MeJA Elicitation on Flavonoid Biosynthesis and Gene Expression in the Hairy Roots of Glycyrrhiza glabra L.
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Inoculation of A. rhizogenes K599
2.3. Induction and Parameter Optimization of the HRs
2.4. PCR Analysis to Screen the Transgenic HRs
2.5. Analysis of the Growth Kinetics of HRs
2.6. HRs Treatments by Elicitors
2.7. Use of HPLC-MS to Quantify the Contents of Flavonoids in the HRs
2.8. Transcriptome Analysis and Gene Annotation
2.9. Validation of Gene Expression by Quantitative Real-Time PCR
2.10. Construction of GgCHS6 That Overexpresses the HRs
2.11. Data Analysis
3. Results
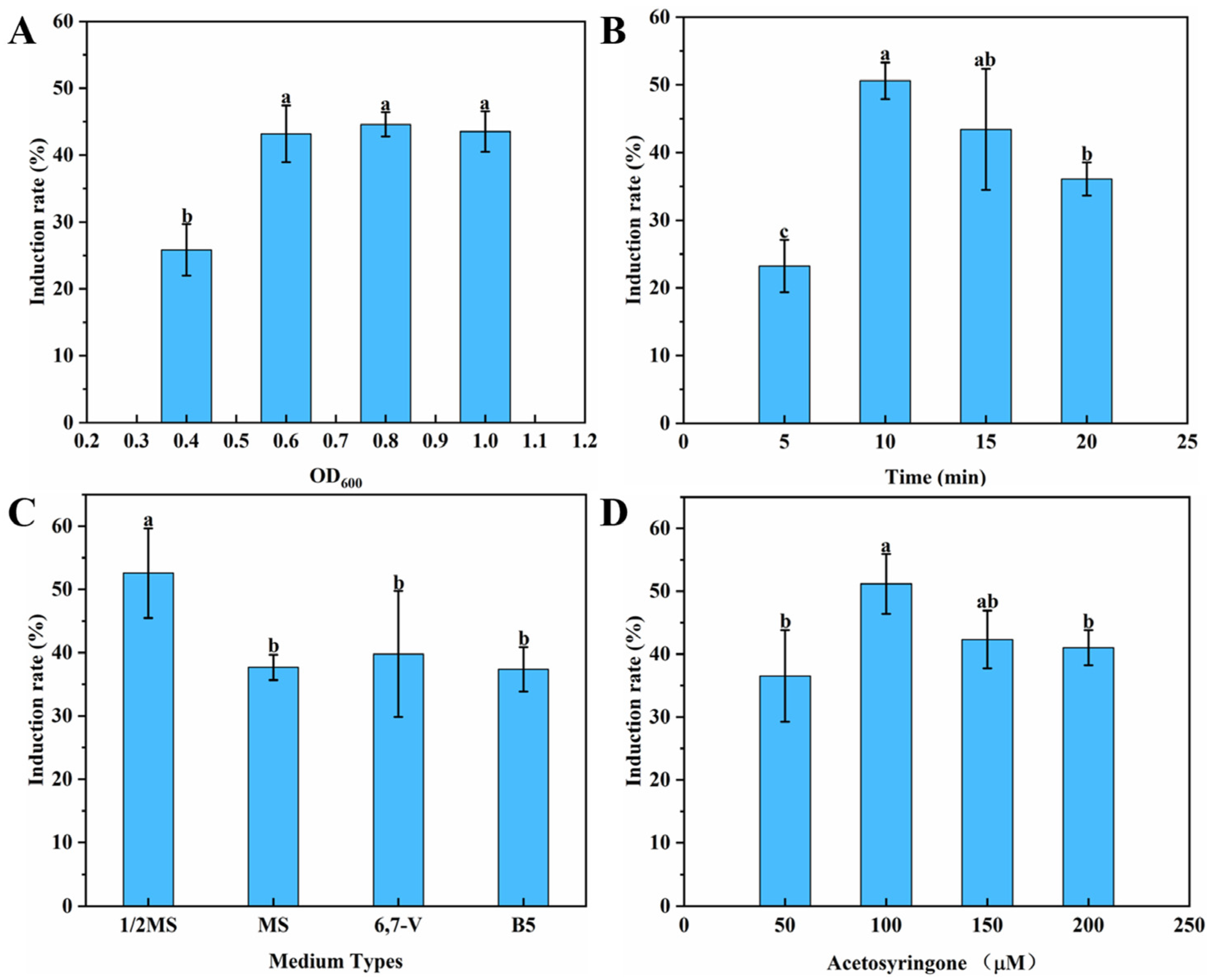
3.1. Optimization of the Induction and Condition of the HRs
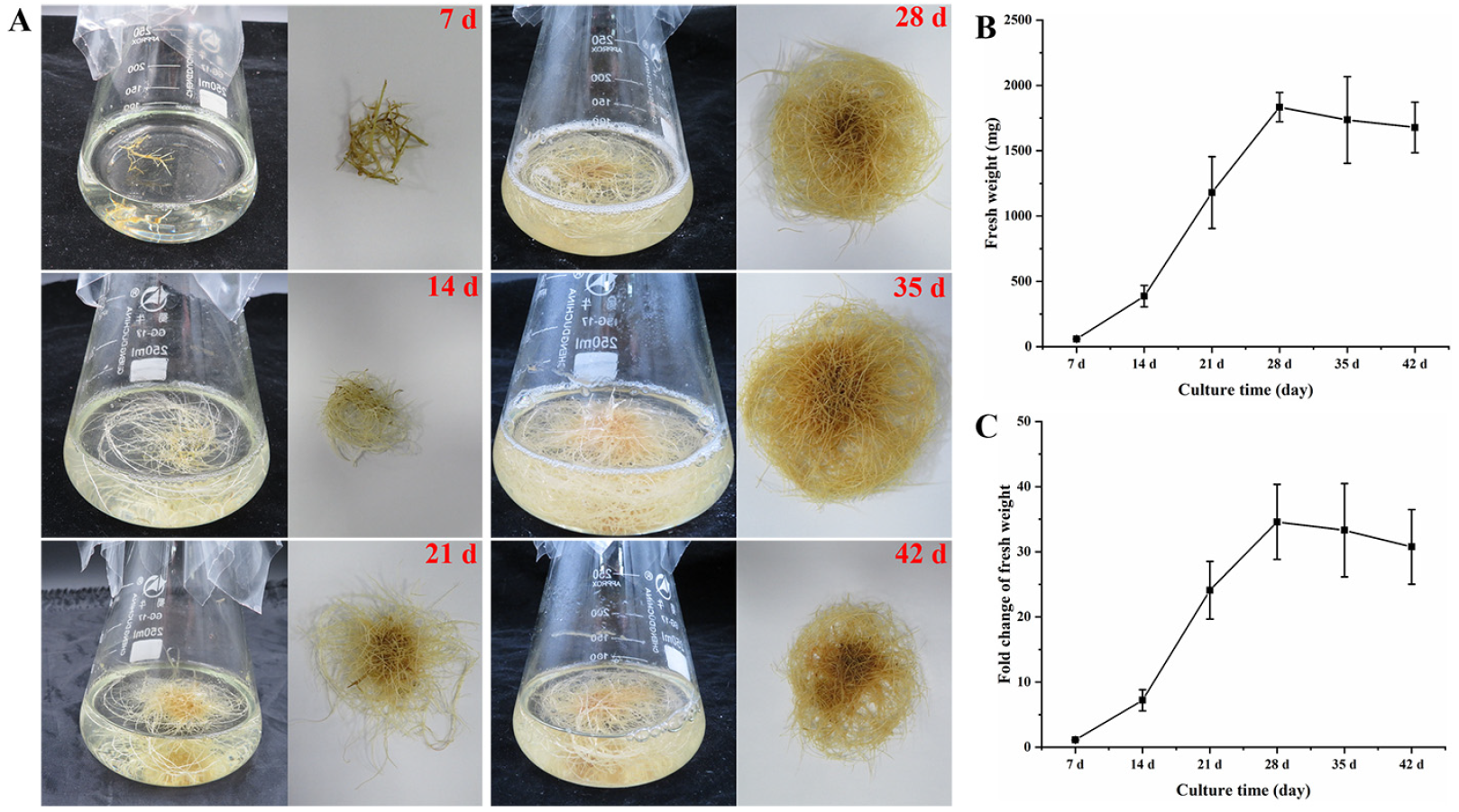
3.2. Growth Kinetics of G. glabra HRs
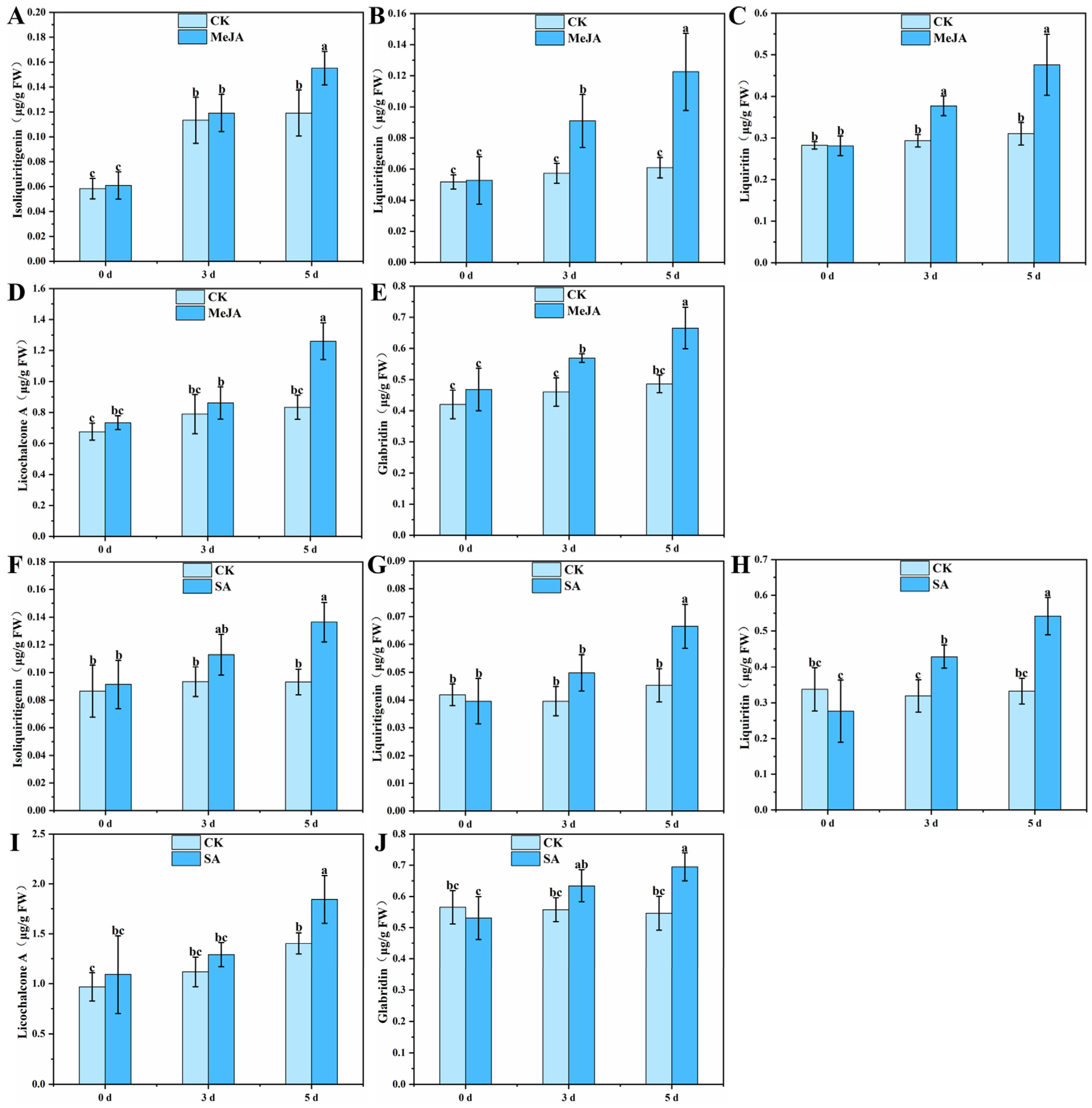
3.3. Effects of MeJA and SA on the Content of Flavonoids in G. glabra HRs
3.4. GO Association and KEGG Enrichment Analysis in Two Comparisons
3.5. Expression Clustering Revealed Patterns of Flavonoids Associated with Genes
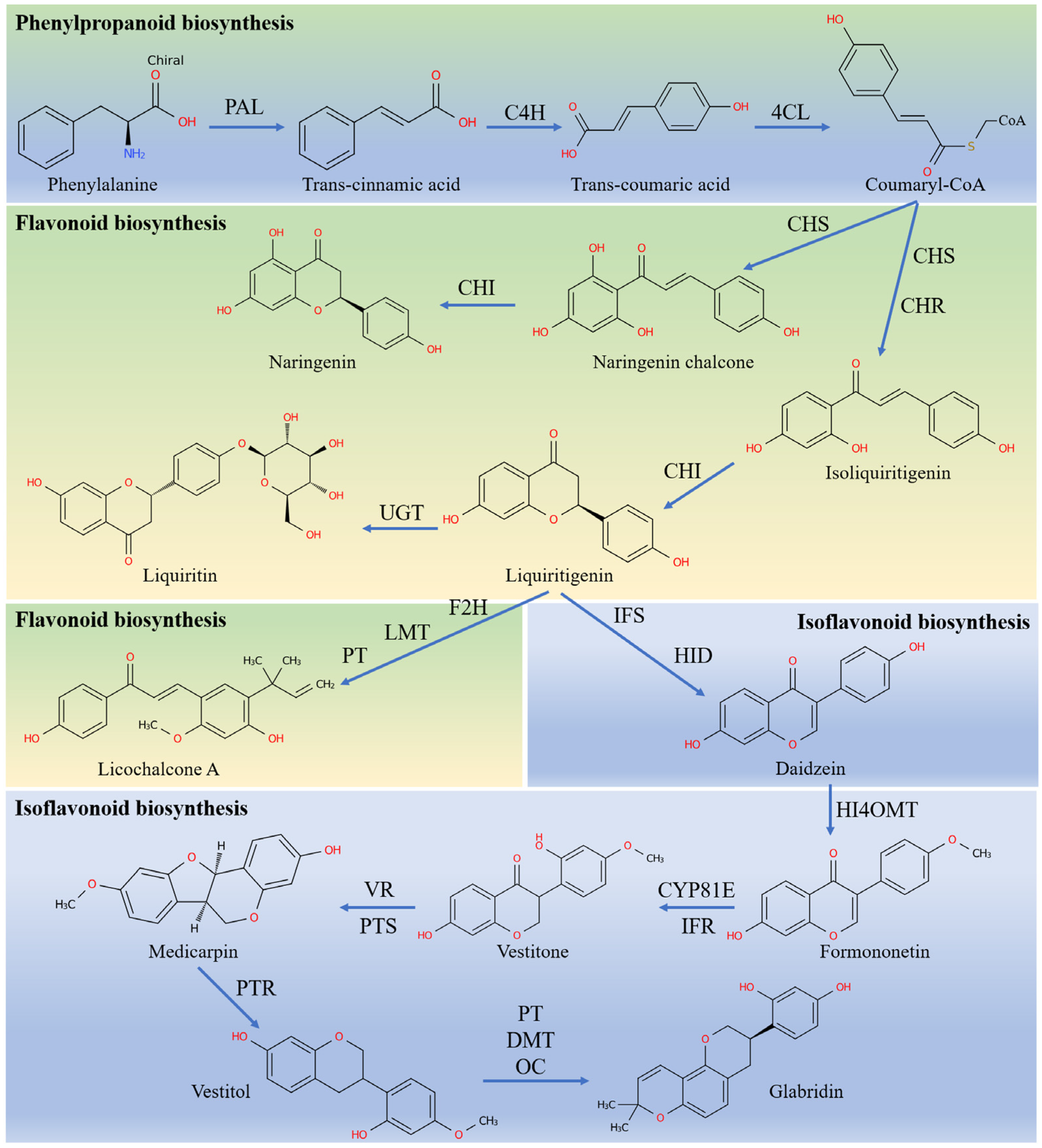
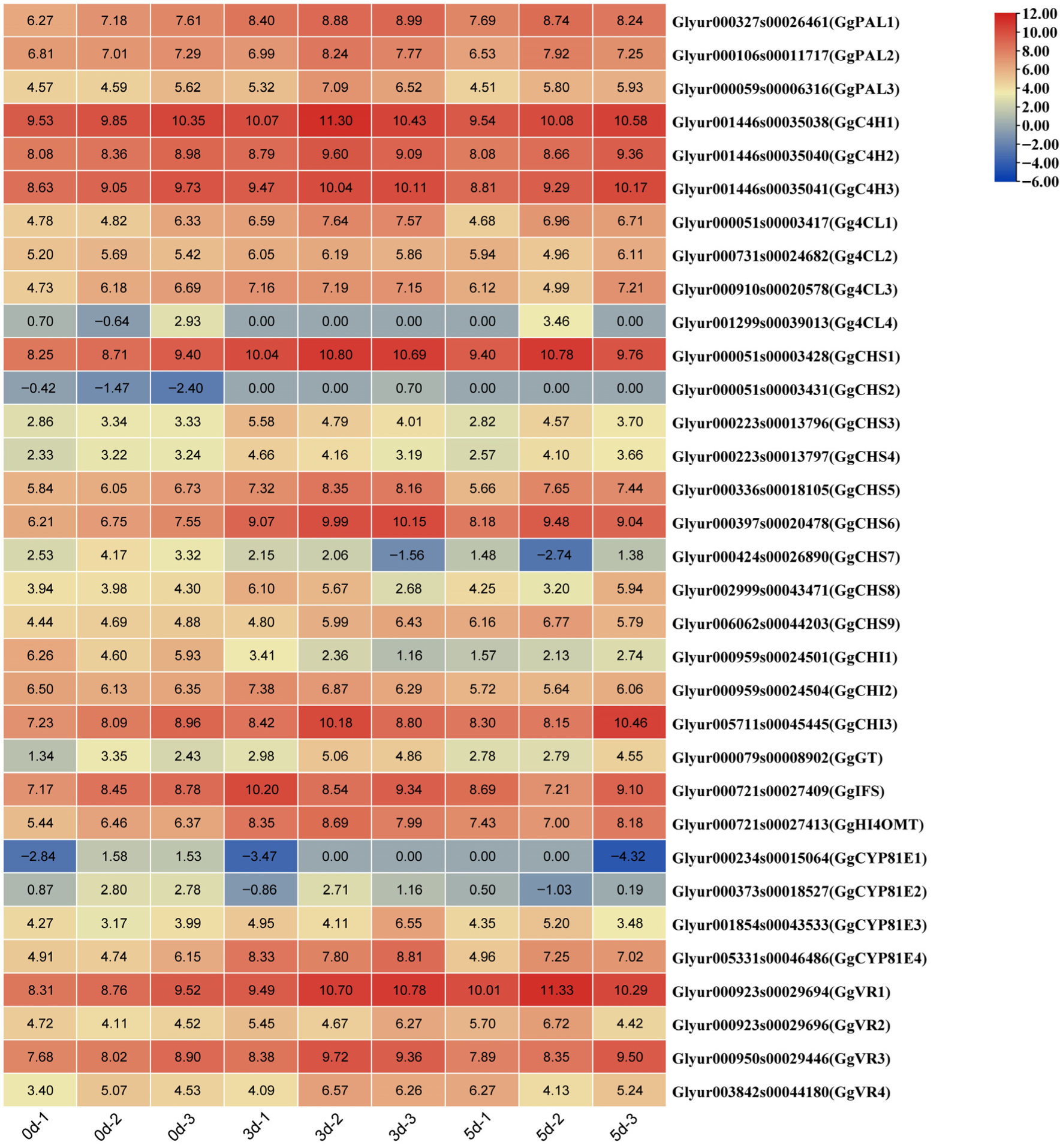
3.6. MeJA Effects the Levels of Transcription of the Genes Related to the Biosynthesis of Flavonoids in the G. glabra HRs
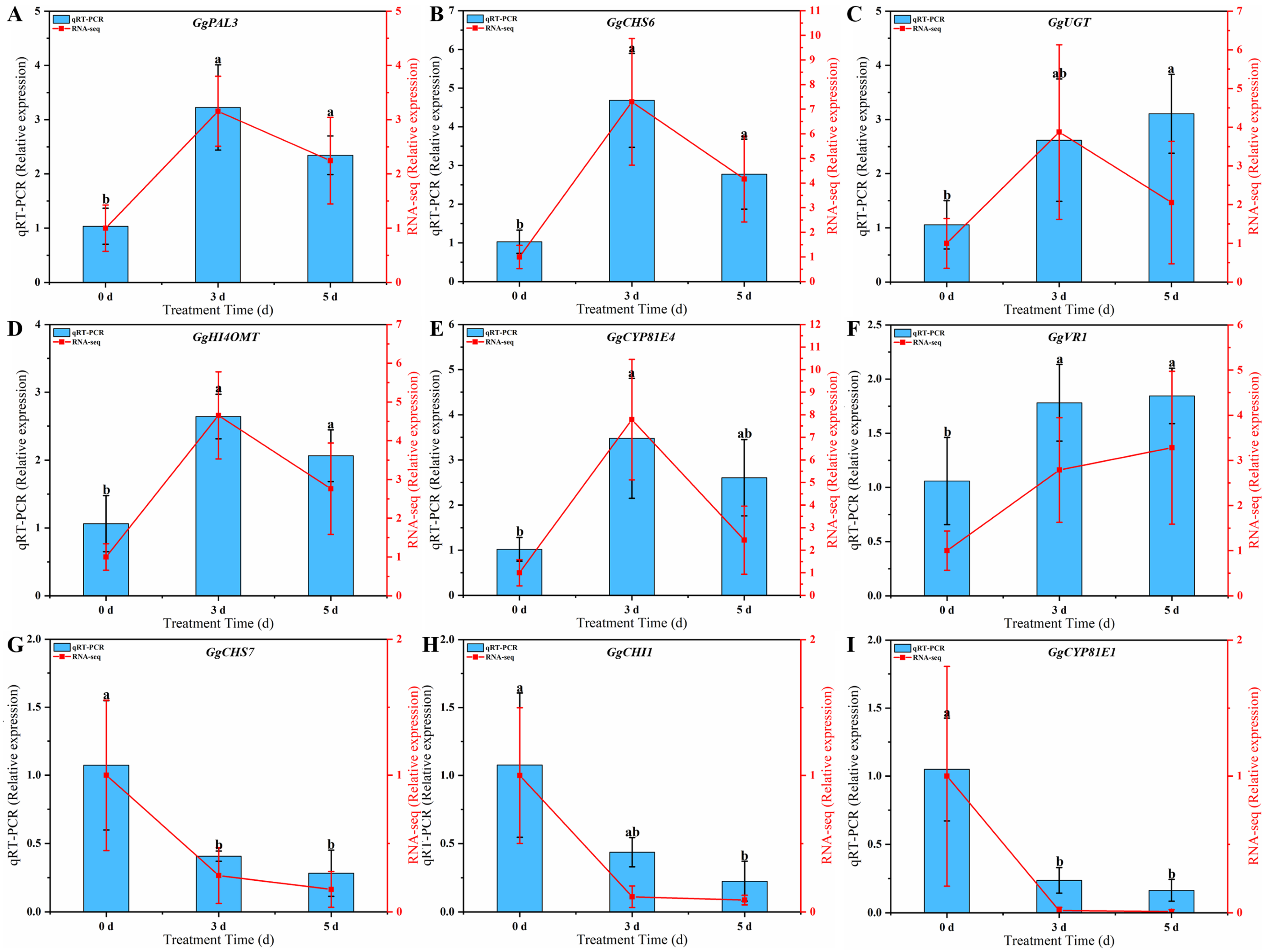
3.7. qRT-PCR Validation of Key Genes in the Biosynthesis of Flavonoids
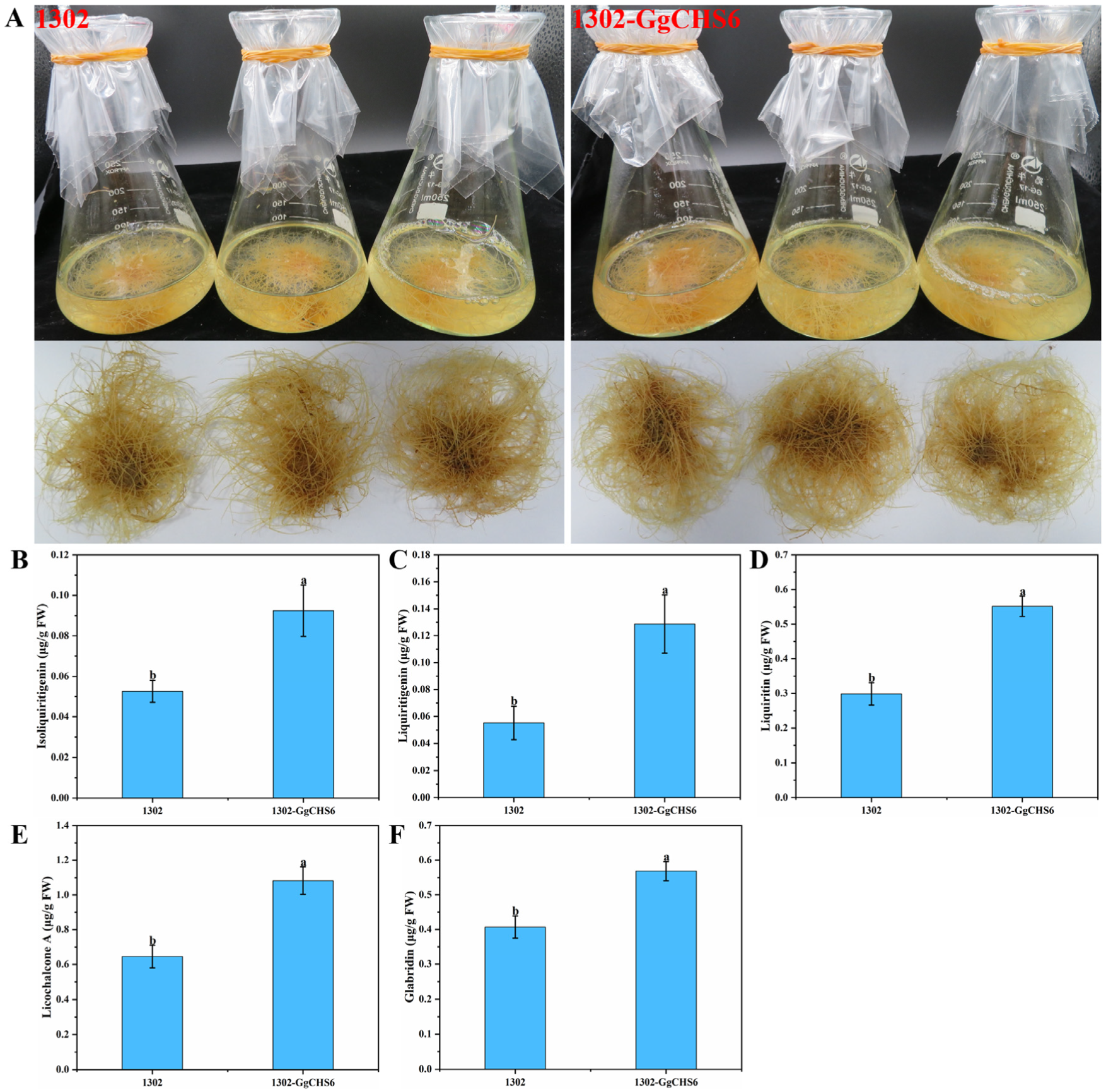
3.8. Determination of the Contents of Flavonoid in GgCHS6 That Overexpressed HRs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, C.; Chen, L.; Xu, C.; Shi, J.; Chen, S.; Tan, M.; Chen, J.; Zou, L.; Chen, C.; Liu, Z.; et al. A comprehensive review for phytochemical, pharmacological, and biosynthesis studies on Glycyrrhiza spp. Am. J. Chin. Med. 2020, 48, 17–45. [Google Scholar] [CrossRef]
- Wahab, S.; Annadurai, S.; Abullais, S.S.; Das, G.; Ahmad, W.; Ahmad, M.F.; Kandasamy, G.; Vasudevan, R.; Ali, M.S.; Amir, M. Glycyrrhiza glabra (Licorice): A comprehensive review on its phytochemistry, biological activities, clinical evidence and toxicology. Plants 2021, 10, 2751–2787. [Google Scholar] [CrossRef]
- Zheng, Y.P. Visual analysis of research progress and trend on hairy roots. Front. Plant Sci. 2025, 16, 1580007–1580023. [Google Scholar] [CrossRef]
- Gerszberg, A.; Wiktorek-Smagur, A. Hairy root cultures as a multitask platform for green biotechnology. Plant Cell Tissue Organ. Cult. 2022, 150, 493–509. [Google Scholar] [CrossRef]
- Gantait, S.; Mukherjee, E. Hairy root culture technology: Applications, constraints and prospect. Appl. Microbiol. Biotechnol. 2021, 105, 35–53. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.S.; Singh, P.; Kumari, R.; Kumar, S. Hairy root cultures for secondary metabolite production. In Genetic Manipulation of Secondary Metabolites in Medicinal Plant; Singh, R., Kumar, N., Eds.; Springer Nature Singapore: Singapore, 2023; pp. 205–223. [Google Scholar]
- Zhu, Y.; Zhu, X.; Wen, Y.; Wang, L.; Wang, Y.; Liao, C.; Zhao, M.; Li, T.; Liu, D.; Li, B.; et al. Plant hairy roots: Induction, applications, limitations and prospects. Ind. Crops Prod. 2024, 219, 119104–119123. [Google Scholar] [CrossRef]
- Afsharzadeh, N.; Paltram, R.; Jungwirth, A.; Tabrizi, L.; Nazeri, V.; Kalantari, H.; Halbwirth, H.; Samiei, L.; Sheehan, H.; Shokrpour, M. Advancing Glycyrrhiza glabra L. cultivation and hairy root transformation and elicitation for future metabolite overexpression. Horticulturae 2025, 11, 62–79. [Google Scholar] [CrossRef]
- Venkatasai, N.N.; Shetty, D.N.; Vinay, C.M.; Sekar, M.; Muthusamy, A.; Rai, P.S. A comprehensive review of factors affecting growth and secondary metabolites in hydroponically grown medicinal plants. Planta 2025, 261, 48–66. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, Q.; Kollie, L.; Dong, J.; Liang, Z. Molecular networks of secondary metabolism accumulation in plants: Current understanding and future challenges. Ind. Crops Prod. 2023, 201, 116901–116917. [Google Scholar] [CrossRef]
- Wongwicha, W.; Tanaka, H.; Shoyama, Y.; Putalun, W. Methyl jasmonate elicitation enhances glycyrrhizin production in Glycyrrhiza inflata hairy roots cultures. Z. Für Naturforschung C 2011, 66, 423–428. [Google Scholar] [CrossRef]
- Li, J.; Li, B.; Luo, L.; Cao, F.; Yang, B.; Gao, J.; Yan, Y.; Zhang, G.; Peng, L.; Hu, B. Increased phenolic acid and tanshinone production and transcriptional responses of biosynthetic genes in hairy root cultures of Salvia przewalskii Maxim. treated with methyl jasmonate and salicylic acid. Mol. Biol. Rep. 2020, 47, 8565–8578. [Google Scholar] [CrossRef]
- Amani, S.; Mohebodini, M.; Khademvatan, S.; Jafari, M.; Kumar, V. Modifications in gene expression and phenolic compounds content by methyl jasmonate and fungal elicitors in Ficus carica. Cv. Siah hairy root cultures. BMC Plant Biol. 2024, 24, 520–534. [Google Scholar] [CrossRef]
- Bao, J.; Lu, X.; Ma, L.; Zhang, X.; Tian, P.; Zhang, X.; Li, S.; Ma, S.; Yang, J.; Lu, Y.; et al. Transcriptome analysis of genes related to glucoraphanin and sulforaphane synthesis in methyl jasmonate treated broccoli (Brassica oleracea var. italica) hairy roots. J. Plant Res. 2022, 135, 757–770. [Google Scholar] [CrossRef] [PubMed]
- Veremeichik, G.N.; Bulgakov, D.V.; Solomatina, T.O.; Makhazen, D.S. In the interkingdom horizontal gene transfer, the small rolA gene is a big mystery. Appl. Microbiol. Biotechnol. 2023, 107, 2097–2109. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, Y.; Sun, J.; Wang, H. Induction and cultivation of hairy roots of Zanthoxylum dissitum. Plant Physiol. J. 2023, 59, 2117–2125. [Google Scholar] [CrossRef]
- Foti, C.; Pavli, O.I. High-efficiency Agrobacterium rhizogenes-mediated transgenic hairy root induction of Lens culinaris. Agronomy 2020, 10, 1170–1181. [Google Scholar] [CrossRef]
- Medina, F.; Condori, J.; Rimando, A.M.; Hubstenberger, J.; Shelton, K.; O’Keefe, S.F.; Bennett, S.; Dolan, M.C. Production and secretion of resveratrol in hairy root cultures of peanut. Phytochemistry 2007, 68, 1992–2003. [Google Scholar] [CrossRef]
- Serino, G.; Clerot, D.; Brevet, J.; Costantino, P.; Cardarelli, M. rol genes of Agrobacterium rhizogenes cucumopine strain: Sequence, effects and pattern of expression. Plant Mol. Biol. 1994, 26, 415–422. [Google Scholar] [CrossRef]
- Grąbkowska, R.; Krzemińska, M.; Gaweda-Walerych, K.; Kiss, A.K.; Pluta, K.; Grzegorczyk-Karolak, I. Enhancement of rosmarinic acid production in hairy root cultures of Perovskia atriplicifolia Benth. Int. J. Mol. Sci. 2025, 26, 3187–3208. [Google Scholar] [CrossRef]
- Su, L.; Li, S.; Qiu, H.; Wang, H.; Wang, C.; He, C.; Xu, M.; Zhang, Z. Full-length transcriptome analyses of genes involved in triterpenoid saponin biosynthesis of Psammosilene tunicoides hairy root cultures with exogenous salicylic acid. Front. Genet. 2021, 12, 657060–657076. [Google Scholar] [CrossRef]
- Shang, Z.; Tian, Y.; Yi, Y.; Li, K.; Qiao, X.; Ye, M. Comparative bioactivity evaluation and chemical profiling of different parts of the medicinal plant Glycyrrhiza uralensis. J. Pharm. Biomed. Anal. 2022, 215, 114793–114803. [Google Scholar] [CrossRef]
- Zhu, L.; Li, M.; Yang, W.; Zhang, J.; Yang, X.; Zhang, Q.; Wang, H. Effects of different drying methods on drying characteristics and quality of Glycyrrhiza uralensis (Licorice). Foods 2023, 12, 1652–1665. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. CUTADAPT removes adapter sequences from high-throughput sequencing reads. EMBnet.J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Mochida, K.; Sakurai, T.; Seki, H.; Yoshida, T.; Takahagi, K.; Sawai, S.; Uchiyama, H.; Muranaka, T.; Saito, K. Draft genome assembly and annotation of Glycyrrhiza uralensis, a medicinal legume. Plant J. 2017, 89, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Love, M.I.; Anders, S.; Huber, W. Differential analysis of count data-the DESeq2 package. Genome Biol. 2015, 15, 10-1186. [Google Scholar]
- Harris, M.A.; Clark, J.; Ireland, A.; Lomax, J.; Ashburner, M.; Foulger, R.; Eilbeck, K.; Lewis, S.; Marshall, B.; Mungall, C.; et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004, 32, D258–D261. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kumar, L.; Futschik, M.E. Mfuzz: A software package for soft clustering of microarray data. Bioinformation 2007, 2, 5–7. [Google Scholar] [CrossRef]
- Maroufi, A. Selection of reference genes for real-time quantitative PCR analysis of gene expression in Glycyrrhiza glabra under drought stress. Biol. Plant. 2016, 60, 645–654. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Jyothishwaran, G.; Kotresha, D.; Selvaraj, T.; Srideshikan, S.; Rajvanshi, M. A modified freeze-thaw method for efficient transformation of Agrobacterium tumefaciens. Curr. Sci. 2007, 93, 770–772. [Google Scholar]
- Kaushik, S.; Ranjan, A.; Singh, A.K.; Sirhindi, G. Methyl jasmonate reduces cadmium toxicity by enhancing phenol and flavonoid metabolism and activating the antioxidant defense system in pigeon pea (Cajanus cajan). Chemosphere 2024, 346, 140681–140694. [Google Scholar] [CrossRef]
- Ninkuu, V.; Aluko, O.O.; Yan, J.; Zeng, H.; Liu, G.; Zhao, J.; Li, H.; Chen, S.; Dakora, F.D. Phenylpropanoids metabolism: Recent insight into stress tolerance and plant development cues. Front. Plant Sci. 2025, 16, 1571825–1571846. [Google Scholar] [CrossRef]
- Li, X.; Xu, Y.; Zhang, J.; Xu, K.; Zheng, X.; Luo, J.; Lu, J. Integrative physiology and transcriptome reveal salt-tolerance differences between two licorice species: Ion transport, Casparian strip formation and flavonoids biosynthesis. BMC Plant Biol. 2024, 24, 272–298. [Google Scholar] [CrossRef]
- Yao, H.; Wang, F.; Bi, Q.; Liu, H.; Liu, L.; Xiao, G.; Zhu, J.; Shen, H.; Li, H. Combined analysis of pharmaceutical active ingredients and transcriptomes of Glycyrrhiza uralensis under PEG6000-induced drought stress revealed glycyrrhizic acid and flavonoids accumulation via JA-mediated signaling. Front. Plant Sci. 2022, 13, 920172–920186. [Google Scholar] [CrossRef]
- Zhong, C.; Chen, C.; Gao, X.; Tan, C.; Bai, H.; Ning, K. Multi-omics profiling reveals comprehensive microbe-plant-metabolite regulation patterns for medicinal plant Glycyrrhiza uralensis Fisch. Plant Biotechnol. J. 2022, 20, 1874–1887. [Google Scholar] [CrossRef]
- Ozyigit, I.I.; Dogan, I.; Hocaoglu-Ozyigit, A.; Yalcin, B.; Erdogan, A.; Yalcin, I.E.; Cabi, E.; Kaya, Y. Production of secondary metabolites using tissue culture-based biotechnological applications. Front. Plant Sci. 2023, 14, 1132555–1132583. [Google Scholar] [CrossRef]
- Mipeshwaree Devi, A.; Khedashwori Devi, K.; Premi Devi, P.; Lakshmipriyari Devi, M.; Das, S. Metabolic engineering of plant secondary metabolites: Prospects and its technological challenges. Front. Plant Sci. 2023, 14, 1171154–1171170. [Google Scholar] [CrossRef]
- Mirmazloum, I.; Slavov, A.K.; Marchev, A.S. The untapped potential of hairy root cultures and their multiple applications. Int. J. Mol. Sci. 2024, 25, 12682–12718. [Google Scholar] [CrossRef]
- Ebrahimi, T.; Piri, K.; Abdoli, A.; Tohidfar, M. Developing an efficient protocol for hairy root induction in Lythrum salicaria L. Biologia 2023, 78, 2667–2677. Biologia 2023, 78, 2667–2677. [Google Scholar] [CrossRef]
- Zhao, S.; Zhao, K.; Yang, J.; Huasong, W. Research and application of synthesis of secondary metabolites in hairy roots induced by Agrobacterium rhizogenes. Mol. Plant Breed. 2023, 21, 7554–7565. [Google Scholar] [CrossRef]
- Hwang, C.; Yan, S.; Choe, Y.; Yun, C.; Xu, S.; Im, M.; Xue, Z. Efficient hairy root induction system of Astragalus membranaceus and significant enhancement of astragalosides via overexpressing AmUGT15. Plant Cell Rep. 2024, 43, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Gai, Q.Y.; Wang, X.; Liu, J.; Lu, Y.; Wang, Z.Y.; Xu, X.J.; Fu, Y.J. Effective production of phenolic compounds with health benefits in pigeon pea [Cajanus cajan (L.) Millsp.] hairy root cultures. J. Agric. Food Chem. 2020, 68, 8350–8361. [Google Scholar] [CrossRef]
- Hussain, M.J.; Abbas, Y.; Nazli, N.; Fatima, S.; Drouet, S.; Hano, C.; Abbasi, B.H. Root cultures, a boon for the production of valuable compounds: A comparative review. Plants 2022, 11, 439–477. [Google Scholar] [CrossRef]
- Miao, Y.; Hu, Y.; Yi, S.; Zhang, X.; Tan, N. Establishment of hairy root culture of Rubia yunnanensis Diels: Production of rubiaceae-type cyclopeptides and quinones. J. Biotechnol. 2021, 341, 21–29. [Google Scholar] [CrossRef]
- Mahendran, G.; Vimolmangkang, S. Effect of carbon source and elicitors on biomass production and accumulation of friedelin and epifriedelanol in hairy roots of hemp (Cannabis sativa L.). Plant Cell Tissue Organ Cult. (PCTOC) 2024, 156, 61–77. [Google Scholar] [CrossRef]
- Ji, H.; Yang, B.; Jing, Y.; Luo, Y.; Li, B.; Yan, Y.; Zhang, G.; Zhao, F.; Wang, B.; Peng, L.; et al. Trehalose and brassinolide enhance the signature ingredient accumulation and anti-oxidant activity in the hairy root cultures of Polygala tenuifolia Willd. Ind. Crops Prod. 2023, 196, 116521–116536. [Google Scholar] [CrossRef]
- Sharma, A.R.; Gajurel, G.; Abdel-Karim, S.; Alam, M.A.; Shields, R.C.; Medina-Bolivar, F. Production of malheuran A, a geranylated flavonoid with antimicrobial and anti-inflammatory activities, in hairy root cultures of Dalea purpurea. Plants 2025, 14, 259–281. [Google Scholar] [CrossRef] [PubMed]
- Verma, P.; Khan, S.A.; Alhandhali, A.J.A.; Parasharami, V.A. Bioreactor upscaling of different tissue of medicinal herbs for extraction of active phytomolecules: A step towards industrialization and enhanced production of phytochemicals. In Plant Growth Regulators: Signalling under Stress Conditions; Aftab, T., Hakeem, K.R., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 455–481. [Google Scholar]
- Liu, C.; Ahmad, N.; Tao, Y.; Hussain, H.; Chang, Y.; Umar, A.W.; Liu, X. Reprogramming hairy root cultures: A synthetic biology framework for precision metabolite biosynthesis. Plants 2025, 14, 1928–1945. [Google Scholar] [CrossRef] [PubMed]
- Allahdou, M.; Mehravaran, L.; Khajeh, H. Improving the production of glycyrrhizin and glycyrrhetinic acid by eliciting hairy root of Glycyrrhiza glabra. Russ. J. Plant Physiol. 2025, 72, 3. [Google Scholar] [CrossRef]
- Behdad, A.; Ganjeali, A. Phytohormones and microbial elicitation on glycyrrhizin production and gene expression in the hairy root of Glycyrrhiza glabra L. Plant Cell Tissue Organ Cult. (PCTOC) 2024, 160, 4–22. [Google Scholar] [CrossRef]
- Yang, B.; Pan, F.; Yasmeen, F.; Shan, L.; Pan, J.; Zhang, M.; Weng, X.; Wang, M.; Li, M.; Wang, Q.; et al. Integrated multi-omic analysis reveals the cytokinin and sucrose metabolism-mediated regulation of flavone glycoside biosynthesis by MeJA exposure in Ficus pandurata Hance. Food Res. Int. 2023, 174, 113680–113695. [Google Scholar] [CrossRef]
- Zeng, J.; Ma, X.; Li, Y.; Zhou, L.; Fu, J.; Wang, H.; Liu, Y.; Yuan, L.; Wang, Y.; Li, Y. Histone deacetylases repress the accumulation of licochalcone A by inhibiting the expression of flavonoid biosynthetic pathway-related genes in licorice (Glycyrrhiza inflata). Mol. Hortic. 2025, 5, 32–49. [Google Scholar] [CrossRef]
- Yu, S.; Zhang, J.; Cao, Y.; Xie, J.; Zhong, C. Combined transcriptome and metabolome analysis reveals that DcWRKY11 promotes methyl jasmonate-induced proanthocyanidin biosynthesis in Dioscorea composita. Plant Cell Environ. 2025, 48, 6782–6795. [Google Scholar] [CrossRef]
- Zheng, F.; Xu, Z.; Deng, X.; Wang, X.; Sun, Y.; Shen, X.; Yu, Z. Genome-wide transcriptome analysis reveals GRF transcription factors involved in methyl jasmonate-induced flavonoid biosynthesis in Hedera helix. Plants 2025, 14, 2094–2103. [Google Scholar] [CrossRef]
- Zhao, H.-H.; Du, R.; Han, Y.-L.; Yang, Z.-H.; Qiu, X.; Li, Y.-Q.; Zhang, J.-G.; Cheng, Z.-W. Effects of methyl jasmonate and salicylhydroxamic acid on the biosynthesis of flavonoids in Glycyrrhiza glabra L. hairy roots. Plant Stress 2025, 15, 100774–100786. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Higashi, Y.; Nakabayashi, R. The origin and evolution of plant flavonoid metabolism. Front. Plant Sci. 2019, 10, 943–959. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Wang, B.; Xue, B.; Wang, R.; Tang, G.; Zhu, T.; Zhao, M.; Li, T.; Liao, C.; Zhang, H.; et al. MeJA Elicitation on Flavonoid Biosynthesis and Gene Expression in the Hairy Roots of Glycyrrhiza glabra L. Genes 2025, 16, 1387. https://doi.org/10.3390/genes16111387
Zhu Y, Wang B, Xue B, Wang R, Tang G, Zhu T, Zhao M, Li T, Liao C, Zhang H, et al. MeJA Elicitation on Flavonoid Biosynthesis and Gene Expression in the Hairy Roots of Glycyrrhiza glabra L. Genes. 2025; 16(11):1387. https://doi.org/10.3390/genes16111387
Chicago/Turabian StyleZhu, Yutao, Bohan Wang, Bingyi Xue, Runqian Wang, Ganlin Tang, Tao Zhu, Mei Zhao, Taotao Li, Chunli Liao, Huamin Zhang, and et al. 2025. "MeJA Elicitation on Flavonoid Biosynthesis and Gene Expression in the Hairy Roots of Glycyrrhiza glabra L." Genes 16, no. 11: 1387. https://doi.org/10.3390/genes16111387
APA StyleZhu, Y., Wang, B., Xue, B., Wang, R., Tang, G., Zhu, T., Zhao, M., Li, T., Liao, C., Zhang, H., Liu, D., Chen, J., & Wang, L. (2025). MeJA Elicitation on Flavonoid Biosynthesis and Gene Expression in the Hairy Roots of Glycyrrhiza glabra L. Genes, 16(11), 1387. https://doi.org/10.3390/genes16111387





