1. Introduction
The genus
Coelogyne is one of the most diverse in the Orchidaceae family, consisting of approximately 200 species found in various regions of Asia and the Pacific Islands [
1]. Orchids of the genus
Coelogyne show considerable morphological diversity and adaptation to different environments, from lowland areas to humid high-altitude forests. Most species are epiphytes that grow on trees, although some are lithophytes growing on rocks or in soil, using their aerial roots to capture moisture [
2,
3,
4]. A characteristic feature of these orchids is pseudobulbs, which store water and vary in shape and size depending on the species [
2,
5]. The flowers are often intensely fragrant, attracting pollinators such as bees and wasps, and their colors range from white to greenish, yellow, or brown. Sexual reproduction occurs via seeds, which require symbiosis with mycorrhizal fungi to germinate, making the process relatively inefficient. Vegetative reproduction by dividing pseudobulbs is common in cultivation [
2,
3].
Orchids of the genus
Coelogyne are valued not only for their beautiful flowers but also for their medicinal properties. They contain phytochemicals such as alkaloids, flavonoids, and terpenoids, which are used in traditional medicine to treat various ailments, including headaches, fever, stomachaches, and burns [
4,
6]. Some species also exhibit anticancer and anti-inflammatory properties [
7,
8,
9].
Despite their ecological and medical importance, accurate identification of species within the genus
Coelogyne remains a challenge due to overlapping morphological characteristics, especially in the absence of reproductive structures, leading to frequent misidentifications [
10,
11]. Difficulties in reliable identification are of significant importance for the conservation of these plants, especially since many
Coelogyne species are seriously threatened by illegal trade driven by their ornamental and medicinal appeal. This exploitation leads to a significant decline in wild populations, hence their inclusion in Appendix II of the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) [
4,
12,
13]. To effectively enforce regulations and curb illegal trade in orchids, customs officers and trade inspectors need fast, reliable, and easy-to-use tools for accurate species identification.
DNA barcoding is an effective solution to these identification problems. It involves sequencing short, standardized regions of DNA that are characteristic of specific species, allowing for quick and accurate identification, especially when traditional methods fail [
11,
14]. This technique is particularly useful when plant material is damaged, incomplete, or difficult to identify based on morphological characteristics. DNA barcoding also plays a key role in the conservation of medicinal plant genetic resources and in the detection of counterfeit plant products on the market [
15]. Research on orchids highlights the importance of this method in the conservation of endangered species [
16].
The history and development of DNA barcoding techniques have been crucial for the identification of both animal and plant species. Hebert and colleagues (2003) have proposed the mitochondrial
CO1 gene as a standard marker for animal identification, which has been successfully used in various animal groups [
17]. In the case of plants, the process was more complex due to lower variability of mitochondrial sequences, which led researchers to focus on plastid and nuclear markers such as
rbcL and
matK [
14]. Research on chloroplast genomes has shown that these markers are effective in identifying plant species, while other regions, such as ITS2, have proven valuable in studies of medicinal plants [
15]. In the case of orchids, markers such as
ycf1b have been particularly useful in species identification [
18].
The selection of molecular markers is crucial for DNA barcoding. Various plastid and nuclear markers, such as ITS,
matK,
rbcL,
trnH-psbA, and
atpF-atpH, are used in plant research. ITS is widely employed to distinguish closely related species, while
matK and
rbcL are commonly used in a two-locus approach to plant barcoding [
14]. The
rbcL gene is conservative and universal but has limited resolution at the species level [
19], while
matK offers greater variability and better species discrimination [
10].
trnH-psbA and
atpF-atpH show interspecific variability, making them useful for identifying plants, including orchids [
19].
Although DNA barcoding has been previously applied to the genus
Coelogyne, existing studies have been limited in scope, resolution, or practical applicability. Ramudu and Khasim (2018) conducted one of the earliest experimental investigations using the
rbcL marker to barcode Indian
Coelogyne species, achieving modest species resolution (36.36% via distance method, 72.72% via phylogenetic tree), but their study focused solely on a single locus and a narrow geographic range (9 Coelogyne species) [
20]. More recently, Pratiwi et al. (2023) employed an in silico approach to compare four loci (
matK,
rbcL,
rpoC1, and nrDNA) across Asian
Coelogyne species (19 species), concluding that nrDNA offered the highest species discrimination [
21]. However, their study lacked experimental validation and did not address practical field applications. To date, no study has experimentally evaluated five barcode regions—
rbcL,
matK,
trnH-psbA,
atpF-atpH, and ITS2—within a single framework, nor proposed multi-locus combinations tailored for enforcement contexts such as CITES. This study fills that gap by integrating laboratory-based amplification, sequencing, and BLAST-based identification across 19
Coelogyne species.
The main objective of this work is to identify effective DNA markers for barcoding orchids of the genus
Coelogyne, with a view to developing a quick and simple diagnostic test for use in the field by customs officers and trade inspectors. Such a tool will significantly improve CITES enforcement by enabling the rapid detection of
Coelogyne species in trade, directly contributing to their conservation. The key results of this work include the identification of suitable DNA barcoding regions with high resolution and reliability, which will form the basis for the subsequent development of practical identification kits. These achievements have great potential to reduce illegal trade in orchids and contribute to biodiversity conservation in a broader sense [
13].
3. Results
Agarose gel electrophoresis was used to evaluate the effectiveness of DNA amplification using suitable primers for loci rbcL, matK, trnH-psbA, and atpF-atpH for each of the 19 Coelogyne orchid DNA samples. According to the electrophoresis results, amplification was successful in 74% of rbcL samples, 74% of matK samples, 84% of trnH-psbA samples, 89% of atpF-atpH samples, and 95% of ITS2 samples.
It is worth noting that reference sequences were not available in the NCBI Gene and NCBI Nucleotide databases for several of the analyzed species. No entries were found for
C. parishii,
C. salmonicolor,
C. pulchella,
C. Lyme Bay, and
C. triplicatula. Only a few sequences were available for
C. celebensis (1),
C. intermedia (2), and
C. assamica (4). Slightly more data were found for
C. barbata (12),
C. rochussenii (23),
C. tomentosa (24),
C. trinervis (28),
C. cumingii (33),
C. asperata (36),
C. pandurata (44), and
C. flaccida (50). The most extensive sequence data were available for
C. fimbriata (753),
C. ovalis (91), and
C. cristata (75), suggesting that they have more genetic documentation than the other species. To fill this gap, we created a database for 19
Coelogyne species (see
Supplementary Table S1).
The lengths of correctly amplified sequences were analyzed as the study’s next step. Following editing and successful amplification, the length of the edited sequences was measured; the results are displayed in
Table 1.
MatK and
trnH-psbA were found to have the longest sequences. The
rbcL sequence had an average length of 543 base pairs (bp), the ITS2 sequence was 505 bp long, and the
atpF-atpH sequence was slightly over 300 bp in length. Significantly,
atpF-atpH sequences shorter than 100 bp were not included in additional analysis, because the identification success rate for these shorter fragments was less than 94%, indicating their ineffectiveness in species identification.
Sequencing analysis results for previously identified species of
Coelogyne are presented in
Supplementary Table S2. Each barcode was analyzed using the BLAST tool available on the NCBI platform, which allowed comparison with the sequence database and assignment of the appropriate results for each locus (
rbcL,
matK,
trnH-psbA,
atpF-atpH, and ITS2).
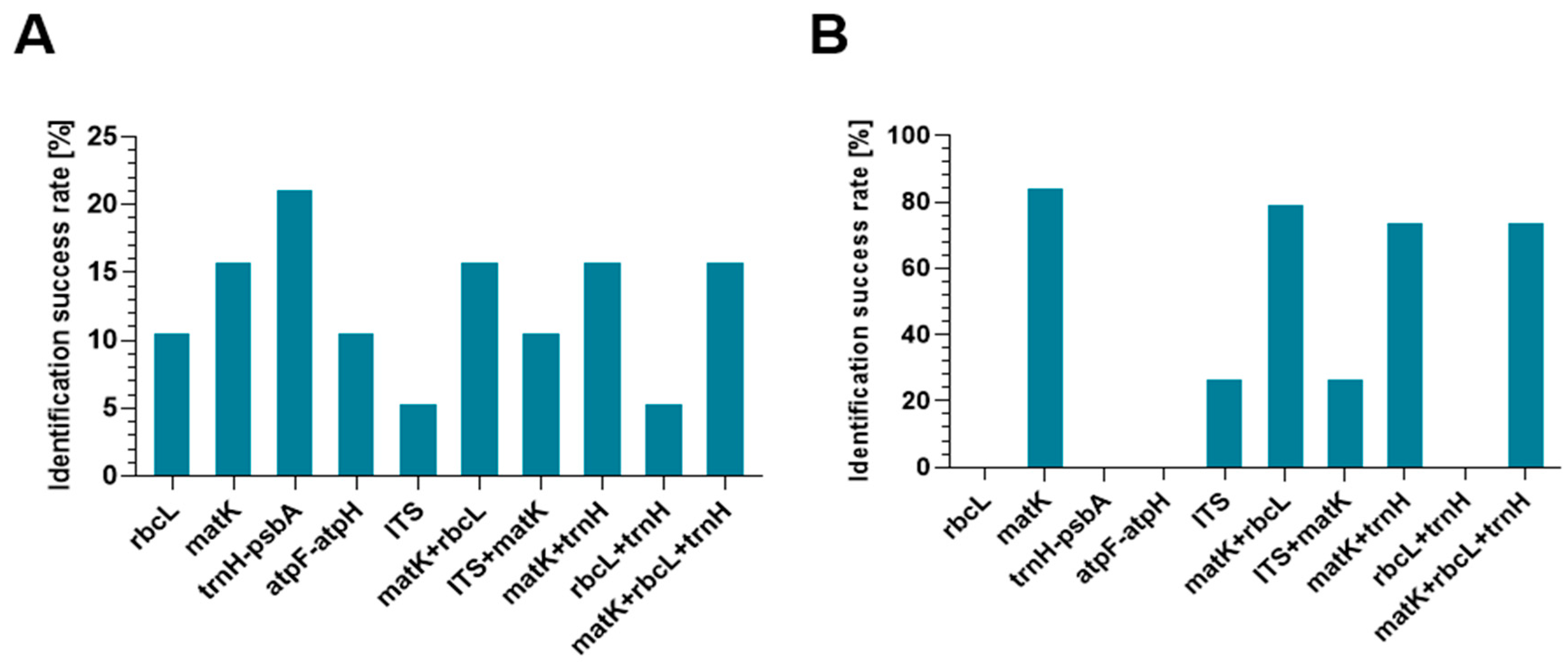
The four species—C. fimbriata, C. cristata, C. rochussenii, and C. assamica—were the most easily identified using the trnH-psbA barcode, according to an analysis of the sequencing results. C. flaccida, C. cristata, and C. rochussenii were the three species that could be unmistakably identified thanks to the matK region. In terms of rbcL, the two species that were correctly identified were C. fimbriata and C. rochussenii. Meanwhile, C. cristata and C. rochussenii were correctly identified using atpF-atpH. Only C. cristata was correctly identified at the species level in the ITS2 region, despite high amplification efficiency.
The results indicate that among the analyzed single barcodes,
trnH-psbA is characterized by the highest efficiency of unambiguous identification of
Coelogyne species (
Figure 2). The other markers, especially ITS2 and
atpF-atpH, showed lower utility, mainly due to insufficient variability for unambiguous identification. The
matK region, although requiring longer sequences, showed good taxonomic resolution and high identification efficiency.
Additional multi-marker analysis demonstrates that combining regions—particularly matK with trnH-psbA—increases the number of correctly identified species. However, identification at the species level was not always achievable, despite the use of multiple markers, which would suggest that barcoding techniques for this diverse orchid genus still require improvement.
In keeping with the examination of the sequencing data, it is important to note that, both with single- and multi-marker methods, the assignment of samples to the genus Coelogyne proved to be far more successful than species-level identification.
The most dependable region for genus-level identification in the case of single markers was matK; all samples for which sequences were obtained were unambiguously assigned to Coelogyne, even in cases where species identification was unclear or impossible (for example, for C. celebensis, C. cumingii, or C. asperata). Despite its decreased efficacy at the species level, ITS2 also made it possible to reliably assign all correctly sequenced samples to the genus.
The atpF-atpH, trnH-psbA, and rbcL markers all failed to provide unambiguous identification at the genus level: while almost all assignments were within Coelogyne, they were ambiguous, indicating low resolution of both markers.
A combined barcode analysis was also performed for
Coelogyne species to determine whether using more than one locus could improve identification efficiency. The results of this analysis are shown in
Figure 2 and
Supplementary Table S3.
Analysis of the results presented in
Supplementary Table S3 indicates that the use of a multi-barcode approach enables more effective identification of species of the genus
Coelogyne than single markers.
Three out of the fifteen species examined—C. flaccida, C. cristata, and C. rochussenii—were correctly identified in the matK + rbcL combination. The results from the remaining samples were unclear or unidentified.
C. cristata and C. parishii were the only two species that could be unambiguously identified using the combination of ITS and matK; the other three species—C. pulchella, C. triplicatula, and C. intermedia—were left unidentified at the species level.
Three species (C. flaccida, C. cristata, and C. rochussenii) were successfully identified through the use of matK + trnH-psbA markers. The results for C. fimbriata and C. assamica were not conclusive.
The best results were obtained for the combination of matK + rbcL + trnH-psbA, where three species were correctly identified: C. flaccida, C. cristata, and C. rochussenii. However, here, too, ambiguous results were obtained for some species, including C. assamica and C. fimbriata.
The combination of rbcL + trnH-psbA was the least effective—only C. rochussenii was correctly identified, while the remaining samples continued to be unidentified or ambiguous.
Assigning samples to their genus was successful when multi-marker strategies were used. In 79% of cases, the analyzed samples could be categorized under the genus Coelogyne using the matK + rbcL combination, and in 74% of cases, the matK + trnH-psbA combination. Even in situations where species identification was still unclear, such as C. assamica, C. fimbriata, or C. ovalis, 74% of the examined samples were assigned to the correct genus thanks to the equally effective combination of matK + rbcL + trnH-psbA.
These findings emphasize the value of employing a multi-marker approach, particularly when it comes to law enforcement and nature conservation (e.g., CITES regulations), where accurate plant material assignment to at least the genus level is crucial. This suggests that the matK barcode, whether alone or in combination with other markers such as trnH-psbA or rbcL, may be useful as a diagnostic tool for studying biodiversity and controlling plant material that could be used illegally.
To further explore the genetic variation underlying the observed differences in discriminatory power, sequence characteristics of the
matK and
rbcL regions were examined in more detail. The frequency of base substitutions in both loci was analyzed, as shown in
Table 2. In both regions, transversions occurred more frequently than transitions. Among all observed substitutions, guanine-to-adenine changes were the most prevalent.
Percentage identity (PID) is a quantitative measure of sequence similarity. Closely related species are generally expected to exhibit higher PID values compared to more distantly related taxa, making this parameter a useful indicator of genetic relatedness. Among the fifteen analyzed
Coelogyne species, the similarity of the
rbcL sequences ranged from 93.5% to 99.8%, with an average of 97.8%. For the
matK sequences, PID ranged from 94.6% to 99.6%, with an average of 97.9%. The highest
rbcL sequence similarity (99.8%) was observed between
C. celebensis and
C. assamica. In the
matK region, the greatest similarity (99.6%) occurred between
C. ovalis and
C. triplicatula, as well as between
C. pulchella and
C. triplicatula. A high similarity was also found between
C. celebensis and
C. Lyme Bay. The results are presented in
Table 3.
4. Discussion
DNA barcoding is a technique that enables species identification based on a short DNA fragment that is common to all organisms but exhibits sufficient variability for individual species to be distinguished. The chosen DNA fragment needs to fulfill two essential requirements in order for this technique to work. To design universal primers for its amplification, it must first have conserved regions. Second, the fragment must be sufficiently variable to allow unambiguous identification of individual species [
28]. In many plant families, including the Orchidaceae, DNA barcoding is widely used to identify species that are hard to differentiate from one another based solely on morphological traits [
29].
The study included 19 species of orchids from the genus Coelogyne with the aim of validating previous morphological analysis-based identifications. An attempt was made to find one or more loci that would be helpful for barcoding this specific plant group and to create a database for the 19 analyzed Coelogyne species. Several chloroplast DNA fragments, such as the rbcL, matK, trnH-psbA, atpF-atpH, ITS2 sequences, and various combinations of these markers, were analyzed to assess their ability to distinguish species within the genus Coelogyne.
One of the key stages of the study was the amplification of isolated DNA using PCR. The results showed that the amplification efficiency varied depending on the loci used. For the
rbcL and
matK sequences, the efficiency was 74%; for
trnH-psbA, 84%; for
atpF-atpH, 89%; and for ITS2, 95%. In comparison, in a study of terrestrial plants conducted by Kress and Erickson (2007), only two loci,
trnH-psbA and
rbcL, showed high amplification efficiency, achieving 95.8% and 92.7%, respectively [
30]. Since amplification of the
matK locus was successful in fewer than 40% of the plant species examined, it was much less effective in this investigation. The low amplification efficiency of
matK was attributed to the high variability of the
matK sequence at the primer binding sites and the size of the PCR product, which averaged 778 base pairs [
14].
In studies conducted by the Consortium for the Barcode of Life (CBOL), the
trnH-psbA locus showed good amplification across the entire group of land plants, achieving 93% efficiency in angiosperms using a single pair of universal primers. For the tested samples, the amplification efficiency with a single pair of primers was 90%, which is regarded as a noteworthy accomplishment [
10]. Li et al. (2016) [
31] studied orchids in the genus
Oberonia and examined a number of loci, including
rbcL,
matK,
trnH-psbA, ITS, and ITS2. The findings demonstrated that
rbcL,
matK, and ITS produced the best PCR amplification and sequencing outcomes, with all samples exhibiting 100% success rates in both procedures. Furthermore, the
trnH-psbA barcode had a high sequencing success rate of 95.12%.
Problems associated with amplification failures when using
matK primers are widely discussed in the literature. Research shows that by creating new, more efficient primers that match the variable sequences of
matK more closely, the low amplification efficiency of this region can be raised [
32]. Therefore, the creation of more effective
matK primers is essential to the potential use of this region in a universal plant barcode. In studies on orchids of the genus
Paphiopedilum, amplification results similar to those for
Coelogyne were obtained, with sequence lengths ranging from 267 to 528 bp for
rbcL, from 834 to 873 bp for
matK, from 550 to 921 bp for
trnH-psbA, and from 262 to 494 bp for
atpF-atpH [
23]. Similarly, in a study on
Oberonia, the average sequence length for
rbcL,
matK, and
trnH-psbA was 1187 bp, 815 bp, and 1001 bp, respectively, indicating the long length of the
matK sequence, which often poses a problem during the amplification of this locus [
31]. Other studies also suggest that
matK is not always an effective marker for species identification, indicating the need for further research and optimization of this sequence [
15,
23].
The trnH locus was the most effective barcode of all those tested in identifying species of the genus Coelogyne, allowing for the identification of 21% of the species under study (four out of 19) and two species that were ambiguously identified (11%). While 16% of species were successfully identified using the matK barcode, a good but marginally lower result than trnH, up to 47% of the results were unclear (nine out of 19). The least successful barcodes were ITS2, which only identified one species correctly, and atpF-atpH and rbcL, which only identified 11% of the species (two out of 19). The low effectiveness of atpF-atpH was mainly due to amplification problems, which failed in as many as 15 samples. These problems may be due to specific difficulties associated with the amplification of this region.
Overall species identification rates in CBOL studies varied between 61% and 69%. For the
trnH-psbA locus, 69% identification efficiency was achieved, 61% for
rbcL, and 66% for
matK. The identification efficiency for
atpF-atpH was roughly 55%, indicating that this region is not as useful as other chloroplast markers. Other loci, like
psbK-psbI, were also examined in the CBOL study and demonstrated remarkable efficacy in identifying terrestrial plants, with a 68% identification rate [
10]. In addition, other sequences, such as
ndhF and
ycf1, also showed high effectiveness in identifying plants of the Orchidaceae family, achieving results of 88.65% and 89.32%, respectively [
29].
The ITS region was found to have the highest percentage of correct species identification (93.2%), the largest barcode gap, and the greatest interspecific and intraspecific variability in studies on orchids of the genus
Cymbidium.
MatK (75.8%),
psbA-trnH (87.1%), and
rbcL (54.2%) were the other regions with lower identification efficiency [
33].
Srivastava and Manjunath studied endemic endangered orchid species from India. A total of 178 sequences were obtained for the ITS,
matK, and
rbcL loci from 62 samples representing 35 species belonging to seven genera. In the BLAST analysis, the ITS locus outperformed
matK (51.61%) and
rbcL (78.69%) with 94.64% correct sequence identifications. ITS,
rbcL, and
matK all had incorrect identification rates of 3.57%, 16.39%, and 38.71%, respectively. These results confirm that ITS is the most effective marker for identifying orchid species [
24].
Considering terrestrial plants, the overall percentage of genera in which species pairs could be distinguished was 45.8% for ITS1, while
trnH-psbA showed the highest resolution (79.1%),
rbcL ranked second (62.5%), and other loci such as
matK,
rpoB1,
rpoC2,
accD,
ndhJ, and
ycf had an effectiveness of less than 50% [
30].
Numerous studies have demonstrated the usefulness of multiple barcodes, such as
matK +
ycf1 and
ndhF + ycf1, which have been effectively used to identify Orchidaceae plants [
26]. Although not all studies support the superiority of this multi-barcode over alternative marker combinations, the combination of
rbcL +
matK has been suggested as a universal barcode for the identification of terrestrial plants [
10]. For instance, research on orchids has demonstrated that the
rbcL locus works better as a barcode on its own than when combined with
matK [
34]. In a
Cymbidium study, Chen (2024) also demonstrated that ITS-containing region combinations did not substantially improve identification efficiency over ITS alone [
33].
Our results suggest that the use of three loci (e.g.,
rbcL,
trnH-psbA, and
matK) does not always translate into a clear improvement in identification efficiency compared to a two-locus approach. One species was correctly identified using a barcode made up of
rbcL +
trnH-psbA, while two had ambiguous results. On the other hand, 16% of species were successfully identified using
matK +
trnH-psbA, while 21% had an ambiguous result, which is comparable to the outcome of using three loci. Accordingly,
matK may be especially helpful in multi-locus codes that contain this locus. In studies of other orchids, such as
Paphiopedilum, the combination of
matK +
atpF-atpH + ITS has been recommended as an effective multiple code for species identification within this genus [
10].
The ability to accurately classify samples to the genus
Coelogyne, which is essential for conservation initiatives and regulatory applications like tracking the illicit orchid trade under CITES, is another significant outcome of this study [
35]. While the results may be simpler to obtain, assignment to the genus
Coelogyne can be just as useful as species identification, because all of its species are legally protected from illegal trade [
13,
36].
Out of all the tested single-locus barcodes,
matK performed best overall in genus-level identification, consistently providing correct genus-level assignment in all the samples tested, even when species-level matches were ambiguous or unsuccessful. Its moderate amplification efficiency (74%) and relatively high discriminatory power make it a robust candidate for genus-level identification in
Coelogyne. Despite occasional issues with primer mismatch and amplicon size (average 778 bp), its performance in genus-level resolution was superior [
14,
33,
34,
37].
Comparatively speaking,
trnH-psbA also achieved high amplification efficiency (84%). However, genus-level results for this marker were ambiguous in all tested samples, suggesting limitations in its taxonomic resolution within the genus
Coelogyne. While it remains useful in broader plant barcoding applications and offers reliable laboratory performance, its effectiveness in assigning samples confidently to
Coelogyne was limited in this study [
10,
30,
31].
With a 74% amplification efficiency, the
matK region demonstrated greater discriminatory power than
rbcL. Even in cases where species-level matches were unclear, it consistently obtained accurate genus-level identification. The larger average amplicon size (778 bp) and frequent primer mismatches, however, are problems with
matK that lower its amplification reliability, particularly for the frequently poor quality or degraded samples frequently found in law enforcement settings [
14,
33,
34,
37].
In summary, studies on DNA barcodes of orchids of the genus
Coelogyne confirm that the
matK locus can be an effective marker, although problems with its amplification require further optimization of primers. Barcodes composed of multiple loci, especially those containing
matK, can significantly improve the effectiveness of species identification, making them a promising tool in research on orchid biodiversity [
14,
33,
34,
37].
Overall performance was lowest in the
atpF-atpH region. Despite its initial amplification success, this marker may not prove appropriate for practical applications in genus-level screening of
Coelogyne orchids, as evidenced by the ambiguity of identification in a number of cases [
23,
38].
Analysis of the ITS2 region showed excellent amplification efficiency (95%). The genus-level identification was consistently achieved for all successfully sequenced samples, highlighting the high discriminatory potential of this region. However, technical challenges associated with ITS2 sequencing limit its practical application as a routine barcode for regulatory purposes [
16,
29].
The
matK +
trnH-psbA combination has shown great promise when it comes to multi-marker strategies. This combination strikes a balance between the robust amplification and trustworthy genus-level identification of
matK and the broad amplification reliability of
trnH-psbA.
TrnH-psbA may permit sequence recovery in cases where
matK amplification is unsuccessful, making this combination a flexible option [
10,
29]. In a similar vein,
rbcL +
trnH-psbA has demonstrated good efficacy in genus assignment, albeit with marginally reduced species-level resolution [
14,
30].
Based on these findings, it seems that
matK, either by itself or in conjunction with
trnH-psbA, offers the best possible balance between amplification efficiency and genus assignment accuracy. Therefore, it is recommended as a key target for further development of rapid diagnostic tests for verifying the identity of
Coelogyne orchids, particularly in the context of CITES enforcement [
13,
23,
39]. However, wider validation across more species and populations is required before customs and wildlife authorities adopt this approach as a standard tool.
Recent advances in plastome sequencing have highlighted that exploring a broader range of chloroplast markers can considerably enhance taxonomic resolution and shed light on evolutionary relationships. For instance, pan-plastome analyses in
Lathyrus oleraceus revealed that certain plastid regions, such as
ycf1,
rpoC2, and
matK, display high variability and could serve as promising barcoding targets for lineage tracking and genetic diversity studies [
40]. Similarly, comprehensive chloroplast genome comparisons in
Chrysanthemum highlighted how plastome-scale data can refine phylogenetic relationships and strengthen the accuracy of molecular classification across closely related taxa [
41].
Although the present study focuses on a limited number of loci rather than complete plastomes, these findings emphasize the potential of chloroplast genome exploration to enhance species discrimination. The patterns observed for matK and trnH-psbA in Coelogyne are consistent with this view, indicating that plastid regions with higher variability could further improve barcoding efficiency. In this context, incorporating additional chloroplast markers such as ycf1 or rpoC2 may represent a logical next step toward achieving higher resolution and more robust identification frameworks for Coelogyne species.
This study provides a valuable foundation for developing DNA barcoding tools to support CITES authorities in species identification and the detection of illegal trade. However, due to the limitations of this study, the approach requires further refinement and validation before broader application. Amplification success varied across loci, with some markers requiring further primer optimization—especially for degraded samples often encountered in law enforcement contexts. Expanding the sampling strategy to include multiple individuals per species from diverse geographic regions would improve the robustness of genetic variability assessments and marker performance. Despite these limitations, genus-level identification using DNA barcoding proves to be a practical and effective solution for CITES enforcement. Reliable markers such as
matK, alone or in combination with
rbcL and/or
trnH-psbA, consistently enable genus-level assignment, which is sufficient for regulatory purposes. This study contributes to biodiversity conservation and the fight against illegal orchid trade by expanding the genetic reference database for
Coelogyne, bridging the gap between academic research and the operational needs of CITES authorities [
13].