An Integrative Multi-Source Evidence Framework for Prioritizing Virulence-Associated Pathways in Metarhizium brunneum
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Source
2.2. Protein Annotation and Completeness Assessment
2.3. KEGG and GO Enrichment Analysis
2.4. Construction of a Virulence-Related Pathway Scoring System
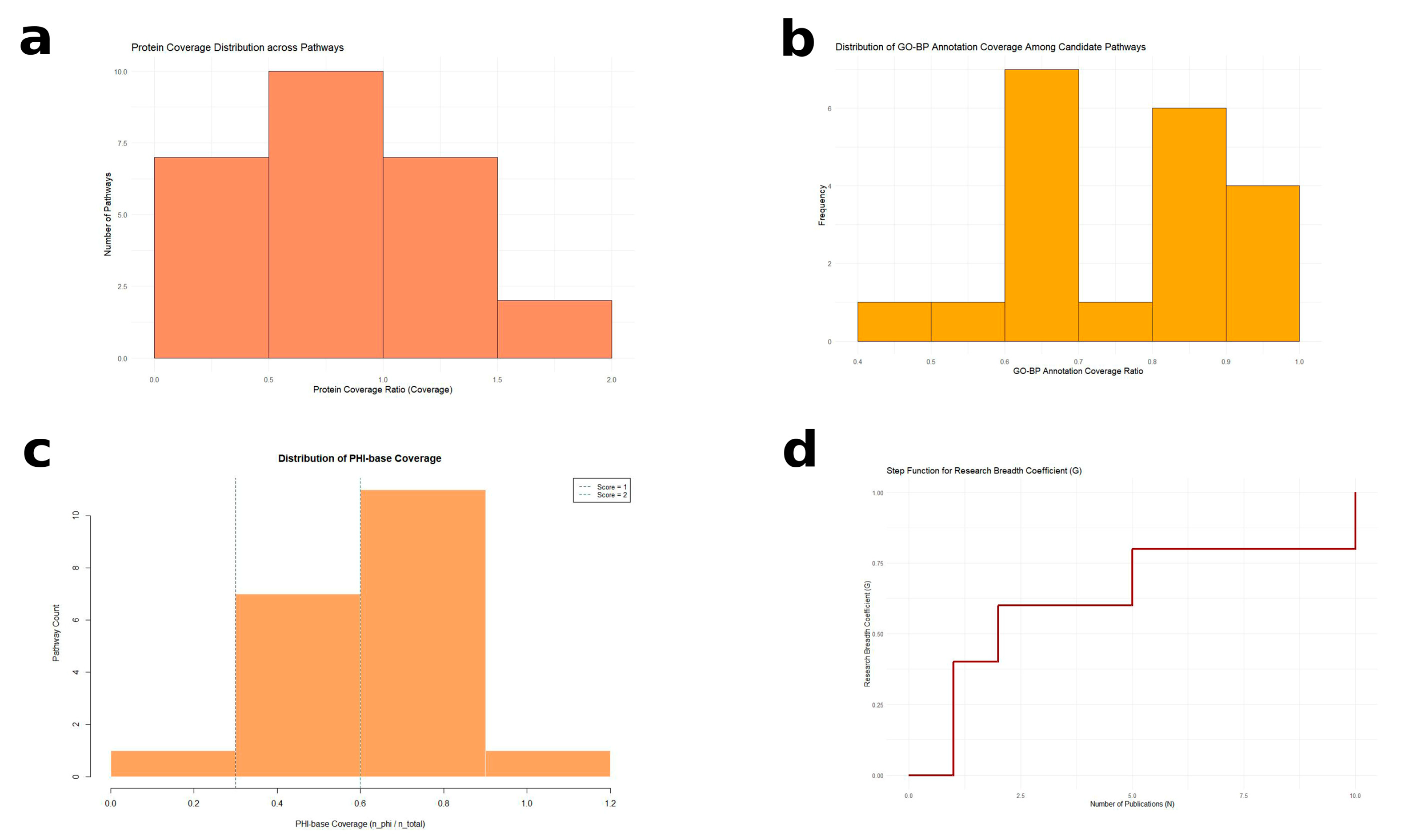
2.4.1. Protein Coverage Scoring (0~2 Points)
2.4.2. GO Functional Support Scoring (0~2 Points)
2.4.3. PHI-Base Hit Scoring (0~3 Points)
2.4.4. Literature Support Scoring (0~3 Points)
3. Results
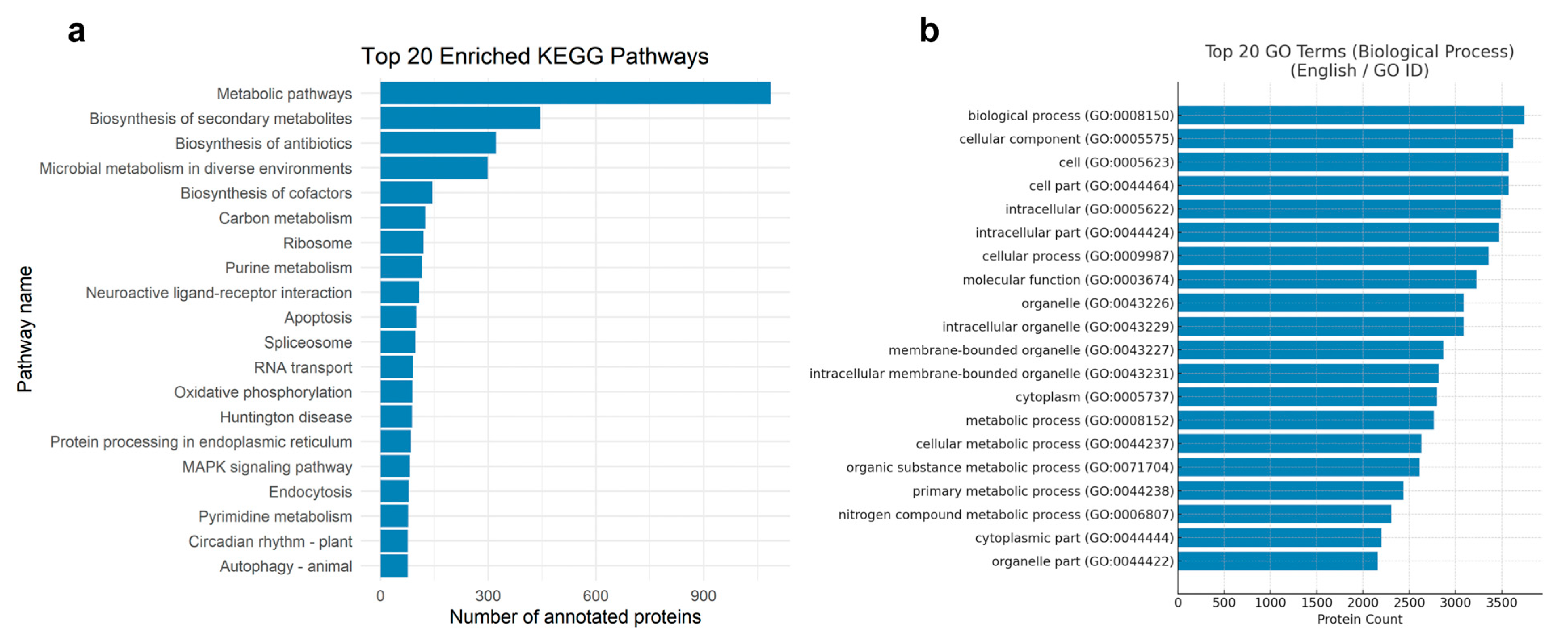
3.1. BUSCO Completeness Assessment and Protein Annotation Overview
3.2. Functional Annotation Overview of the M. brunneum Proteome (KEGG and GO)
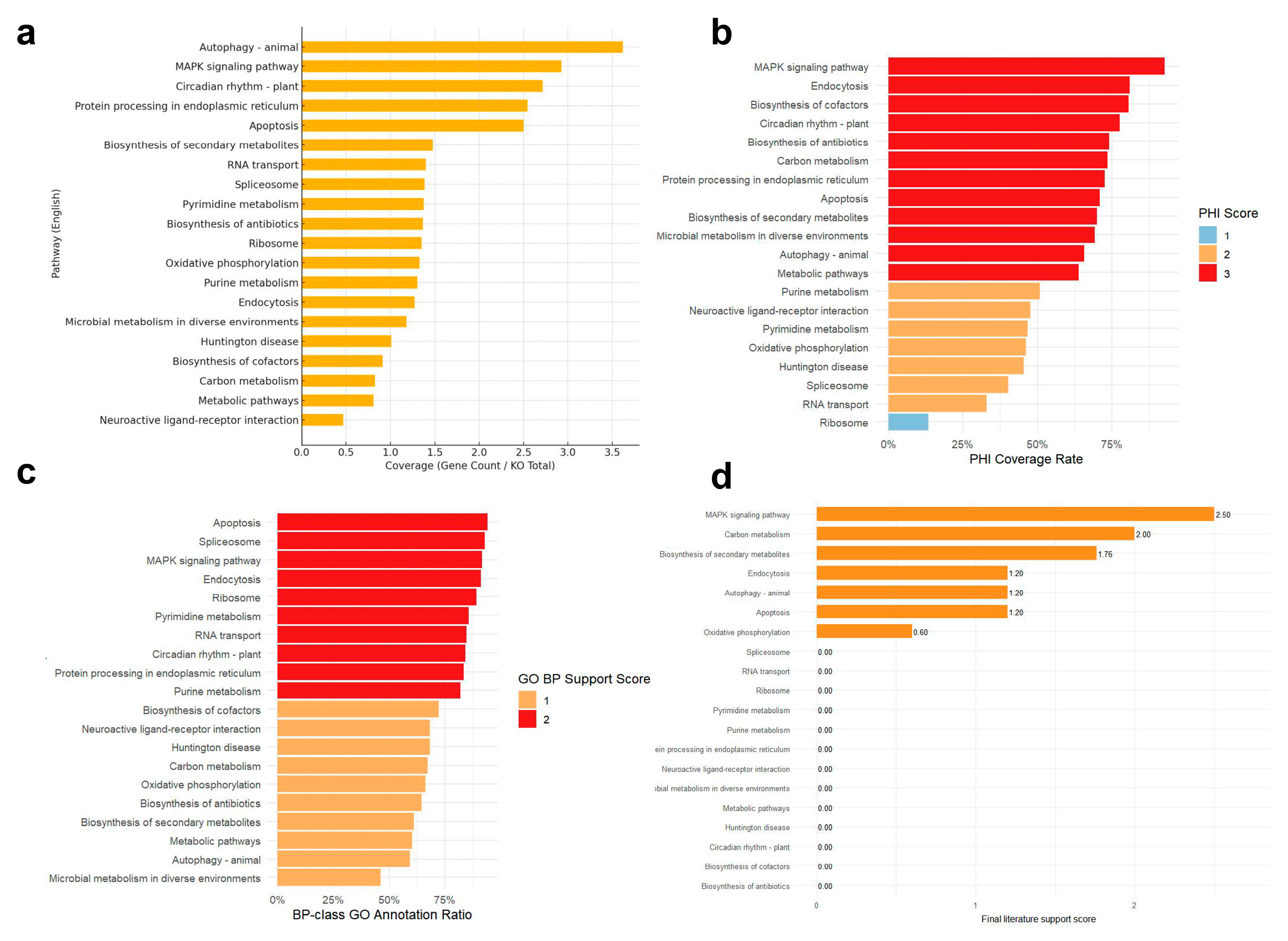
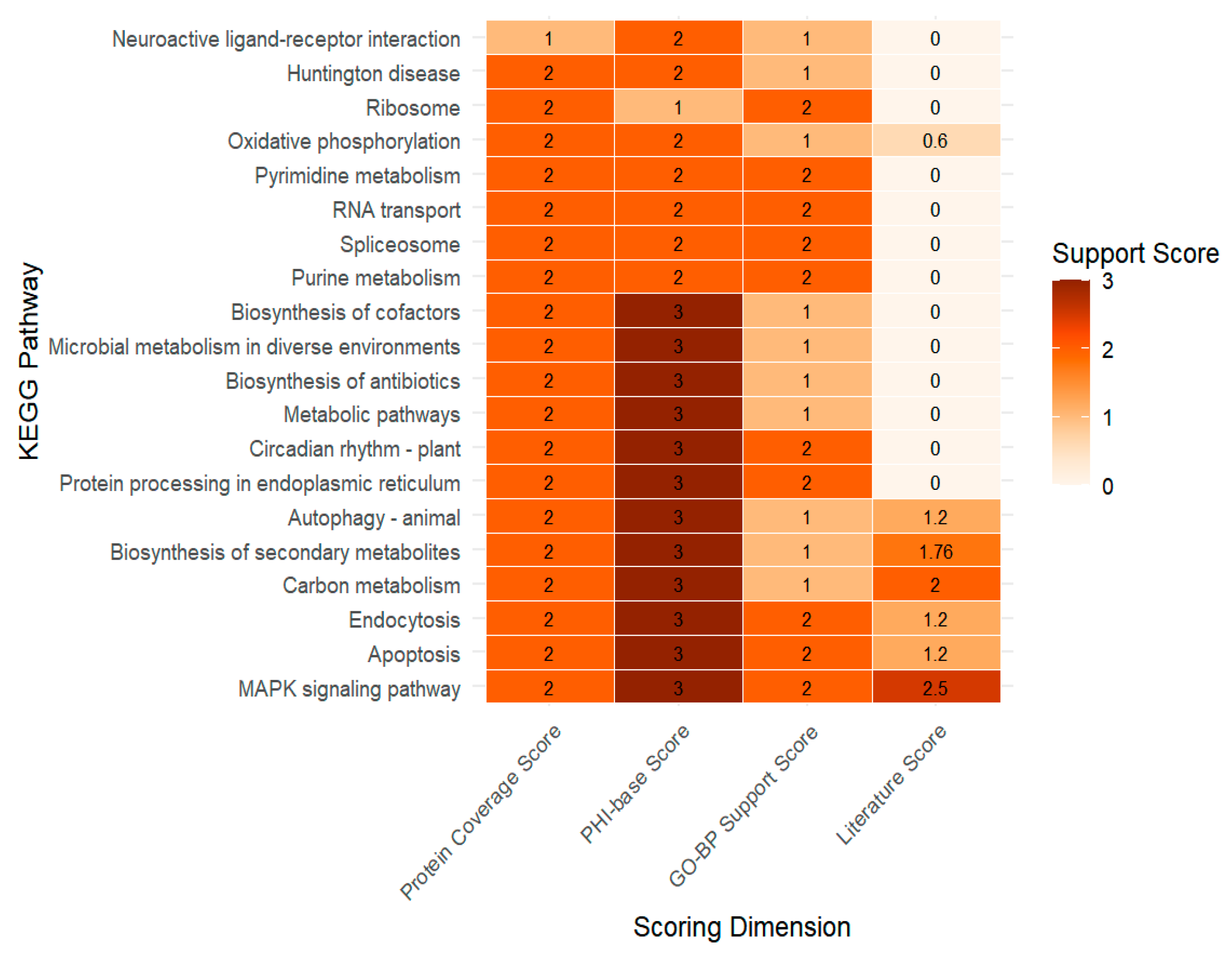
3.3. Functional Evidence-Based Prioritization of Candidate Pathways
4. Discussion
4.1. Biological Significance of the Top Five Pathways in Fungal Virulence
4.2. Applicability and Limitations of the Multidimensional Pathway Scoring Framework
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| M. brunneum | Metarhizium brunneum |
| GO | Gene Ontology |
| KO | KEGG Ortholog |
| BP | Biological Process |
References
- Perumalsamy, N.; Sharma, R.; Subramanian, M.; Nagarajan, S.A. Hard Ticks as Vectors: The Emerging Threat of Tick-Borne Diseases in India. Pathogens 2024, 13, 556. [Google Scholar] [CrossRef] [PubMed]
- Machtinger, E.T.; Weeks, E.N.I.; Geden, C.J. Oviposition Deterrence and Immature Survival of Filth Flies (Diptera: Muscidae) When Exposed to Commercial Fungal Products. J. Insect Sci. 2016, 16, 54. [Google Scholar] [CrossRef] [PubMed]
- Viglietta, M.; Bellone, R.; Blisnick, A.A.; Failloux, A.-B. Vector Specificity of Arbovirus Transmission. Front. Microbiol. 2021, 12, 773211. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.T.; Lindahl, J.F.; Bett, B.; Nguyen-Viet, H.; Lâm, S.; Nguyen-Tien, T.; Unger, F.; Dang-Xuan, S.; Bui, T.X.; Le, H.T.; et al. Understanding Zoonotic Pathogens and Risk Factors from Wildlife in Southeast Asia: A Systematic Literature Review. Vet. Q. 2025, 45, 1–17. [Google Scholar] [CrossRef]
- Howard, A.F.; Koenraadt, C.J.; Farenhorst, M.; Knols, B.G.; Takken, W. Pyrethroid Resistance in Anopheles Gambiae Leads to Increased Susceptibility to the Entomopathogenic Fungi Metarhizium Anisopliae and Beauveria Bassiana. Malar. J. 2010, 9, 168. [Google Scholar] [CrossRef]
- Bittencourt, V.R. Trials to Control South American Ticks with Entomopathogenic Fungi. Ann. N. Y. Acad. Sci. 2000, 916, 555–558. [Google Scholar] [CrossRef]
- Cafarchia, C.; Pellegrino, R.; Romano, V.; Friuli, M.; Demitri, C.; Pombi, M.; Benelli, G.; Otranto, D. Delivery and Effectiveness of Entomopathogenic Fungi for Mosquito and Tick Control: Current Knowledge and Research Challenges. Acta Trop. 2022, 234, 106627. [Google Scholar] [CrossRef]
- Andrade, G.C.R.M.; Monteiro, S.H.; Pereira, J.R.; Duarte, F.C.; Mendes, M.C.; De Almeida, J.E.M.; Nicodemos, F.G. Tick Biocontrol Strategy in Dairy Cattle: Drone-Applied Metarhizium anisopliae Fungus as an Alternative to Conventional Acaricides for Reducing Pesticide Residues in Milk. ACS Agric. Sci. Technol. 2025, 5, 2021–2029. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Mubeen, S.; Kodamullil, A.T.; Hofmann-Apitius, M.; Domingo-Fernández, D. On the Influence of Several Factors on Pathway Enrichment Analysis. Brief. Bioinform. 2022, 23, bbac143. [Google Scholar] [CrossRef]
- Lin, S.-J.; Lu, T.-P.; Yu, Q.-Y.; Hsiao, C.K. Probabilistic Prioritization of Candidate Pathway Association with Pathway Score. BMC Bioinform. 2018, 19, 391. [Google Scholar] [CrossRef]
- Kortsinoglou, A.M.; Wood, M.J.; Myridakis, A.I.; Andrikopoulos, M.; Roussis, A.; Eastwood, D.; Butt, T.; Kouvelis, V.N. Comparative Genomics of Metarhizium Brunneum Strains V275 and ARSEF 4556: Unraveling Intraspecies Diversity. G3 Genes Genomes Genet. 2024, 14, jkae190. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-Mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Tegenfeldt, F.; Kuznetsov, D.; Manni, M.; Berkeley, M.; Zdobnov, E.M.; Kriventseva, E.V. OrthoDB and BUSCO Update: Annotation of Orthologs with Wider Sampling of Genomes. Nucleic Acids Res. 2025, 53, D516–D522. [Google Scholar] [CrossRef]
- Franzosa, E.A.; McIver, L.J.; Rahnavard, G.; Thompson, L.R.; Schirmer, M.; Weingart, G.; Lipson, K.S.; Knight, R.; Caporaso, J.G.; Segata, N.; et al. Species-Level Functional Profiling of Metagenomes and Metatranscriptomes. Nat. Methods 2018, 15, 962–968. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive Functional Profiling of Microbial Communities Using 16S rRNA Marker Gene Sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Urban, M.; Cuzick, A.; Seager, J.; Nonavinakere, N.; Sahoo, J.; Sahu, P.; Iyer, V.L.; Khamari, L.; Martinez, M.C.; Hammond-Kosack, K.E. PHI-Base—The Multi-Species Pathogen–Host Interaction Database in 2025. Nucleic Acids Res. 2025, 53, D826–D838. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Wen, Z.; Fan, Y.; Xia, Y.; Jin, K. MaOpy2, a Transmembrane Protein, Is Involved in Stress Tolerances and Pathogenicity and Negatively Regulates Conidial Yield by Shifting the Conidiation Pattern in Metarhizium Acridum. J. Fungi 2022, 8, 587. [Google Scholar] [CrossRef]
- Qu, J.; Tong, Y.; Yang, L.; Wu, H.; Chen, M.; Huang, B. Dissecting the Multifaceted Influence of the MrBHLH1 Transcription Factor on Development, Stress Response, and Virulence in Metarhizium Robertsii. Pest Manag. Sci. 2025, 81, 6229–6240. [Google Scholar] [CrossRef]
- Zhang, L.; Yang, T.; Yu, W.; Wang, X.; Zhou, X.; Zhou, X. Genome-Wide Study of Conidiation-Related Genes in the Aphid-Obligate Fungal Pathogen Conidiobolus Obscurus (Entomophthoromycotina). J. Fungi 2022, 8, 389. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Wen, Z.; Xia, Y.; Jin, K. The Transmembrane Protein MaSho1 Negatively Regulates Conidial Yield by Shifting the Conidiation Pattern in Metarhizium Acridum. Appl. Microbiol. Biotechnol. 2020, 104, 4005–4015. [Google Scholar] [CrossRef]
- Deng, J.; Huang, S.; Kan, Y.; Song, Y.; Zhao, X.; Li, N.; Yao, X.; Luo, Z.; Zhang, Y. A Transcription Factor-Mediated Regulatory Network Controls Fungal Pathogen Colonization of Insect Body Cavities. mBio 2024, 15, e0350423. [Google Scholar] [CrossRef]
- Kan, Y.; He, Z.; Keyhani, N.O.; Li, N.; Huang, S.; Zhao, X.; Liu, P.; Zeng, F.; Li, M.; Luo, Z.; et al. A Network of Transcription Factors in Complex with a Regulating Cell Cycle Cyclin Orchestrates Fungal Oxidative Stress Responses. BMC Biol. 2024, 22, 81. [Google Scholar] [CrossRef]
- Zhao, X.; Jiang, Y.; Wang, H.; Lu, Z.; Huang, S.; Luo, Z.; Zhang, L.; Lv, T.; Tang, X.; Zhang, Y. Fus3/Kss1-MAP Kinase and Ste12-like Control Distinct Biocontrol-Traits besides Regulation of Insect Cuticle Penetration via Phosphorylation Cascade in a Filamentous Fungal Pathogen. Pest Manag. Sci. 2023, 79, 2611–2624. [Google Scholar] [CrossRef]
- Tang, D.; Tang, X.; Fang, W. New Downstream Signaling Branches of the Mitogen-Activated Protein Kinase Cascades Identified in the Insect Pathogenic and Plant Symbiotic Fungus Metarhizium Robertsii. Front. Fungal Biol. 2022, 3, 911366. [Google Scholar] [CrossRef]
- Meng, Y.; Zhang, X.; Guo, N.; Fang, W. MrSt12 Implicated in the Regulation of Transcription Factor AFTF1 by Fus3-MAPK during Cuticle Penetration by the Entomopathogenic Fungus Metarhizium Robertsii. Fungal Genet. Biol. 2019, 131, 103244. [Google Scholar] [CrossRef]
- Wang, Z.-K.; Cai, Q.; Tong, S.-M.; Ying, S.-H.; Feng, M.-G. C-terminal Ser/Thr Residues Are Vital for the Regulatory Role of Ste7 in the Asexual Cycle and Virulence of Beauveria Bassiana. Appl. Microbiol. Biotechnol. 2018, 102, 6973–6986. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sun, H.-H.; Ying, S.-H.; Feng, M.-G. Characterization of Three Mitogen-Activated Protein Kinase Kinase-like Proteins in Beauveria Bassiana. Fungal Genet. Biol. 2018, 113, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Tong, S.-M.; Qiu, L.; Ying, S.-H.; Feng, M.-G. Two Histidine Kinases Can Sense Different Stress Cues for Activation of the MAPK Hog1 in a Fungal Insect Pathogen. Environ. Microbiol. 2017, 19, 4091–4102. [Google Scholar] [CrossRef]
- Liu, J.; Sun, H.-H.; Ying, S.-H.; Feng, M.-G. The Hog1-like MAPK Mpk3 Collaborates with Hog1 in Response to Heat Shock and Functions in Sustaining the Biological Control Potential of a Fungal Insect Pathogen. Appl. Microbiol. Biotechnol. 2017, 101, 6941–6949. [Google Scholar] [CrossRef]
- Liu, J.; Wang, Z.-K.; Sun, H.-H.; Ying, S.-H.; Feng, M.-G. Characterization of the Hog1 MAPK Pathway in the Entomopathogenic Fungus Beauveria Bassiana. Environ. Microbiol. 2017, 19, 1808–1821. [Google Scholar] [CrossRef]
- Song, Z.; Zhong, Q.; Yin, Y.; Shen, L.; Li, Y.; Wang, Z. The High Osmotic Response and Cell Wall Integrity Pathways Cooperate to Regulate Morphology, Microsclerotia Development, and Virulence in Metarhizium Rileyi. Sci. Rep. 2016, 6, 38765. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xu, C.; Qian, Y.; Liu, R.; Zhang, Q.; Zeng, G.; Zhang, X.; Zhao, H.; Fang, W. MAPK Cascade-Mediated Regulation of Pathogenicity, Conidiation and Tolerance to Abiotic Stresses in the Entomopathogenic Fungus Metarhizium Robertsii. Environ. Microbiol. 2016, 18, 1048–1062. [Google Scholar] [CrossRef]
- Wang, Z.-L.; Li, F.; Li, C.; Feng, M.-G. Bbssk1, a Response Regulator Required for Conidiation, Multi-Stress Tolerance, and Virulence of Beauveria Bassiana. Appl. Microbiol. Biotechnol. 2014, 98, 5607–5618. [Google Scholar] [CrossRef]
- Chen, Y.; Zhu, J.; Ying, S.-H.; Feng, M.-G. Three Mitogen-Activated Protein Kinases Required for Cell Wall Integrity Contribute Greatly to Biocontrol Potential of a Fungal Entomopathogen. PLoS ONE 2014, 9, e87948. [Google Scholar] [CrossRef] [PubMed]
- Jin, K.; Ming, Y.; Xia, Y.X. MaHog1, a Hog1-Type Mitogen-Activated Protein Kinase Gene, Contributes to Stress Tolerance and Virulence of the Entomopathogenic Fungus Metarhizium Acridum. Microbiol. Read. Engl. 2012, 158, 2987–2996. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Keyhani, N.O.; Yu, X.; He, Z.; Luo, Z.; Pei, Y.; Zhang, Y. The MAP Kinase Bbslt2 Controls Growth, Conidiation, Cell Wall Integrity, and Virulence in the Insect Pathogenic Fungus Beauveria Bassiana. Fungal Genet. Biol. 2012, 49, 544–555. [Google Scholar] [CrossRef]
- Zhou, G.; Wang, J.; Qiu, L.; Feng, M.-G. A Group III Histidine Kinase (Mhk1) Upstream of High-Osmolarity Glycerol Pathway Regulates Sporulation, Multi-Stress Tolerance and Virulence of Metarhizium Robertsii, a Fungal Entomopathogen. Environ. Microbiol. 2012, 14, 817–829. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.; Jiang, X.; Wang, G.; Luo, Z.; Fan, Y.; Wu, Z.; Pei, Y. Requirement of a Mitogen-Activated Protein Kinase for Appressorium Formation and Penetration of Insect Cuticle by the Entomopathogenic Fungus Beauveria Bassiana. Appl. Environ. Microbiol. 2010, 76, 2262–2270. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, J.; Fang, W.; Zhang, J.; Luo, Z.; Zhang, M.; Fan, Y.; Pei, Y. Mitogen-Activated Protein Kinase Hog1 in the Entomopathogenic Fungus Beauveria Bassiana Regulates Environmental Stress Responses and Virulence to Insects. Appl. Environ. Microbiol. 2009, 75, 3787–3795. [Google Scholar] [CrossRef]
- Singh, D.; Kaur, G. Production, HPLC Analysis, and in Situ Apoptotic Activities of Swainsonine toward Lepidopteran, Sf-21 Cell Line. Biotechnol. Prog. 2014, 30, 1196–1205. [Google Scholar] [CrossRef]
- Jirakkakul, J.; Roytrakul, S.; Srisuksam, C.; Swangmaneecharern, P.; Kittisenachai, S.; Jaresitthikunchai, J.; Punya, J.; Prommeenate, P.; Senachak, J.; So, L.; et al. Culture Degeneration in Conidia of Beauveria Bassiana and Virulence Determinants by Proteomics. Fungal Biol. 2018, 122, 156–171. [Google Scholar] [CrossRef]
- Xie, X.; Wang, Y.; Yu, D.; Xie, R.; Liu, Z.; Huang, B. DNM1, a Dynamin-Related Protein That Contributes to Endocytosis and Peroxisome Fission, Is Required for the Vegetative Growth, Sporulation, and Virulence of Metarhizium Robertsii. Appl. Environ. Microbiol. 2020, 86, e01217-20. [Google Scholar] [CrossRef]
- Wang, Z.; Jiang, Y.; Li, Y.; Feng, J.; Huang, B. MrArk1, an Actin-Regulating Kinase Gene, Is Required for Endocytosis and Involved in Sustaining Conidiation Capacity and Virulence in Metarhizium Robertsii. Appl. Microbiol. Biotechnol. 2019, 103, 4859–4868. [Google Scholar] [CrossRef]
- Ying, S.-H.; Feng, M.-G.; Keyhani, N.O. A Carbon Responsive G-Protein Coupled Receptor Modulates Broad Developmental and Genetic Networks in the Entomopathogenic Fungus, Beauveria Bassiana. Environ. Microbiol. 2013, 15, 2902–2921. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Li, M.; Liu, Q.; Huang, Q.; Yang, S.; Chen, B.; Peng, Y. The Virulence of Metarhizium Rileyi to Locusta Migratoria Is Determined by the Ability of the Fungus to Respond to Carbon and Nitrogen Sources. Int. J. Mol. Sci. 2025, 26, 4156. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Zhang, X.; Tang, D.; Chen, X.; Zhang, D.; Chen, J.; Fang, W. A Novel Nitrogen and Carbon Metabolism Regulatory Cascade Is Implicated in Entomopathogenicity of the Fungus Metarhizium Robertsii. Msystems 2021, 6, e0049921. [Google Scholar] [CrossRef]
- Yang, Y.-T.; Lee, S.J.; Nai, Y.-S.; Kim, S.; Kim, J.S. Up-Regulation of Carbon Metabolism-Related Glyoxylate Cycle and Toxin Production in Beauveria Bassiana JEF-007 during Infection of Bean Bug, Riptortus Pedestris (Hemiptera: Alydidae). Fungal Biol. 2016, 120, 1236–1248. [Google Scholar] [CrossRef]
- Ming, Y.; Wei, Q.; Jin, K.; Xia, Y. MaSnf1, a Sucrose Non-Fermenting Protein Kinase Gene, Is Involved in Carbon Source Utilization, Stress Tolerance, and Virulence in Metarhizium Acridum. Appl. Microbiol. Biotechnol. 2014, 98, 10153–10164. [Google Scholar] [CrossRef]
- Wang, X.-X.; He, P.-H.; Feng, M.-G.; Ying, S.-H. BbSNF1 Contributes to Cell Differentiation, Extracellular Acidification, and Virulence in Beauveria Bassiana, a Filamentous Entomopathogenic Fungus. Appl. Microbiol. Biotechnol. 2014, 98, 8657–8673. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Lu, Y.; Xu, W.; Du, Q.; Sui, L.; Zhao, Y.; Li, Q. RNA Sequencing Analysis of Beauveria Bassiana Isolated from Ostrinia Furnacalis Identifies the Pathogenic Genes. Microb. Pathog. 2019, 130, 190–195. [Google Scholar] [CrossRef]
- Filippou, C.; Diss, R.M.; Daudu, J.O.; Coutts, R.H.A.; Kotta-Loizou, I. The Polymycovirus-Mediated Growth Enhancement of the Entomopathogenic Fungus Beauveria Bassiana Is Dependent on Carbon and Nitrogen Metabolism. Front. Microbiol. 2021, 12, 606366. [Google Scholar] [CrossRef]
- Lai, Y.; Liu, K.; Zhang, X.; Zhang, X.; Li, K.; Wang, N.; Shu, C.; Wu, Y.; Wang, C.; Bushley, K.E.; et al. Comparative Genomics and Transcriptomics Analyses Reveal Divergent Lifestyle Features of Nematode Endoparasitic Fungus Hirsutella Minnesotensis. Genome Biol. Evol. 2014, 6, 3077–3093. [Google Scholar] [CrossRef] [PubMed]
- Singkaravanit, S.; Kinoshita, H.; Ihara, F.; Nihira, T. Cloning and Functional Analysis of the Second Geranylgeranyl Diphosphate Synthase Gene Influencing Helvolic Acid Biosynthesis in Metarhizium Anisopliae. Appl. Microbiol. Biotechnol. 2010, 87, 1077–1088. [Google Scholar] [CrossRef]
- Wang, J.-J.; Bai, W.-W.; Zhou, W.; Liu, J.; Chen, J.; Liu, X.-Y.; Xiang, T.-T.; Liu, R.-H.; Wang, W.-H.; Zhang, B.-L.; et al. Transcriptomic Analysis of Two Beauveria Bassiana Strains Grown on Cuticle Extracts of the Silkworm Uncovers Their Different Metabolic Response at Early Infection Stage. J. Invertebr. Pathol. 2017, 145, 45–54. [Google Scholar] [CrossRef] [PubMed]
| Score | Evidence Level | Evidence Description |
|---|---|---|
| 3 | Strong support | Functional validation (gene knockout/overexpression affects virulence) |
| 2 | Moderate support | Mechanistic association with virulence suggested by experimental data |
| 1 | Weak support | Expression changes only; indirect evidence |
| 0 | No support | No evidence of virulence relevance |
| Pathway | No. of Studies (N) | Mean Evidence Score (Avg) | Breadth Coefficient (G) | Final Literature Support Score |
|---|---|---|---|---|
| Metabolic pathways | 0 | 0 | 0 | 0 |
| Biosynthesis of secondary metabolites | 5 | 2.2 | 0.8 | 1.76 |
| Biosynthesis of antibiotics | 0 | 0 | 0 | 0 |
| Microbial metabolism in diverse environments | 0 | 0 | 0 | 0 |
| Biosynthesis of cofactors | 0 | 0 | 0 | 0 |
| Carbon metabolism | 10 | 2 | 1.0 | 2.0 |
| Ribosome | 0 | 0 | 0 | 0 |
| Purine metabolism | 0 | 0 | 0 | 0 |
| Neuroactive ligand-receptor interaction | 0 | 0 | 0 | 0 |
| Apoptosis | 3 | 2 | 0.6 | 1.2 |
| Spliceosome | 0 | 0 | 0 | 0 |
| RNA transport | 0 | 0 | 0 | 0 |
| Oxidative phosphorylation | 2 | 1 | 0.6 | 0.6 |
| Huntington disease | 0 | 0 | 0 | 0 |
| Protein processing in endoplasmic reticulum | 0 | 0 | 0 | 0 |
| MAPK signaling pathway | 26 | 2.5 | 1.0 | 2.5 |
| Endocytosis | 2 | 2 | 0.6 | 1.2 |
| Pyrimidine metabolism | 0 | 0 | 0 | 0 |
| Circadian rhythm—plant | 0 | 0 | 0 | 0 |
| Autophagy—animal | 1 | 3 | 0.4 | 1.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, J.; Wei, W.; Li, J.; Bai, H.; Narisu; Wang, R. An Integrative Multi-Source Evidence Framework for Prioritizing Virulence-Associated Pathways in Metarhizium brunneum. Genes 2025, 16, 1363. https://doi.org/10.3390/genes16111363
Wen J, Wei W, Li J, Bai H, Narisu, Wang R. An Integrative Multi-Source Evidence Framework for Prioritizing Virulence-Associated Pathways in Metarhizium brunneum. Genes. 2025; 16(11):1363. https://doi.org/10.3390/genes16111363
Chicago/Turabian StyleWen, Jingyi, Wei Wei, Jing Li, Hua Bai, Narisu, and Rui Wang. 2025. "An Integrative Multi-Source Evidence Framework for Prioritizing Virulence-Associated Pathways in Metarhizium brunneum" Genes 16, no. 11: 1363. https://doi.org/10.3390/genes16111363
APA StyleWen, J., Wei, W., Li, J., Bai, H., Narisu, & Wang, R. (2025). An Integrative Multi-Source Evidence Framework for Prioritizing Virulence-Associated Pathways in Metarhizium brunneum. Genes, 16(11), 1363. https://doi.org/10.3390/genes16111363






