Structural Conservation and Transcriptional Plasticity of atp2a1 in Acrossocheilus fasciatus Under Temperature and Flow Acclimation
Abstract
1. Introduction
2. Materials and Methods
2.1. Research Background
2.2. Bioinformatic Analysis of the atp2a1 Gene
2.3. atp2a1 Gene Tissue Expression Analysis
2.4. Promoter Region Prediction of atp2a1
2.5. Expression Profiling of atp2a1 Under Various Stress and Acclimation Conditions
3. Results
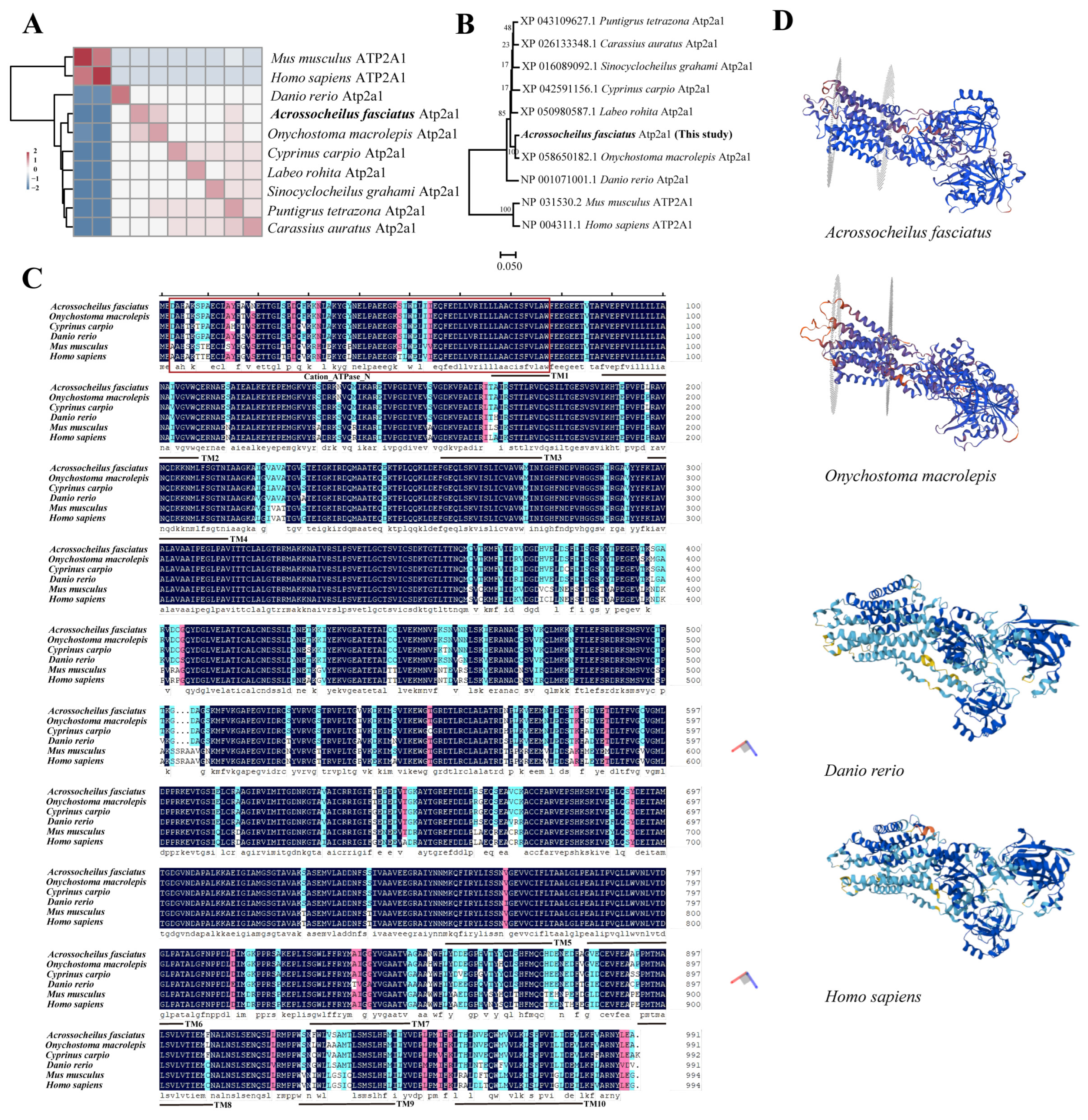
3.1. Bioinformatic Analysis and Phylogenetic Analysis
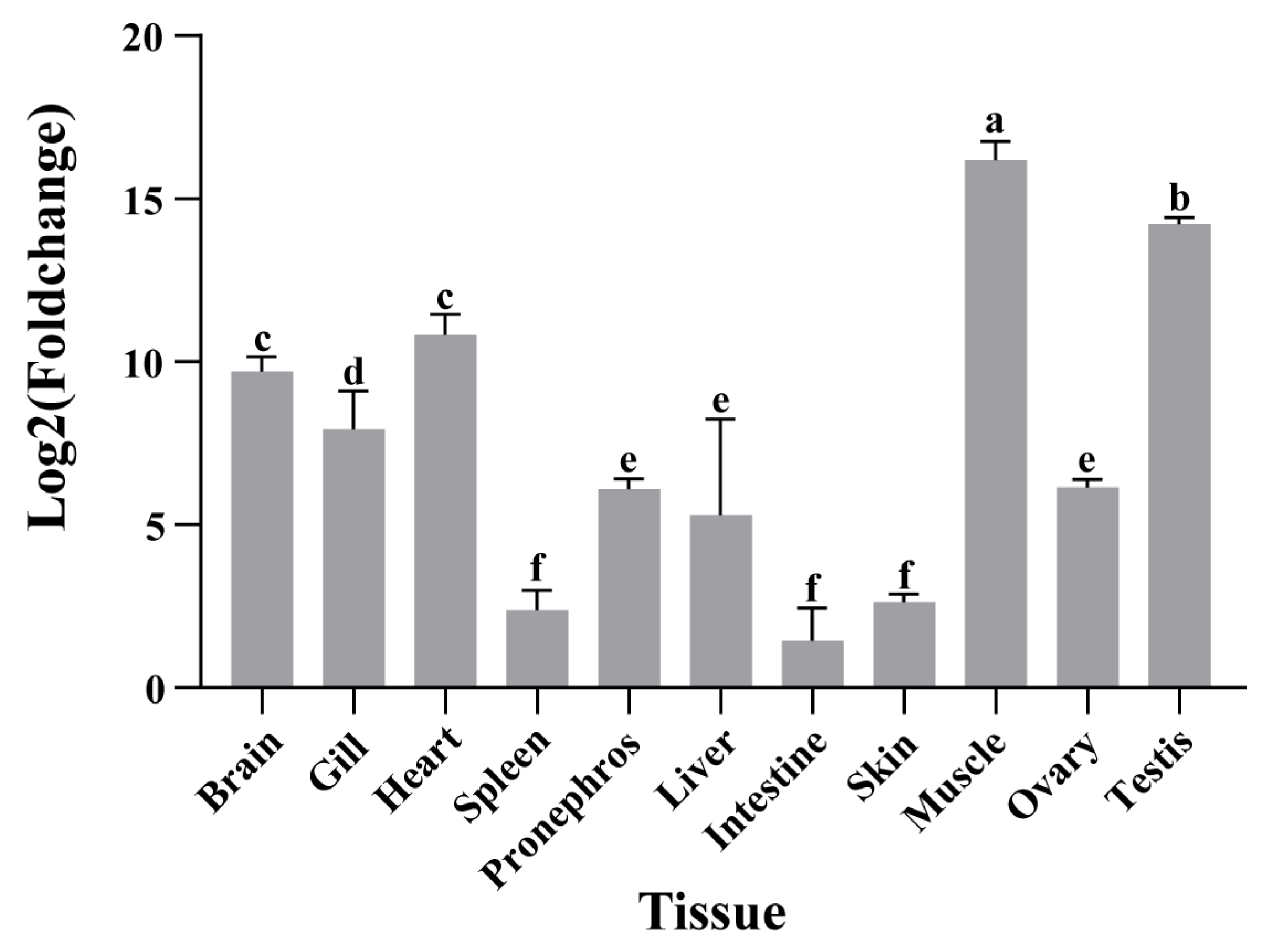
3.2. Expression of A. fasciatus atp2a1 Under Different Tissues
3.3. Promoter Prediction
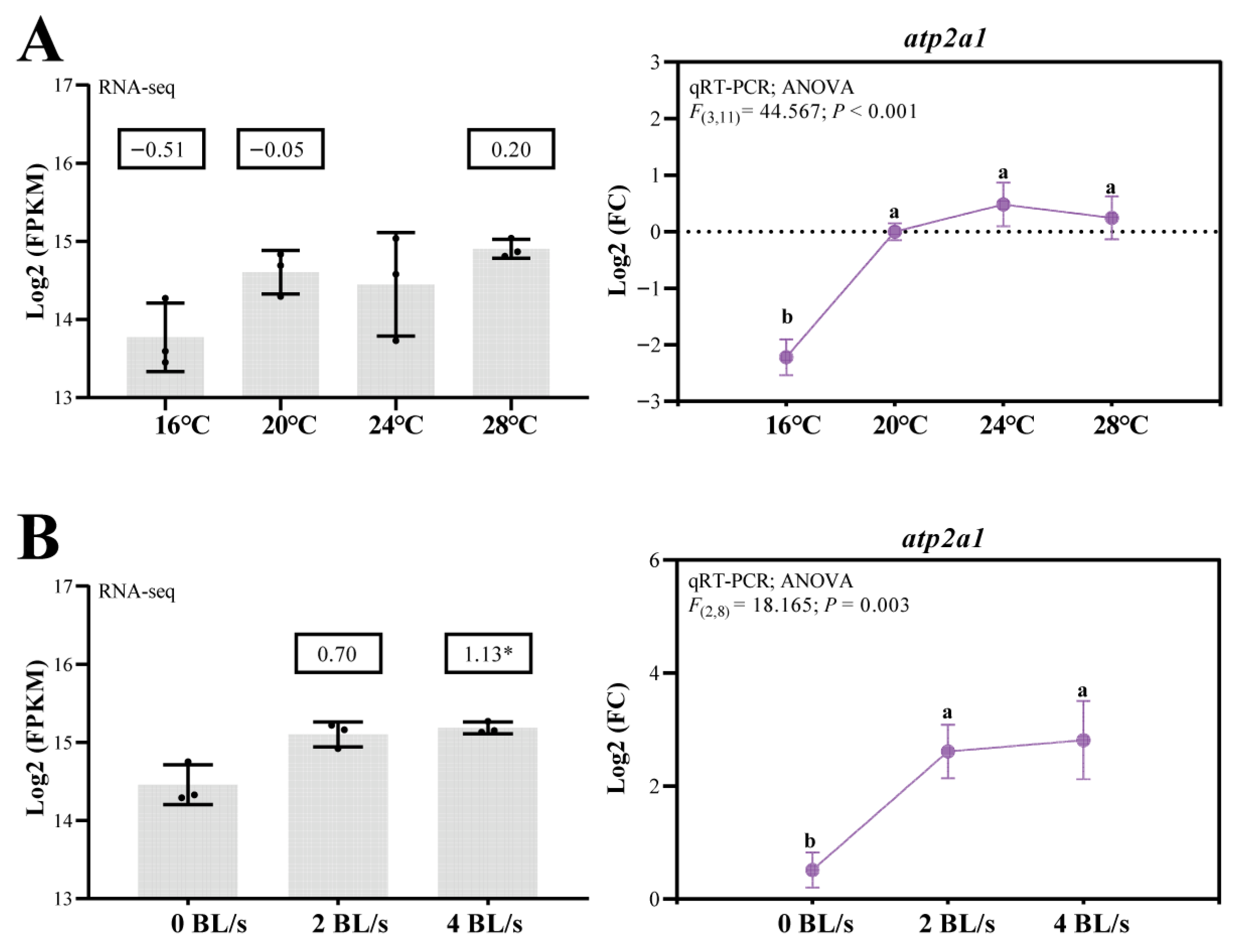
3.4. Regulation of A. fasciatus atp2a1 Expression and SE Alternative Splicing Under Temperature and Flow Velocity Treatments
4. Discussion
4.1. Phylogenetic Conservation, Structural Divergence of atp2a1
4.2. Tissue Specificity of atp2a1
4.3. Promoter Prediction and Transcriptional Regulation of atp2a1
4.4. Thermal and Hydrodynamic Stress Response
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AS | Alternative splicing |
| CBP | CREB-binding protein |
| CpG | Cytosine-phosphate-Guanine dinucleotide |
| DEG | Differentially expressed gene |
| ER | Endoplasmic reticulum |
| FDR | False discovery rate |
| FPKM | Fragments per kilobase of transcript per million mapped reads |
| GC | Guanine-Cytosine content |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MW | Molecular weight |
| NJ | Neighbor-joining (phylogenetic method) |
| ORF | Open reading frame |
| PCR | Polymerase chain reaction |
| pI | Isoelectric point |
| PSI | Percent spliced in (value for AS events) |
| qRT-PCR | Quantitative reverse transcription polymerase chain reaction |
| SE | Skipped exon |
| ATP2A1 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 () |
| TMR | Transmembrane region |
| TF | Transcription factor |
| TFBS | Transcription factor binding site |
| TSS | Transcription start site |
| Note: The fully uppercase form (ATP2A1) refers to the mammalian ortholog; the capitalized form (Atp2a1) refers to the fish ortholog; and the lowercase italicized form (atp2a1) denotes the gene. | |
References
- Palmgren, M.G.; Nissen, P. P-Type ATPases. Annu. Rev. Biophys. 2011, 40, 243–266. [Google Scholar] [CrossRef]
- Periasamy, M.; Kalyanasundaram, A. SERCA Pump Isoforms: Their Role in Calcium Transport and Disease. Muscle Nerve 2007, 35, 430–442. [Google Scholar] [CrossRef] [PubMed]
- Hirata, H.; Saint-Amant, L.; Waterbury, J.; Cui, W.; Zhou, W.; Li, Q.; Goldman, D.; Granato, M.; Kuwada, J.Y. Accordion, a Zebrafish Behavioral Mutant, Has a Muscle Relaxation Defect Due to a Mutation in the ATPase Ca2+ Pump SERCA1. Development 2004, 131, 5457–5468. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.-Y.; Pai, C.-W.; Tsai, I.-T.; Chou, C.-Y.; Tsai, C.-T.; Chen, Y.-H. Molecular Structure and Developmental Expression of Zebrafish atp2a Genes. Genes Genom. 2011, 33, 541–548. [Google Scholar] [CrossRef]
- Toyoshima, C.; Nakasako, M.; Nomura, H.; Ogawa, H. Crystal Structure of the Calcium Pump of Sarcoplasmic Reticulum at 2.6 Å Resolution. Nature 2000, 405, 647–655. [Google Scholar] [CrossRef]
- Arruda, A.P.; Nigro, M.; Oliveira, G.M.; de Meis, L. Thermogenic Activity of Ca2+-ATPase from Skeletal Muscle Heavy Sarcoplasmic Reticulum: The Role of Ryanodine Ca2+ Channel. Biochim. Biophys. Acta Biomembr. 2007, 1768, 1498–1505. [Google Scholar] [CrossRef]
- Launikonis, B.S.; Murphy, R.M. From Muscle-Based Nonshivering Thermogenesis to Malignant Hyperthermia in Mammals. Annu. Rev. Physiol. 2025, 87, 131–150. [Google Scholar] [CrossRef]
- Xu, H.; Van Remmen, H. The Sarco/Endoplasmic Reticulum Calcium ATPase (SERCA) Pump: A Potential Target for Intervention in Aging and Skeletal Muscle Pathologies. Skelet. Muscle 2021, 11, 25. [Google Scholar] [CrossRef]
- Robinson, S.; Hechter, D.; Almoumen, F.; Franck, J.P. Sarcolipin (sln) and Sarcoplasmic Reticulum Calcium ATPase Pump (serca1) Expression Increase in Japanese Medaka (Oryzias latipes) Skeletal Muscle Tissue Following Cold Challenge. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2024, 287, 111534. [Google Scholar] [CrossRef]
- Robinson, S.; Wegner, N.C.; Sepulveda, C.A.; Franck, J.P. Relative Sarcolipin (SLN) and Sarcoplasmic Reticulum Ca2+ ATPase (SERCA1) Transcript Levels in Closely Related Endothermic and Ectothermic Scombrid Fishes: Implications for Molecular Basis of Futile Calcium Cycle Non-Shivering Thermogenesis (NST). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2024, 295, 111667. [Google Scholar] [CrossRef]
- Chou, M.-Y.; Hsiao, C.-D.; Chen, S.-C.; Chen, I.-W.; Liu, S.-T.; Hwang, P.-P. Effects of Hypothermia on Gene Expression in Zebrafish Gills: Upregulation in Differentiation and Function of Ionocytes as Compensatory Responses. J. Exp. Biol. 2008, 211, 3077–3084. [Google Scholar] [CrossRef]
- Pan, Y.; Zvaritch, E.; Tupling, A.R.; Rice, W.J.; de Leon, S.; Rudnicki, M.; McKerlie, C.; Banwell, B.L.; MacLennan, D.H. Targeted Disruption of the ATP2A1 Gene Encoding the Sarco (Endo)plasmic Reticulum Ca2+ ATPase Isoform 1 (SERCA1) Impairs Diaphragm Function and Is Lethal in Neonatal Mice. J. Biol. Chem. 2003, 278, 13367–13375. [Google Scholar] [CrossRef]
- Zhao, Y.; Ogawa, H.; Yonekura, S.-I.; Mitsuhashi, H.; Mitsuhashi, S.; Nishino, I.; Toyoshima, C.; Ishiura, S. Functional Analysis of SERCA1b, a Highly Expressed SERCA1 Variant in Myotonic Dystrophy Type 1 Muscle. Biochim. Biophys. Acta Mol. Basis Dis. 2015, 1852, 2042–2047. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Shi, W.; Guo, Y.; Chen, Y.; Wang, H.; He, J.; Chu, Z. Effect of Temperature on the Growth, Feeding Performance, Gonadal Development, and Nutritive Compositions in the Muscle of Fry Stream Groupers, Acrossocheilus fasciatus. J. World Aquac. Soc. 2024, 55, e13024. [Google Scholar] [CrossRef]
- Guo, Y.; Dong, C.; Peng, H.; Zhang, J.; He, J.; Gao, Y.; Dai, X.; Zhao, S.; Chu, Z.; Zhao, B.; et al. Behavioral Responses and Transcriptional Dynamics of the Stream Fish (Acrossocheilus fasciatus) under Temperature Change. Water Biol. Secur. 2025, 5, 100413. [Google Scholar] [CrossRef]
- He, J.; Wang, H.; Guo, Y.; Chu, Z.; Zhao, B. Molecular Mechanism of Extreme Hypoxia Tolerance Difference between Male and Female Adult Fish and Juvenile Fish of Acrossocheilus fasciatus by Transcriptomics. Indian. J. Anim. Res. 2022, 56, 7–14. [Google Scholar] [CrossRef]
- Huang, J.; Tong, H.; Gao, B.; Wu, Y.; Li, W.; Xiao, P. Long-Term Exposure to Dimefluthrin Inhibits the Growth of Acrossocheilus fasciatus. Environ. Res. 2024, 260, 119617. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, S.; Niyompano, F.; Li, T.; Chen, J.; Luo, Z.; Jiang, X.; Chen, Y.; Zhao, B. Identification and Characterization of hsp70 Gene Family in Acrossocheilus fasciatus Based on Genome and Full-Length Transcripts. Comp. Biochem. Physiol. D Genom. Proteom. 2025, 55, 101480. [Google Scholar] [CrossRef]
- Wei, Z.; Fang, Y.; Shi, W.; Chu, Z.; Zhao, B. Transcriptional Modulation Reveals Physiological Responses to Temperature Adaptation in Acrossocheilus fasciatus. Int. J. Mol. Sci. 2023, 24, 11622. [Google Scholar] [CrossRef]
- Zheng, J.; Jiang, J.; Rui, Q.; Li, F.; Liu, S.; Cheng, S.; Chi, M.; Jiang, W. Chromosome-Level Genome Assembly of Acrossocheilus fasciatus Using PacBio Sequencing and Hi-C Technology. Sci. Data 2024, 11, 166. [Google Scholar] [CrossRef]
- Yuan, X.; Tao, L.; Hu, X.; Lin, R.; Yang, J.; Feng, M.; Peng, M.; Liu, W.; Xiao, Y. Expression Profile Analysis of Muscle Regulation Genes under Growth and Water Flow Stress in Zebrafish. Reprod. Breed. 2024, 4, 5–9. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.E.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar] [CrossRef]
- Thumuluri, V.; Almagro Armenteros, J.J.; Johansen, A.R.; Nielsen, H.; Winther, O. DeepLoc 2.0: Multi-label subcellular localization prediction using protein language models. Nucleic Acids Res. 2022, 50, W228–W234. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. SMART v10: Three decades of the protein domain annotation resource. Nucleic Acids Res 2025, 53, gkaf1023. [Google Scholar] [CrossRef]
- Hallgren, J.; Tsirigos, K.D.; Pedersen, M.D.; Almagro Armenteros, J.J.; Marcatili, P.; Nielsen, H.; Winther, O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. bioRxiv 2022. [Google Scholar] [CrossRef]
- Nielsen, H.; Teufel, F.; Brunak, S.; von Heijne, G. SignalP: The evolution of a web server. In Protein Bioinformatics; Kihara, D., Ed.; Springer: New York, NY, USA, 2024; pp. 331–367. [Google Scholar] [CrossRef]
- Buchan, D.W.; Moffat, L.; Lau, A.; Kandathil, S.M.; Jones, D.T. Deep learning for the PSIPRED protein analysis workbench. Nucleic Acids Res. 2024, 52, W287–W293. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Fleming, J.; Magana, P.; Nair, S.; Tsenkov, M.; Bertoni, D.; Pidruchna, I.; Querino Lima Afonso, M.; Midlik, A.; Paramval, U.; Žídek, A.; et al. AlphaFold Protein Structure Database and 3D-Beacons: New data and capabilities. J. Mol. Biol 2025, 168967. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Madeira, F.; Madhusoodanan, N.; Lee, J.; Eusebi, A.; Niewielska, A.; Tivey, A.R.N.; Lopez, R.; Butcher, S. The EMBL-EBI Job Dispatcher sequence analysis tools framework in 2024. Nucleic Acids Res. 2024, 52, W521–W525. [Google Scholar] [CrossRef]
- Lalitha, S. Primer Premier 5. Biotech Softw. Internet Rep. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- Ren, Y.; Mu, Y.; Zhao, B.; Gao, Y.; Dai, X.; Chu, Z. dmrt3, nom1, abce1, and pkmyt1 Play Key Roles in Gonadal Sex Determination in Acrossocheilus fasciatus. Aquac. Int. 2023, 31, 317–332. [Google Scholar] [CrossRef]
- Reese, M.G. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 2001, 26, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Li, L.C.; Dahiya, R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics 2002, 18, 1427–1431. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Shen, S.; Park, J.W.; Lu, Z.-X.; Lin, L.; Henry, M.D.; Wu, Y.N.; Zhou, Q.; Xing, Y. rMATS: Robust and Flexible Detection of Differential Alternative Splicing from Replicate RNA-Seq Data. Proc. Natl. Acad. Sci. USA 2014, 111, E5593–E5601. [Google Scholar] [CrossRef]
- Zhao, B.; Huang, H.; Guo, Y.; Uzanyinema, T.; Yu, J.; Zhang, Q.; Chu, Z. Morphological and transcriptomic analysis of testes in juvenile Acrossocheilus fasciatus under different water flow conditions. Oceanol. Limnol. Sin. 2025; in press. (In Chinese) [Google Scholar]
- Raguimova, O.N.; Smolin, N.; Blackwell, D.; Bovo, E.; Zima, A.; Robia, S. A Discrete Loop of the SERCA N-Domain Interacts with Phospholamban and Stabilizes a Compact Conformation of the SERCA Cytosolic Headpiece. Biophys. J. 2017, 112 (Suppl. 1), 47a. [Google Scholar] [CrossRef]
- Fernández-de Gortari, E.; Aguayo-Ortiz, R.; Autry, J.M.; Espinoza-Fonseca, L.M. A Hallmark of Phospholamban Functional Divergence Is Located in the N-terminal Phosphorylation Domain. Comput. Struct. Biotechnol. J. 2020, 18, 705–713. [Google Scholar] [CrossRef]
- Olesen, C.; Picard, M.; Winther, A.-M.L.; Gyrup, C.; Morth, J.P.; Oxvig, C.; Møller, J.V.; Nissen, P. The Structural Basis of Calcium Transport by the Calcium Pump. Nature 2007, 450, 1036–1042. [Google Scholar] [CrossRef]
- Toyoshima, C. How Ca2+-ATPase Pumps Ions across the Sarcoplasmic Reticulum Membrane. Biochim. Biophys. Acta Mol. Cell Res. 2009, 1793, 941–946. [Google Scholar] [CrossRef]
- Garriga, F.; Martínez-Hernández, J.; Parra-Balaguer, A.; Llavanera, M.; Yeste, M. The Sarcoplasmic/Endoplasmic Reticulum Ca2+-ATPase (SERCA) Is Present in Pig Sperm and Modulates Their Physiology over Liquid Preservation. Sci. Rep. 2025, 15, 4184. [Google Scholar] [CrossRef] [PubMed]
- Lawson, C.; Dorval, V.; Goupil, S.; Leclerc, P. Identification and Localisation of SERCA 2 Isoforms in Mammalian Sperm. Mol. Hum. Reprod. 2007, 13, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Liao, B.-K.; Deng, A.-N.; Chen, S.-C.; Chou, M.-Y.; Hwang, P.-P. Expression and Water Calcium Dependence of Calcium Transporter Isoforms in Zebrafish Gill Mitochondrion-Rich Cells. BMC Genom. 2007, 8, 354. [Google Scholar] [CrossRef] [PubMed]
- Pinto, P.I.; Matsumura, H.; Thorne, M.A.; Power, D.M.; Terauchi, R.; Reinhardt, R.; Canário, A.V.M. Gill Transcriptome Response to Changes in Environmental Calcium in the Green Spotted Puffer Fish. BMC Genom. 2010, 11, 476. [Google Scholar] [CrossRef] [PubMed]
- Salem, M.; Al-Tobasei, R.; Ali, A.; Kenney, B. Integrated Analyses of DNA Methylation and Gene Expression of Rainbow Trout Muscle under Variable Ploidy and Muscle Atrophy Conditions. Genes 2022, 13, 1151. [Google Scholar] [CrossRef]
- Chami, M.; Oulès, B.; Szabadkai, G.; Tacine, R.; Rizzuto, R.; Paterlini-Bréchot, P. Role of SERCA1 Truncated Isoform in the Proapoptotic Calcium Transfer from ER to Mitochondria during ER Stress. Mol. Cell 2008, 32, 641–651. [Google Scholar] [CrossRef]
- Jin, S.; Kim, J.; Willert, T.; Klein-Rodewald, T.; Garcia-Dominguez, M.; Mosqueira, M.; Fink, R.; Esposito, I.; Hofbauer, L.C.; Charnay, P.; et al. Ebf Factors and MyoD Cooperate to Regulate Muscle Relaxation via Atp2a1. Nat. Commun. 2014, 5, 3793. [Google Scholar] [CrossRef]
- Hayashi, S.; Manabe, I.; Suzuki, Y.; Relaix, F.; Oishi, Y. Klf5 regulates muscle differentiation by directly targeting muscle-specific genes in cooperation with MyoD in mice. eLife 2016, 5, e17462. [Google Scholar] [CrossRef]
- Pei, H.; Yao, Y.; Yang, Y.; Liao, K.; Wu, J.R. Krüppel-like factor KLF9 regulates PPARγ transactivation at the middle stage of adipogenesis. Cell Death Differ. 2011, 18, 315–327. [Google Scholar] [CrossRef]
- Gans, I.M.; Grendler, J.; Babich, R.; Jayasundara, N.; Coffman, J.A. Glucocorticoid-responsive transcription factor Krüppel-like factor 9 regulates fkbp5 and metabolism. Front. Cell Dev. Biol. 2021, 9, 727037. [Google Scholar] [CrossRef]
- Bossone, S.A.; Asselin, C.; Patel, A.J.; Marcu, K.B. MAZ, a zinc finger protein, binds to c-MYC and C2 gene sequences regulating transcriptional initiation and termination. Proc. Natl. Acad. Sci. USA 1992, 89, 7452–7456. [Google Scholar] [CrossRef]
- Little, A.G.; Seebacher, F. Thyroid hormone regulates muscle function during cold acclimation in zebrafish (Danio rerio). J. Exp. Biol. 2013, 216, 3514–3521. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, Y.; Wang, Z.; Luo, T.; Huang, J.; Shao, J. Analysis of Differential Alternative Splicing in Largemouth Bass after High Temperature Exposure. Animals 2024, 14, 3005. [Google Scholar] [CrossRef] [PubMed]
- Suriyampola, P.S.; Zúñiga-Vega, J.J.; Jayasundara, N.; Flores, J.; Lopez, M.; Bhat, A.; Martins, E.P. River Zebrafish Combine Behavioral Plasticity and Generalized Morphology with Specialized Sensory and Metabolic Physiology to Survive in a Challenging Environment. Sci. Rep. 2023, 13, 16398. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Feng, W.; Chen, X.; Song, C.; Su, S.; Ge, J.; Tang, Y. Molecular Cloning and Functional Characterization of Sarco/Endoplasmic Reticulum Ca2+-ATPase from Chinese Mitten Crab (Eriocheir sinensis). Aquac. Res. 2022, 53, 4676–4688. [Google Scholar] [CrossRef]
- Mandal, A.; Arunachalam, S.C.; Meleshkevitch, E.A.; Mandal, P.K.; Boudko, D.Y.; Ahearn, G.A. Cloning of Sarco-Endoplasmic Reticulum Ca2+-ATPase (SERCA) from Caribbean Spiny Lobster Panulirus argus. J. Comp. Physiol. B 2009, 179, 205–214. [Google Scholar] [CrossRef][Green Version]
- Wang, Y.; Luo, P.; Zhang, L.; Hu, C.; Ren, C.; Xia, J. Cloning of Sarco/Endoplasmic Reticulum Ca2+-ATPase (SERCA) Gene from White Shrimp Litopenaeus vannamei and Its Expression Level Analysis under Salinity Stress. Mol. Biol. Rep. 2013, 40, 6213–6221. [Google Scholar] [CrossRef]
- Roegner, M.E.; Chen, H.Y.; Watson, R.D. Molecular Cloning and Characterization of a Sarco/Endoplasmic Reticulum Ca2+-ATPase (SERCA) from Y-Organs of the Blue Crab Callinectes sapidus. Gene 2018, 673, 12–21. [Google Scholar] [CrossRef]
| Start | End | Score | Promoter Sequence (5′−3′) |
|---|---|---|---|
| 201 | 251 | 0.97 | ggagggcactgtatatacacacacacacacacacacacacacacacacac |
| 255 | 305 | 0.88 | aaaaacatataataaaaaggagacaagtagatagaatagagaaagctagt |
| 1038 | 1088 | 0.82 | tcatataaattatatgaattcgccaaacgtagttataagttgtcacgaga |
| 1480 | 1530 | 0.83 | cagcactcctctaactgccccctcccatcagaggccacttggcccatcac |
| 1921 | 1971 | 0.95 | agggacaatcaataaaaggcacggatagacctgtttaactaagctttcgc |
| Transcription Factor | Star/bp | End/bp | Strand | Score | q-Value | Matched Sequence |
|---|---|---|---|---|---|---|
| KLF9 | 216 | 226 | + | 10.4065 | 6.88 × 10−5 | tacacacacac |
| 218 | 228 | + | 12.2033 | 2.57 × 10−5 | cacacacacac | |
| CTCF | 1485 | 1517 | + | 12.2752 | 7 × 10−6 | ctcctctaactgccccctcccatcagaggccac |
| MAZ | 1498 | 1505 | + | 16.0816 | 8.13 × 10−6 | cccctccc |
| KLF5 | 1497 | 1506 | + | 13.4082 | 2.25 × 10−5 | ccccctccca |
| ONECUT3 | 1924 | 1935 | + | 13.3708 | 1.2 × 10−5 | gacaatcaataa |
| HOXB13 | 1929 | 1937 | + | 13.0873 | 2.65 × 10−5 | tcaataaaa |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Guo, Y.; Cai, P.; Chu, Z.; Zhao, B. Structural Conservation and Transcriptional Plasticity of atp2a1 in Acrossocheilus fasciatus Under Temperature and Flow Acclimation. Genes 2025, 16, 1385. https://doi.org/10.3390/genes16111385
Chen Y, Guo Y, Cai P, Chu Z, Zhao B. Structural Conservation and Transcriptional Plasticity of atp2a1 in Acrossocheilus fasciatus Under Temperature and Flow Acclimation. Genes. 2025; 16(11):1385. https://doi.org/10.3390/genes16111385
Chicago/Turabian StyleChen, Ye, Yongyao Guo, Peihao Cai, Zhangjie Chu, and Bo Zhao. 2025. "Structural Conservation and Transcriptional Plasticity of atp2a1 in Acrossocheilus fasciatus Under Temperature and Flow Acclimation" Genes 16, no. 11: 1385. https://doi.org/10.3390/genes16111385
APA StyleChen, Y., Guo, Y., Cai, P., Chu, Z., & Zhao, B. (2025). Structural Conservation and Transcriptional Plasticity of atp2a1 in Acrossocheilus fasciatus Under Temperature and Flow Acclimation. Genes, 16(11), 1385. https://doi.org/10.3390/genes16111385







