences. Future research in larger, multi-ethnic cohorts with functional analyses is warranted to clarify the biological role of VEGFA variants in DN.
5. Conclusions
In conclusion, while the precise mechanisms require further elucidation, the data suggest that VEGFA polymorphisms, particularly rs833070 and rs2146323, contribute to genetic susceptibility to DN. The genetic model-free approach takes advantage of the full genotype distribution and provides a straightforward interpretation of genetic associations. However, future research should aim for larger, multi-ethnic, prospective cohort studies with detailed phenotypic characterization and functional validation to clarify the impact of these genetic variants, individually and in combination with other genetic or environmental factors. Such comprehensive efforts could ultimately enable the identification of individuals at increased risk for DN, paving the way for targeted screening and personalized therapeutic interventions.
Author Contributions
Conceptualization, I.S.; Methodology, I.S., E.D. and E.E.T.; Software, M.T.; Validation, M.D., T.E. and C.Z.; Formal Analysis, M.T.; Investigation, M.T. and C.C.; Resources, M.T., C.C. and M.D.; Data Curation, M.T. and C.Z.; Writing—Original Draft Preparation, M.T.; Writing—Review And Editing, M.T., I.S., T.E., E.E.T. and E.D.; Visualization, M.T.; Supervision, I.S.; Project Administration, I.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was approved by the University of Thessaly Ethics Committee, and informed consent was received from all participants (3/17-03-2014).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Hussain, S.; Chand Jamali, M.; Habib, A.; Hussain, M.S.; Akhtar, M.; Najmi, A.K. Diabetic Kidney Disease: An Overview of Prevalence, Risk Factors, and Biomarkers. Clin. Epidemiol. Glob. Health 2021, 9, 2–6. [Google Scholar] [CrossRef]
- Thomas, M.C.; Brownlee, M.; Susztak, K.; Sharma, K.; Jandeleit-Dahm, K.A.M.; Zoungas, S.; Rossing, P.; Groop, P.-H.; Cooper, M.E. Diabetic Kidney Disease. Nat. Rev. Dis. Primers 2015, 1, 15070. [Google Scholar] [CrossRef]
- Navarro-González, J.F.; Mora-Fernández, C.; Muros de Fuentes, M.; García-Pérez, J. Inflammatory Molecules and Pathways in the Pathogenesis of Diabetic Nephropathy. Nat. Rev. Nephrol. 2011, 7, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Sulaiman, M.K. Diabetic Nephropathy: Recent Advances in Pathophysiology and Challenges in Dietary Management. Diabetol. Metab. Syndr. 2019, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Dronavalli, S.; Duka, I.; Bakris, G. The Pathogenesis of Diabetic Nephropathy. Nat. Clin. Pract. Endocrinol. Metab. 2008, 4, 444–452. [Google Scholar] [CrossRef]
- Fernandez-Fernandez, B.; Ortiz, A.; Gomez-Guerrero, C.; Egido, J. Therapeutic Approaches to Diabetic Nephropathy—Beyond the RAS. Nat. Rev. Nephrol. 2014, 10, 325–346. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Cao, Y.; Ma, L. New Insights in the Treatment of DKD: Recent Advances and Future Prospects. BMC Nephrol. 2025, 26, 72. [Google Scholar] [CrossRef]
- Thomas, M.C.; Groop, P.-H.; Tryggvason, K. Towards Understanding the Inherited Susceptibility for Nephropathy in Diabetes. Curr. Opin. Nephrol. Hypertens. 2012, 21, 195–202. [Google Scholar] [CrossRef]
- Cole, J.B.; Florez, J.C. Genetics of Diabetes Mellitus and Diabetes Complications. Nat. Rev. Nephrol. 2020, 16, 377–390. [Google Scholar] [CrossRef]
- Haukka, J.; Sandholm, N.; Valo, E.; Forsblom, C.; Harjutsalo, V.; Cole, J.B.; McGurnaghan, S.J.; Colhoun, H.M.; Groop, P.-H.; on behalf of the FinnDiane Study Group. Novel Linkage Peaks Discovered for Diabetic Nephropathy in Individuals with Type 1 Diabetes. Diabetes 2021, 70, 986–995. [Google Scholar] [CrossRef]
- Dawn Teare, M.; Barrett, J.H. Genetic Linkage Studies. Lancet 2005, 366, 1036–1044. [Google Scholar] [CrossRef]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; de Vries, J.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-Wide Association Studies. Nat. Rev. Methods Primers 2021, 1, 59. [Google Scholar] [CrossRef]
- Salem, R.M.; Todd, J.N.; Sandholm, N.; Cole, J.B.; Chen, W.-M.; Andrews, D.; Pezzolesi, M.G.; McKeigue, P.M.; Hiraki, L.T.; Qiu, C.; et al. Genome-Wide Association Study of Diabetic Kidney Disease Highlights Biology Involved in Renal Basement Membrane Collagen. bioRxiv 2018, 499616. [Google Scholar] [CrossRef]
- Tziastoudi, M.; Stefanidis, I.; Zintzaras, E. The Genetic Map of Diabetic Nephropathy: Evidence from a Systematic Review and Meta-Analysis of Genetic Association Studies. Clin. Kidney J. 2020, 13, 768–781. [Google Scholar] [CrossRef]
- Tziastoudi, M.; Stefanidis, I.; Stravodimos, K.; Zintzaras, E. Identification of Chromosomal Regions Linked to Diabetic Nephropathy: A Meta-Analysis of Genome-Wide Linkage Scans. Genet. Test. Mol. Biomark. 2019, 23, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Tziastoudi, M.; Stefanidis, I.; Hadjigeorgiou, G.M.; Stravodimos, K.; Zintzaras, E. A Systematic Review and Meta-Analysis of Genetic Association Studies for the Role of Inflammation and the Immune System in Diabetic Nephropathy. Clin. Kidney J. 2017, 10, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Tufro, A.; Veron, D. VEGF and Podocytes in Diabetic Nephropathy. Semin. Nephrol. 2012, 32, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Schrijvers, B.F.; Flyvbjerg, A.; De Vriese, A.S. The Role of Vascular Endothelial Growth Factor (VEGF) in Renal Pathophysiology. Kidney Int. 2004, 65, 2003–2017. [Google Scholar] [CrossRef]
- Ziyadeh, F.N. Different Roles for TGF-β And VEGF in the Pathogenesis of the Cardinal Features of Diabetic Nephropathy. Diabetes Res. Clin. Pract. 2008, 82, 38–41. [Google Scholar] [CrossRef]
- Majumder, S.; Advani, A. VEGF and the Diabetic Kidney: More than Too Much of a Good Thing. J. Diabetes Complicat. 2017, 31, 273–279. [Google Scholar] [CrossRef]
- Sung, S.H.; Ziyadeh, F.N.; Wang, A.; Pyagay, P.E.; Kanwar, Y.S.; Chen, S. Blockade of Vascular Endothelial Growth Factor Signaling Ameliorates Diabetic Albuminuria in Mice. J. Am. Soc. Nephrol. 2006, 17, 3093–3104. [Google Scholar] [CrossRef]
- Chen, S.; Ziyadeh, F.N. Vascular Endothelial Growth Factor and Diabetic Nephropathy. Curr. Diabetes Rep. 2008, 8, 470–476. [Google Scholar] [CrossRef]
- Njeim, R.; Braych, K.; Ghadieh, H.E.; Azar, N.S.; Azar, W.S.; Dia, B.; Leone, A.; Cappello, F.; Kfoury, H.; Harb, F.; et al. VEGF-A: A Novel Mechanistic Link Between CYP2C-Derived EETs and Nox4 in Diabetic Kidney Disease. Diabetes 2023, 72, 947–957. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Li, J.; Zhang, J.; Chen, S. Serum VEGF as a Predictive Marker of Glycemic Control and Diabetic Nephropathy in Chinese Older Adults with Type 2 Diabetes Mellitus. Front. Endocrinol. 2023, 14, 1274025. [Google Scholar] [CrossRef]
- Mooyaart, A.L.; Valk, E.J.J.; van Es, L.A.; Bruijn, J.A.; de Heer, E.; Freedman, B.I.; Dekkers, O.M.; Baelde, H.J. Genetic Associations in Diabetic Nephropathy: A Meta-Analysis. Diabetologia 2011, 54, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Nazir, N.; Siddiqui, K.; Al-Qasim, S.; Al-Naqeb, D. Meta-Analysis of Diabetic Nephropathy Associated Genetic Variants in Inflammation and Angiogenesis Involved in Different Biochemical Pathways. BMC Med. Genet. 2014, 15, 103. [Google Scholar] [CrossRef]
- Luo, Y.; Luo, J.; Peng, H. Associations Between Genetic Polymorphisms in the VEGFA, ACE, and SOD2 Genes and Susceptibility to Diabetic Nephropathy in the Han Chinese. Genet. Test. Mol. Biomark. 2019, 23, 644–651. [Google Scholar] [CrossRef] [PubMed]
- Tziastoudi, M.; Theoharides, T.C.; Nikolaou, E.; Efthymiadi, M.; Eleftheriadis, T.; Stefanidis, I. Key Genetic Components of Fibrosis in Diabetic Nephropathy: An Updated Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2022, 23, 15331. [Google Scholar] [CrossRef]
- Siokas, V.; Dardiotis, E.; Sokolakis, T.; Kotoula, M.; Tachmitzi, S.V.; Chatzoulis, D.Z.; Almpanidou, P.; Stefanidis, I.; Hadjigeorgiou, G.M.; Tsironi, E.E. Plasminogen Activator Inhibitor Type-1 Tag Single-Nucleotide Polymorphisms in Patients with Diabetes Mellitus Type 2 and Diabetic Retinopathy. Curr. Eye Res. 2017, 42, 1048–1053. [Google Scholar] [CrossRef]
- Zintzaras, E. The Power of Generalized Odds Ratio in Assessing Association in Genetic Studies. J. Appl. Stat. 2012, 39, 2569–2581. [Google Scholar] [CrossRef]
- Zintzaras, E. The Generalized Odds Ratio as a Measure of Genetic Risk Effect in the Analysis and Meta-Analysis of Association Studies. Stat. Appl. Genet. Mol. Biol. 2010, 9. [Google Scholar] [CrossRef]
- DerSimonian, R.; Laird, N. Meta-Analysis in Clinical Trials. Control. Clin. Trials 1986, 7, 177–188. [Google Scholar] [CrossRef]
- Cochran, W.G. The Combination of Estimates from Different Experiments. Biometrics 1954, 10, 101–129. [Google Scholar] [CrossRef]
- Higgins, J.P.T.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring Inconsistency in Meta-Analyses. BMJ 2003, 327, 557–560. [Google Scholar] [CrossRef] [PubMed]
- Stefanidis, I.; Tziastoudi, M.; Tsironi, E.E.; Dardiotis, E.; Tachmitzi, S.V.; Fotiadou, A.; Pissas, G.; Kytoudis, K.; Sounidaki, M.; Ampatzis, G.; et al. The Contribution of Genetic Variants of SLC2A1 Gene in T2DM and T2DM-Nephropathy: Association Study and Meta-Analysis. Ren. Fail. 2018, 40, 561–576. [Google Scholar] [CrossRef]
- Nikzamir, A.; Esteghamati, A.; Hammedian, A.A.; Mahmoudi, T. The Role of Vascular Endothelial Growth Factor +405 G/C Polymorphism and Albuminuria in Patients with Type 2 Diabetes Mellitus. Mol. Biol. Rep. 2012, 39, 881–886. [Google Scholar] [CrossRef]
- McKnight, A.-J.; Maxwell, A.P.; Patterson, C.C.; Brady, H.R.; Savage, D.A. Association of VEGF-1499C→T Polymorphism with Diabetic Nephropathy in Type 1 Diabetes Mellitus. J. Diabetes Complicat. 2007, 21, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Buraczynska, M.; Ksiazek, P.; Baranowicz-Gaszczyk, I.; Jozwiak, L. Association of the VEGF Gene Polymorphism with Diabetic Retinopathy in Type 2 Diabetes Patients. Nephrol. Dial. Transplant. 2007, 22, 827–832. [Google Scholar] [CrossRef][Green Version]
- Fathi, M.; Nikzamir, A.R.; Esteghamati, A.; Nakhjavani, M.; Yekaninejad, M.S. Combination of Angiotensin Converting Enzyme Insertion/Deletion (I/D) (Rs4646994) and VEGF Polymorphism (+405G/C.; Rs2010963) Synergistically Associated with the Development, of Albuminuria in Iranian Patients with Type 2 Diabetes. Iran. Red Crescent Med. J. 2015, 17, e19469. [Google Scholar] [CrossRef][Green Version]
- Tiwari, A.K.; Prasad, P.; Kumar, K.P.; Ammini, A.C.; Gupta, A.; Gupta, R. Oxidative Stress Pathway Genes and Chronic Renal Insufficiency in Asian Indians with Type 2 Diabetes. J. Diabetes Complicat. 2009, 23, 102–111. [Google Scholar] [CrossRef]
- Yang, B.; Cross, D.F.; Ollerenshaw, M.; Millward, B.A.; Demaine, A.G. Polymorphisms of the Vascular Endothelial Growth Factor and Susceptibility to Diabetic Microvascular Complications in Patients with Type 1 Diabetes Mellitus. J. Diabetes Complicat. 2003, 17, 1–6. [Google Scholar] [CrossRef]
- Dabhi, B.; Mistry, K.N.; Patel, H.; Lal, S. Vascular Endothelial Growth Factor Insertion/Deletion Gene Polymorphism in West Indian Patients of Type 2 Diabetes and Diabetic Nephropathy. Indian J. Biochem. Biophys. 2015, 52, 209–212. [Google Scholar]
- Amle, D.; Mir, R.; Khaneja, A.; Agarwal, S.; Ahlawat, R.; Ray, P.C.; Saxena, A. Association of 18bp Insertion/Deletion Polymorphism, at −2549 Position of VEGF Gene, with Diabetic Nephropathy in Type 2 Diabetes Mellitus Patients of North Indian Population. J. Diabetes Metab. Disord. 2015, 14, 19. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bonnefond, A.; Saulnier, P.-J.; Stathopoulou, M.G.; Grarup, N.; Ndiaye, N.C.; Roussel, R.; Nezhad, M.A.; Dechaume, A.; Lantieri, O.; Hercberg, S.; et al. What Is the Contribution of Two Genetic Variants Regulating VEGF Levels to Type 2 Diabetes Risk and to Microvascular Complications? PLoS ONE 2013, 8, e55921. [Google Scholar] [CrossRef]
- Nussdorfer, P.; Petrovič, D.; Alibegović, A.; Cilenšek, I.; Petrovič, D. The KDR Gene Rs2071559 and the VEGF Gene Rs6921438 May Be Associated with Diabetic Nephropathy in Caucasians with Type 2 Diabetes Mellitus. Int. J. Mol. Sci. 2024, 25, 9439. [Google Scholar] [CrossRef]
- Trégouet, D.-A.; Groop, P.-H.; McGinn, S.; Forsblom, C.; Hadjadj, S.; Marre, M.; Parving, H.-H.; Tarnow, L.; Telgmann, R.; Godefroy, T.; et al. G/T Substitution in Intron 1 of the UNC13B Gene Is Associated with Increased Risk of Nephropathy in Patients with Type 1 Diabetes. Diabetes 2008, 57, 2843–2850. [Google Scholar] [CrossRef]
- Ma, R.C.W.; Cooper, M.E. Genetics of Diabetic Kidney Disease-From the Worst of Nightmares to the Light of Dawn? J. Am. Soc. Nephrol. 2017, 28, 389–393. [Google Scholar] [CrossRef]
- Jourde-Chiche, N.; Fakhouri, F.; Dou, L.; Bellien, J.; Burtey, S.; Frimat, M.; Jarrot, P.-A.; Kaplanski, G.; Le Quintrec, M.; Pernin, V.; et al. Endothelium Structure and Function in Kidney Health and Disease. J. Am. Soc. Nephrol. 2019, 15, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Kim, M.-J.; Kumar, A.; Lee, H.-W.; Yang, Y.; Kim, Y. Vascular Endothelial Growth Factor Signaling in Health and Disease: From Molecular Mechanisms to Therapeutic Perspectives. Signal Transduct. Target. Ther. 2025, 10, 170. [Google Scholar] [CrossRef] [PubMed]
- Flicek, P.; Ahmed, I.; Amode, M.R.; Barrell, D.; Beal, K.; Brent, S.; Carvalho-Silva, D.; Clapham, P.; Coates, G.; Fairley, S.; et al. Ensembl 2013. Nucleic Acids Res. 2013, 41, D48–D55. [Google Scholar] [CrossRef]
- Hagstrom, S.A.; Ying, G.; Pauer, G.J.; Sturgill-Short, G.M.; Huang, J.; Maguire, M.G.; Martin, D.F. VEGF-A and VEGFR-2 Gene Polymorphisms and Response to Anti-VEGF Therapy in the Comparison of AMD Treatments Trials (CATT). JAMA Ophthalmol. 2014, 132, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.-Y.; Chiou, J.-J.; Tseng, W.-H.; Liu, C.-H.; Liu, C.-K.; Lin, Y.-J.; Wang, H.-H.; Yao, A.; Chen, Y.-T.; Hsu, C.-N. FASTSNP: An Always up-to-Date and Extendable Service for SNP Function Analysis and Prioritization. Nucleic Acids Res. 2006, 34, W635–641. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Xin, K.; Chen, K.; Tang, H.; Chen, H.; Zhi, L.; Liu, H. Relationship of Common Variants in VEGFA Gene with Osteonecrosis of the Femoral Head: A Han Chinese Population Based Association Study. Sci. Rep. 2018, 8, 16221. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).