Genome-Wide Association Study of First-Parity Reproductive Traits in Suzi Pig
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals
2.2. Measurement of Reproductive Performance
2.3. Sample Collection and Genomic DNA Extraction
2.4. Whole-Genome Snp Detection
2.5. Genome-Wide Association Study (Gwas)
3. Results
3.1. Results of First-Parity Reproductive Performance
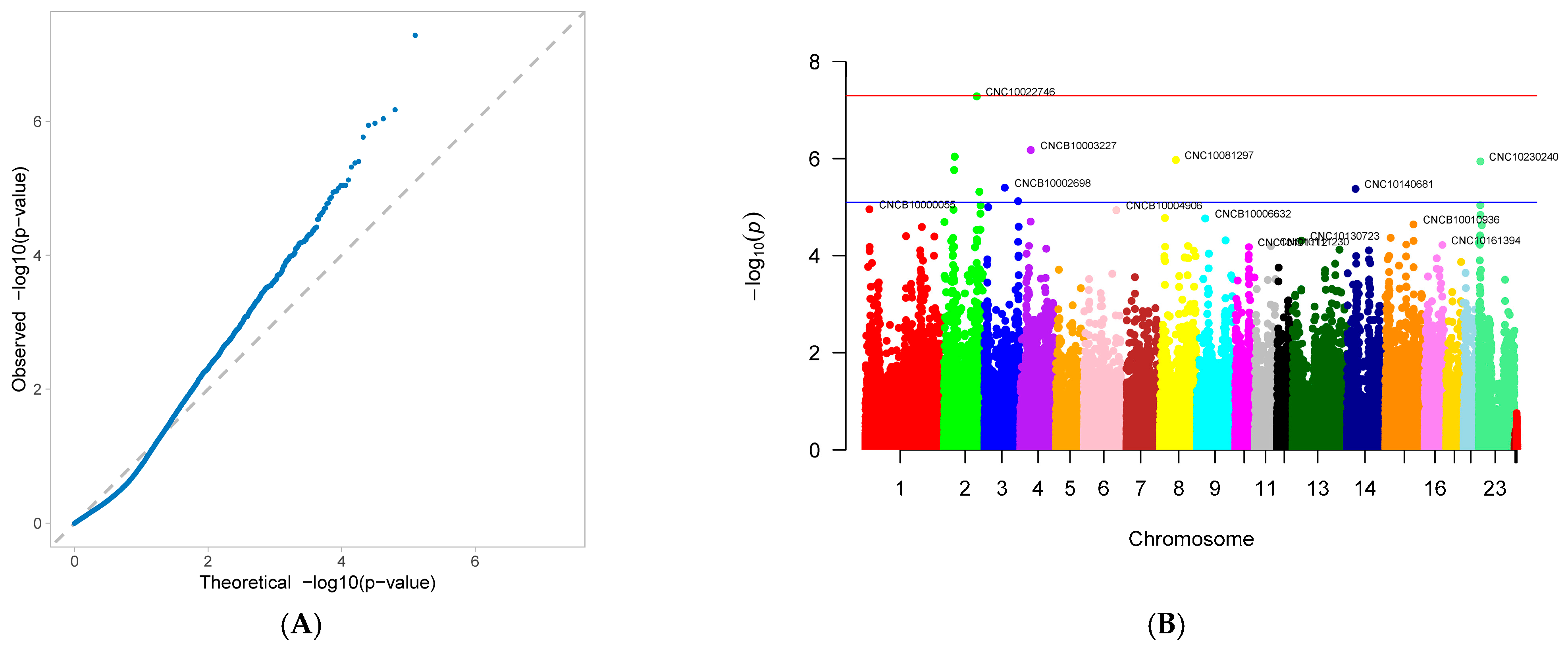
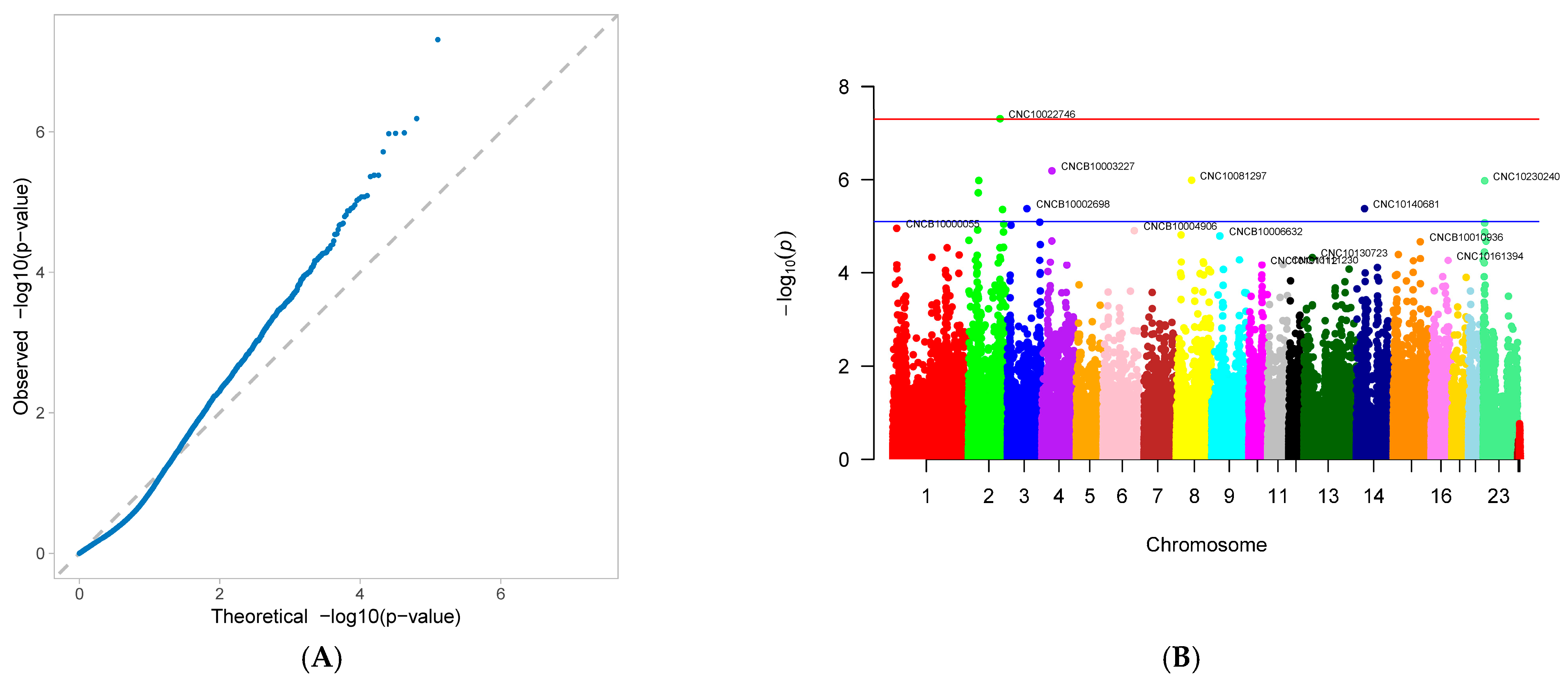
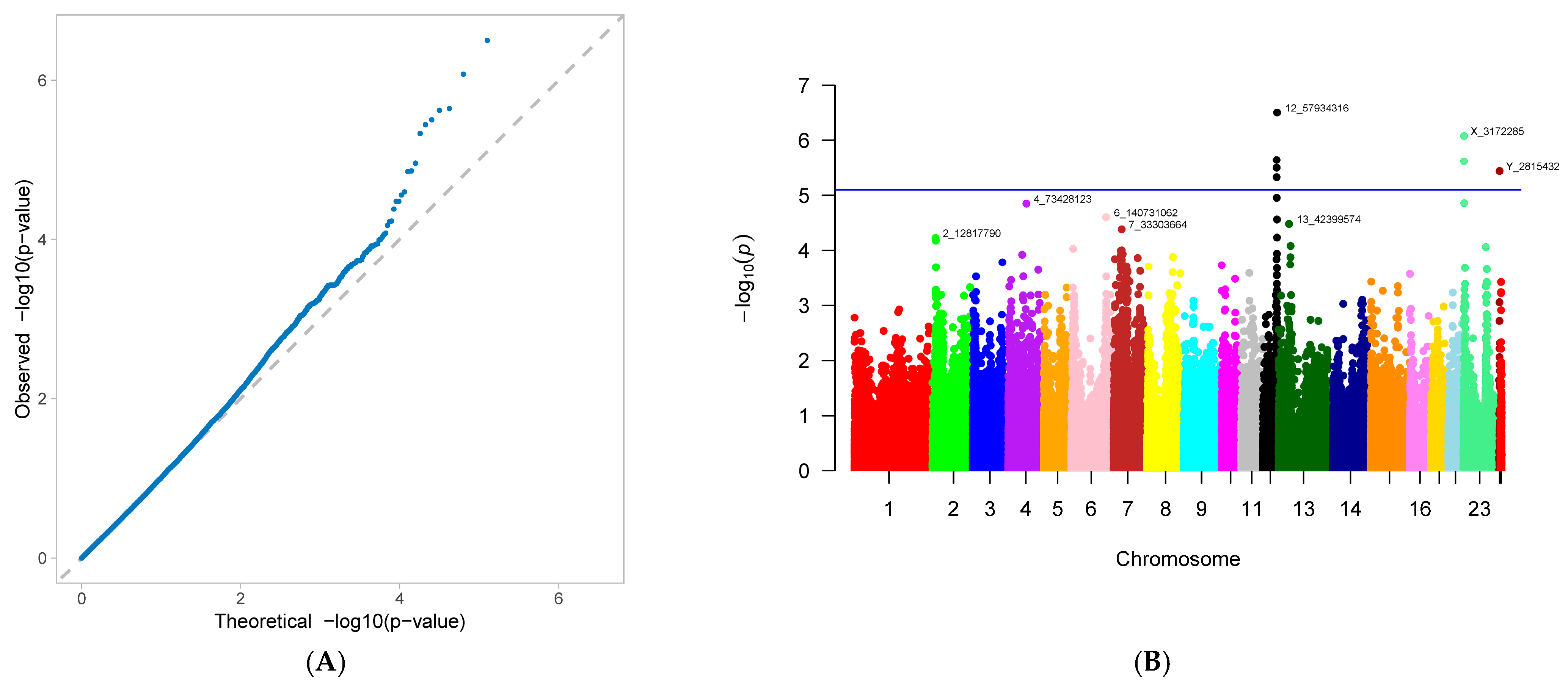
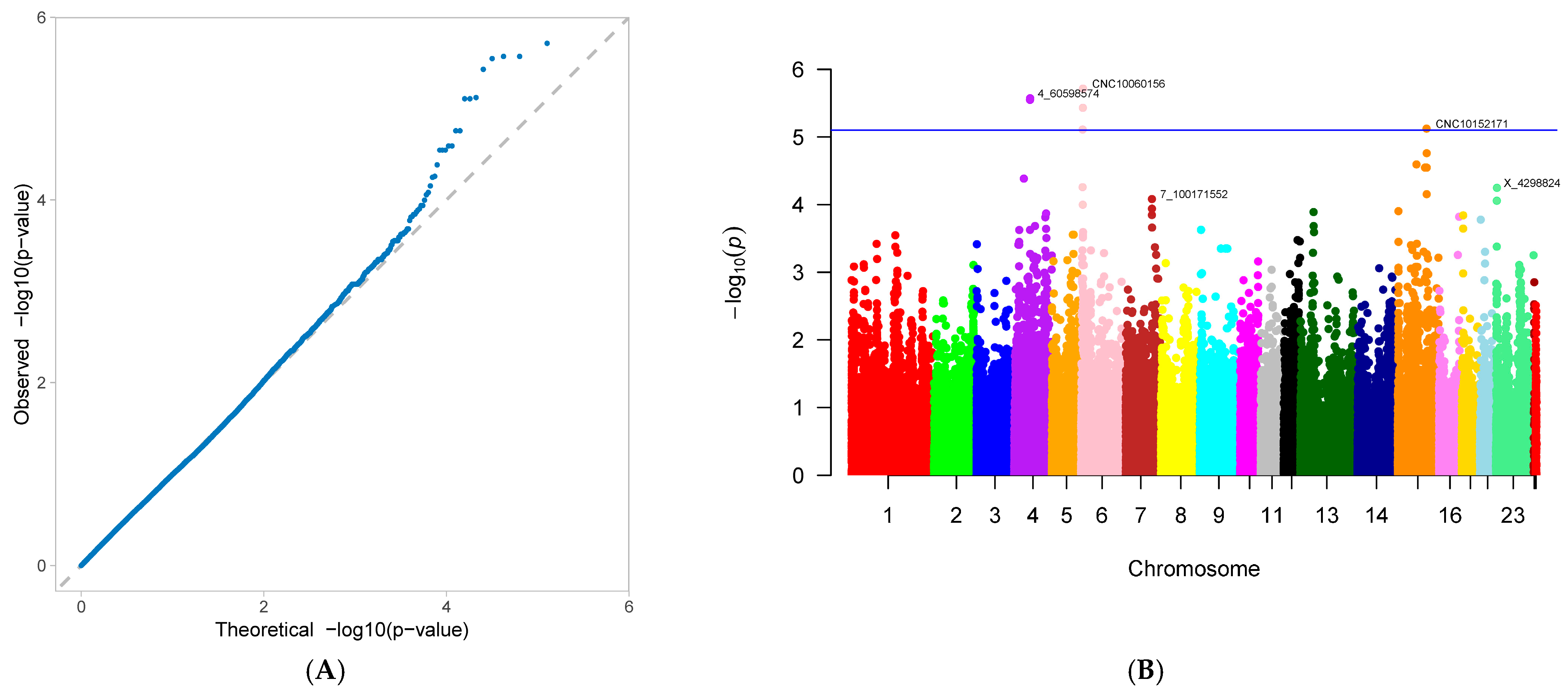
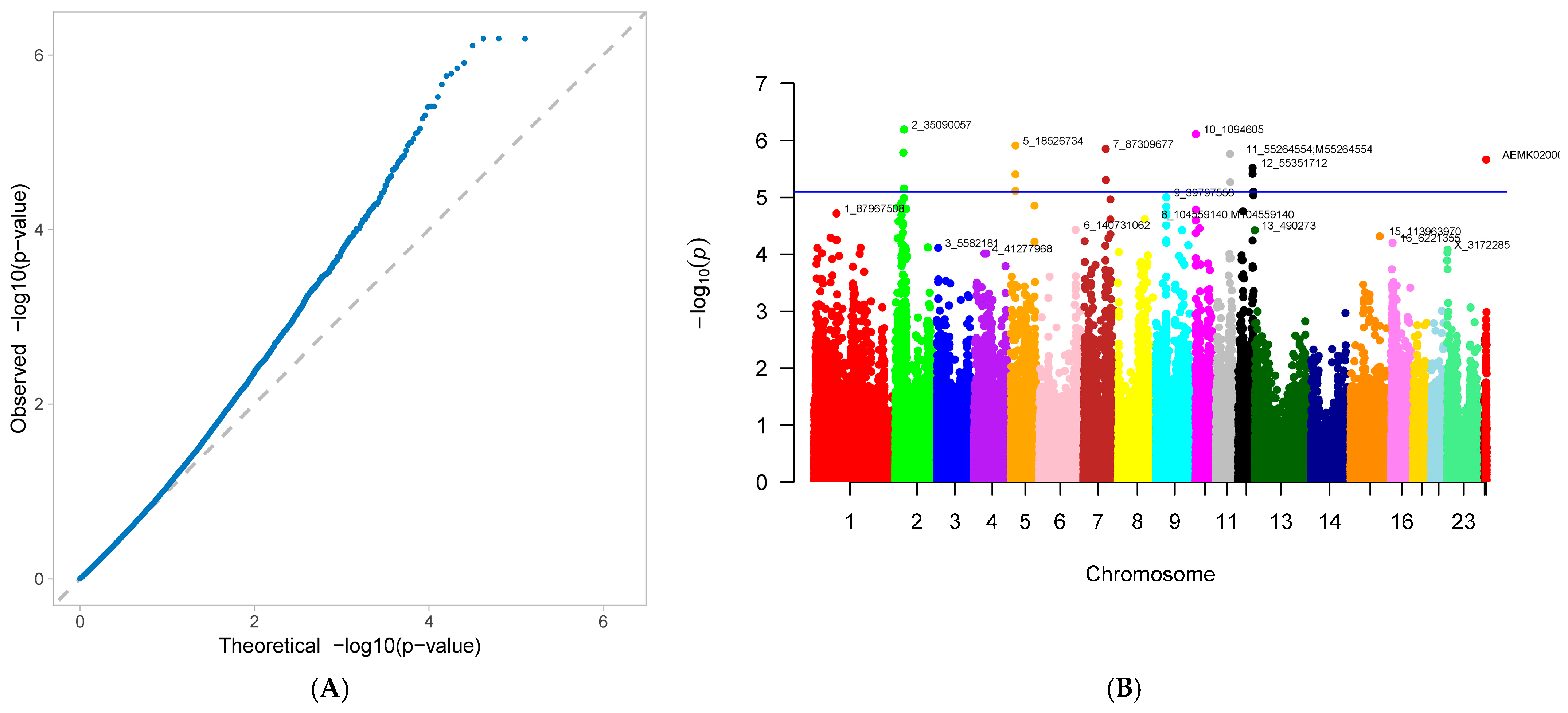
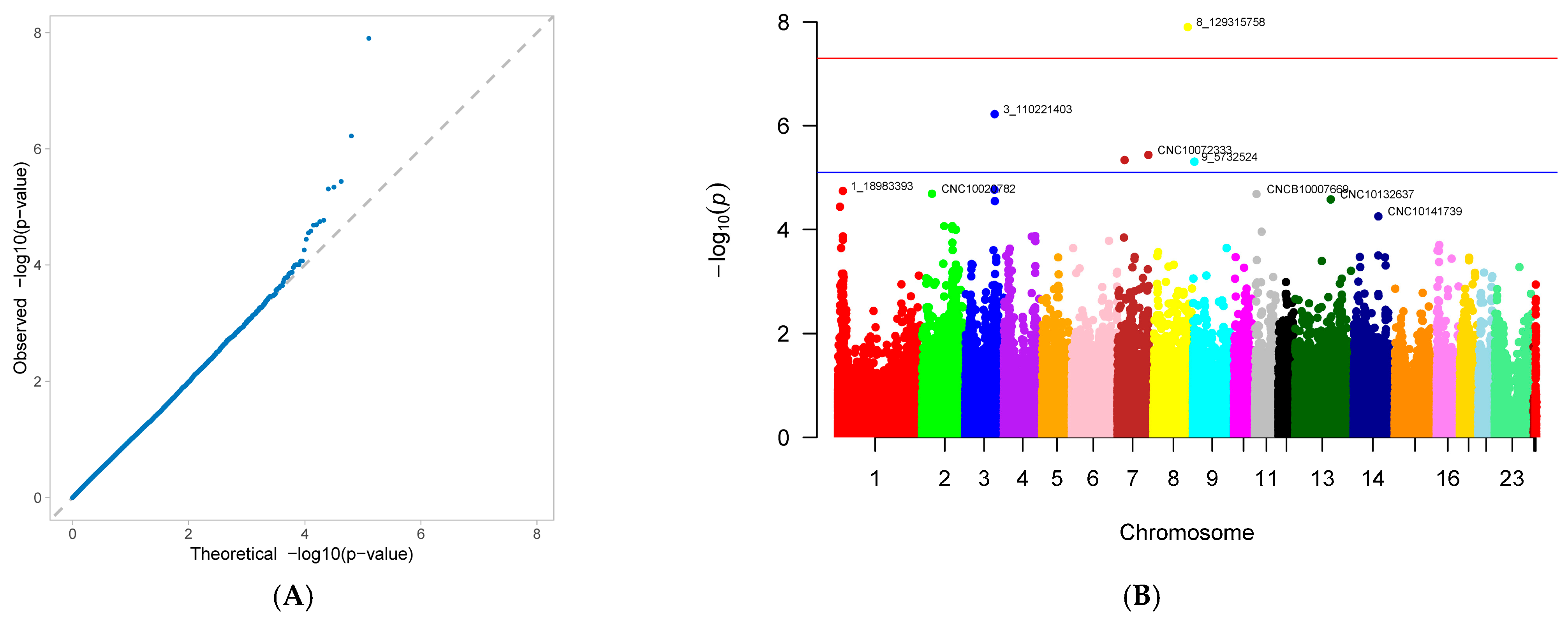
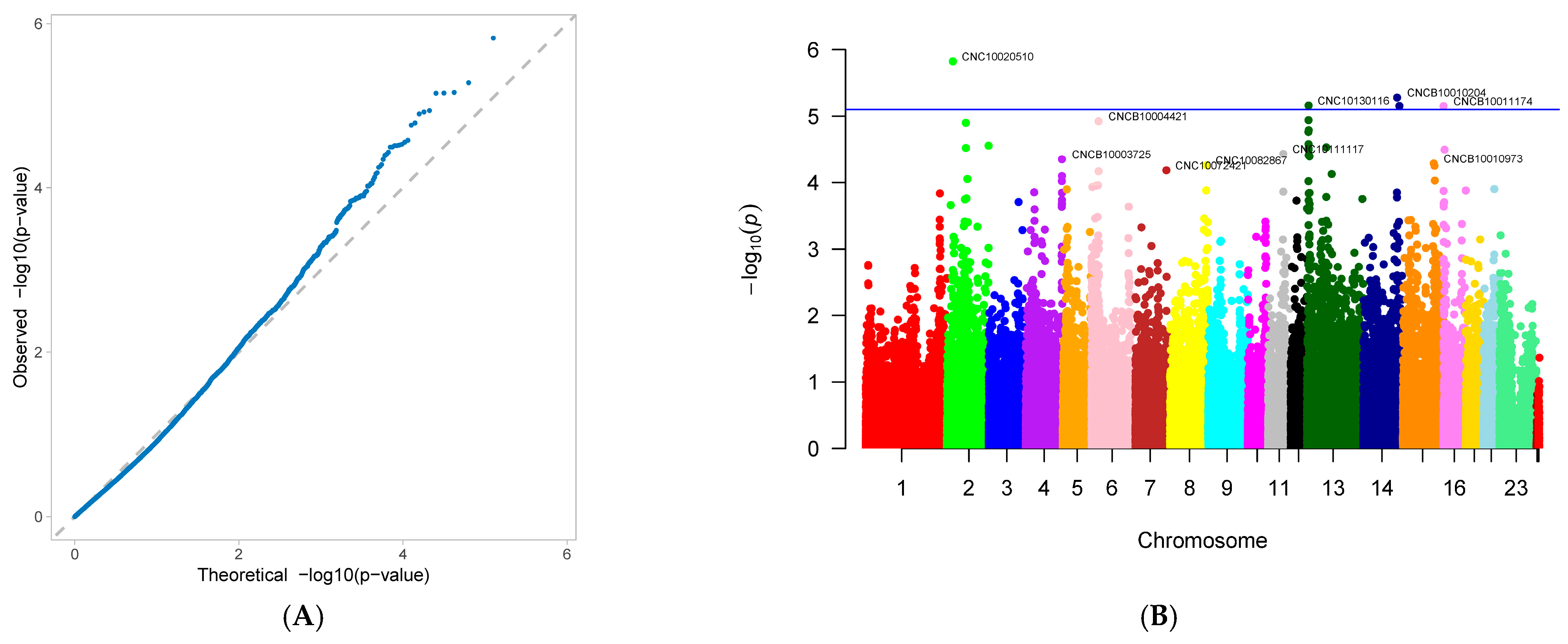
3.2. Genome-Wide Association Study
3.3. Genome-Wide Variance Component Analysis
3.4. Gene Annotation
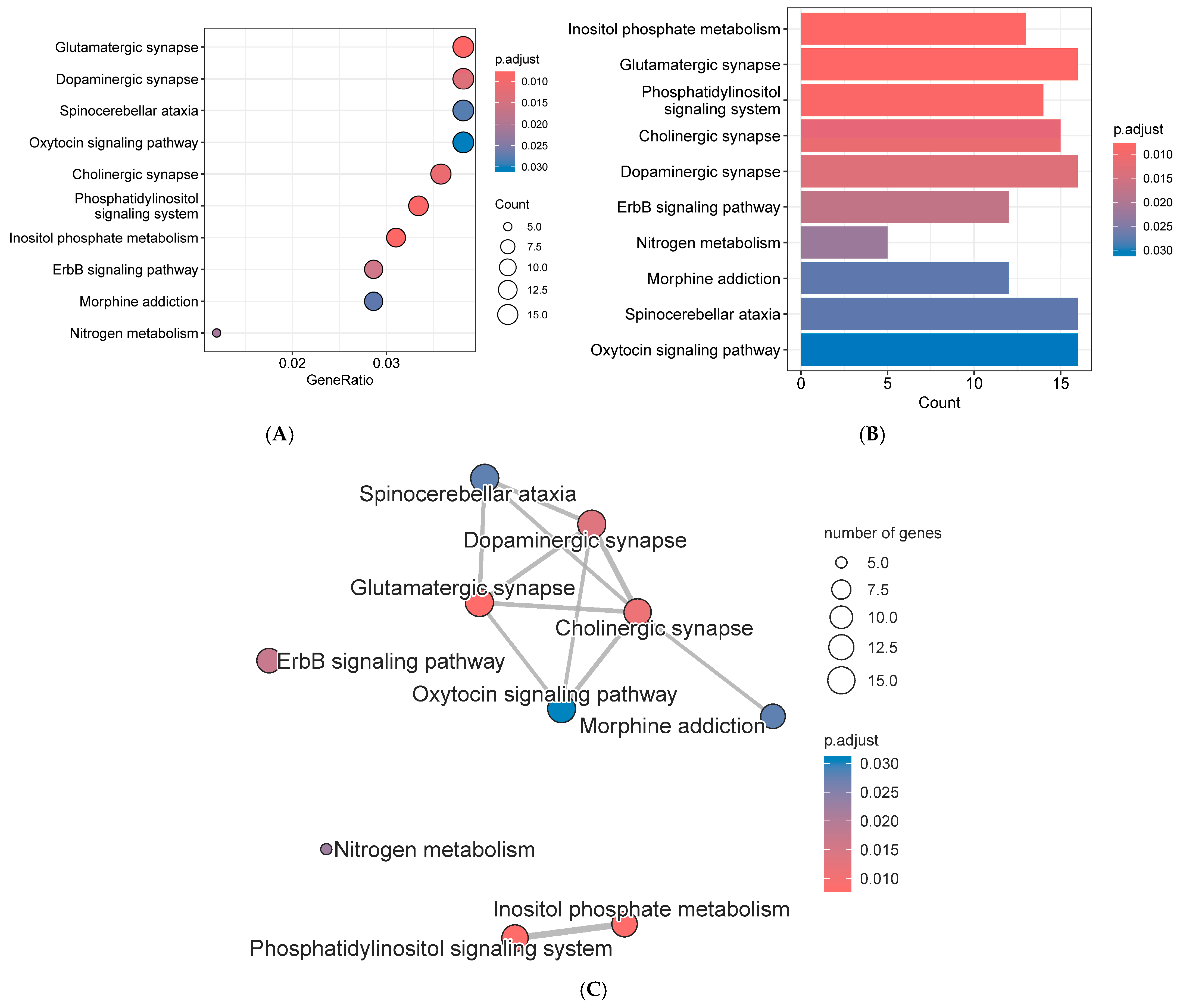
3.5. Go Analysis of Annotated Genes
3.6. Kegg Analysis of Annotated Genes
4. Discussion
4.1. Results of Reproductive Performance Assessment in First-Parity Sows
4.2. Gwas Results for Reproductive Traits in First-Parity Sows
4.3. Genome-Wide Variance Component Analysis
4.4. Go and Kegg Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GWAS | Genome-wide association study |
| SNP | Single-nucleotide polymorphism |
| QTL | Quantitative trait loci |
| AFS | Age at first service |
| AFF | Age at first farrowing |
| cEBV | Comprehensive estimated breeding value |
| GP | Gestation period |
| CNB | Comprehensive number of piglets born |
| TNB | Total number of piglets at birth |
| NBA | Number of piglets born alive at birth |
| NNB | Number of non-viable piglets born |
| NH | Number of healthy offspring |
| NW | Number of weak offspring |
| NS | Number of stillborn piglets |
| NM | Number of mummified piglets |
| ND | Number of deformed piglets |
| mNBA | Number of male piglets born alive at birth |
| fNBA | Number of female piglets born alive at birth |
| LBW | Litter birth weight |
| IBW | Individual birth weight |
| NLT | Number of left teats in piglets |
| NRT | Number of right teats in piglets |
| TNT | Total number of teats in piglets |
| NP | Number of spotted piglets |
| PB | Percentage of pure black piglets |
| WA | Weaning age |
| NW | Number of weaned piglets |
| IWW | Individual weaning weight |
| LWW | Litter weaning weight |
| DGBW | Daily gain from birth to weaning |
| AN | Age at transfer to nursery |
| BWN | Body weight at transfer to nursery |
| BLN | Body length at transfer to nursery |
| CCN | Chest circumference at transfer to nursery |
| ACN | Abdominal circumference at transfer to nursery |
| LCN | Leg circumference at transfer to nursery |
| TNB | Total number of piglets born |
| NBA | Number of born piglets alive |
| BP | Biological process |
| MF | Molecular function |
| CC | Cellular component |
References
- Yuan, H.; Wei, W.; Zhang, Y.; Li, C.; Zhao, S.; Chao, Z.; Xia, C.; Quan, J.; Gao, C. Unveiling the Influence of Copy Number Variations on Genetic Diversity and Adaptive Evolution in China’s Native Pig Breeds via Whole-Genome Resequencing. Int. J. Mol. Sci. 2024, 25, 5843. [Google Scholar] [CrossRef]
- Li, W.; Li, R.; Wei, Y.; Meng, X.; Wang, B.; Zhang, Z.; Wu, W.; Liu, H. Effect of MSTN Mutation on Growth and Carcass Performance in Duroc x Meishan Hybrid Population. Animals 2020, 10, 932. [Google Scholar] [CrossRef]
- Luo, J.; Yang, Y.; Liao, K.; Liu, B.; Chen, Y.; Shen, L.; Chen, L.; Jiang, A.; Liu, Y.; Li, Q.; et al. Genetic parameter estimation for reproductive traits in QingYu pigs and comparison of carcass and meat quality traits to Berkshire × QingYu crossbred pigs. Asian-Australas J. Anim. Sci. 2020, 33, 1224–1232. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhao, A.; Pan, J.; Tan, K.; Zhu, Z.; Zhang, L.; Yu, F.; Liu, R.; Zhong, L.; Huang, J. Statistical Analysis of Reproductive Traits in Jinwu Pig and Identification of Genome-Wide Association Loci. Genes 2025, 16, 550. [Google Scholar] [CrossRef]
- Fu, Y.; Li, L.; Li, B.; Fang, X.; Ren, S. Long form leptin receptor and SNP effect on reproductive traits during embryo attachment in Suzhong sows. Anim. Reprod. Sci. 2016, 168, 57–65. [Google Scholar] [CrossRef]
- Li, X.; Lu, L.; Tong, X.; Li, R.; Jin, E.; Ren, M.; Gao, Y.; Gu, Y.; Li, S. Transcriptomic Profiling of Meat Quality Traits of Skeletal Muscles of the Chinese Indigenous Huai Pig and Duroc Pig. Genes 2023, 14, 1548. [Google Scholar] [CrossRef]
- Chen, B.; Liufu, S.; Wen, S.; Wang, K.; Chen, W.; Xiao, L.; Liu, X.; Yi, L.; Liu, J.; Xu, X.; et al. Identification and Functional Validation of ACSL1 and FABP3 as Muscle-Related Genes Screened by Transcriptomics in Crossbred Duroc × Berkshire × Diannan Small-Eared Pigs. Genes 2025, 16, 520. [Google Scholar] [CrossRef]
- Fu, Y.; Li, L.; Fang, X.; Li, B.; Zhao, W.; Zhou, L.; Ren, S. Investigation of Eph-ephrin A1 in the regulation of embryo implantation in sows. Reprod. Domest. Anim. 2018, 53, 1563–1574. [Google Scholar] [CrossRef]
- Ajay, A.; Chauhan, A.; Vaishnav, S.; Rani, C.; Kumar, B.; De, U.K.; Verma, M.R.; Singh, M.; Gaur, G.K. Impact of body condition on sow and litter performance, postpartum physiological, hematological, and biochemical parameters in Landlly crossbred pigs. Trop. Anim. Health Prod. 2023, 55, 393. [Google Scholar] [CrossRef]
- Cieleń, G.; Derks, M.F.L.; Knol, E.F.; Sell-Kubiak, E. The impact of Box-Cox transformation on phenotypic and genomic characteristics of litter size variability in Landrace pigs. Animal 2023, 17, 100784. [Google Scholar] [CrossRef]
- Martins, T.F.; Braga Magalhães, A.F.; Verardo, L.L.; Santos, G.C.; Silva Fernandes, A.A.; Gomes Vieira, J.I.; Irano, N.; Dos Santos, D.B. Functional analysis of litter size and number of teats in pigs: From GWAS to post-GWAS. Theriogenology 2022, 193, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Ding, R.; Qiu, Y.; Zhuang, Z.; Ruan, D.; Wu, J.; Zhou, S.; Ye, J.; Cao, L.; Hong, L.; Xu, Z.; et al. Genome-wide association studies reveals polygenic genetic architecture of litter traits in Duroc pigs. Theriogenology 2021, 173, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, S.; Yuan, M.; Zhan, F.; Song, M.; Shang, P.; Yang, F.; Li, X.; Qiao, R.; Han, X. Genome-Wide Association Studies and Runs of Homozygosity to Identify Reproduction-Related Genes in Yorkshire Pig Population. Genes 2023, 14, 2133. [Google Scholar] [CrossRef]
- Li, M.; Li, X.; Liu, C.; Han, J.; Han, X.; Wang, K.; Qiao, R.; Yang, F.; Li, X.L.; Li, X.J. Genome-wide association study reveals the candidate genes for reproduction traits in Yunong black pigs. Anim. Genet. 2023, 54, 403–407. [Google Scholar] [CrossRef]
- Turner, S.D. qqman: An R package for visualizing GWAS results using Q-Q and manhattan plots. Biorxiv 2014, 5, 1–2. [Google Scholar]
- Duan, H.; Li, J.; Sun, Y.; Xiong, X.; Sun, L.; Li, W.; Gao, J.; Li, N.; Zhang, J.; Cui, J. Candidate loci for leaf angle in maize revealed by a combination of genome-wide association study and meta-analysis. Front. Genet. 2022, 13, 1004211. [Google Scholar] [CrossRef]
- de Leeuw, C.A.; Stringer, S.; Dekkers, I.A.; Heskes, T.; Posthuma, D. Conditional and interaction gene-set analysis reveals novel functional pathways for blood pressure. Nat. Commun. 2018, 9, 3768. [Google Scholar] [CrossRef]
- Usui, S.; Koketsu, Y. Lifetime reproductive performance and survival of English Berkshire female pigs raised in commercial herds in subtropical Japan. Trop. Anim. Health Prod. 2015, 47, 479–482. [Google Scholar] [CrossRef]
- Lee, S.; Hosseindoust, A.; Choi, Y.; Kim, M.; Kim, K.; Lee, J.; Kim, Y.; Chae, B. Age and weight at first mating affects plasma leptin concentration but no effects on reproductive performance of gilts. J. Anim. Sci. Technol. 2019, 61, 285–293. [Google Scholar] [CrossRef]
- Ju, M.; Wang, X.; Li, X.; Zhang, M.; Shi, L.; Hu, P.; Zhang, B.; Han, X.; Wang, K.; Li, X.; et al. Effects of Litter Size and Parity on Farrowing Duration of Landrace × Yorkshire Sows. Animals 2021, 12, 94. [Google Scholar] [CrossRef]
- Faccin, J.E.G.; Tokach, M.D.; Allerson, M.W.; Woodworth, J.C.; DeRouchey, J.M.; Dritz, S.S.; Bortolozzo, F.P.; Goodband, R.D. Relationship between weaning age and antibiotic usage on pig growth performance and mortality. J. Anim. Sci. 2020, 98, skaa363. [Google Scholar] [CrossRef]
- Archer, C.; Garcia, A.; Henderson, M.; McGlone, J.J. Olfactory enrichment using a maternal pheromone improved post-weaning pig performance and behavior. Front. Vet. Sci. 2022, 9, 965370. [Google Scholar] [CrossRef]
- Wu, P.; Wang, K.; Zhou, J.; Yang, Q.; Yang, X.; Jiang, A.; Jiang, Y.; Li, M.; Zhu, L.; Bai, L.; et al. A genome wide association study for the number of animals born dead in domestic pigs. BMC Genet. 2019, 20, 4. [Google Scholar] [CrossRef]
- Bhatia, V.; Stevens, T.; Derks, M.F.L.; Dunkelberger, J.; Knol, E.F.; Ross, J.W.; Dekkers, J.C.M. Identification of the genetic basis of sow pelvic organ prolapse. Front. Genet. 2023, 14, 1154713. [Google Scholar] [CrossRef]
- Chen, Z.; Ye, S.; Teng, J.; Diao, S.; Yuan, X.; Chen, Z.; Zhang, H.; Li, J.; Zhang, Z. Genome-wide association studies for the number of animals born alive and dead in duroc pigs. Theriogenology 2019, 139, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Corredor, F.A.; Sanglard, L.P.; Ross, J.W.; Keating, A.F.; Leach, R.J.; Serão, N.V.L. Phenotypic and genomic relationships between vulva score categories and reproductive performance in first-parity sows. J. Anim. Sci. Biotechnol. 2021, 12, 7. [Google Scholar] [CrossRef]
- An, M.; Zhou, G.; Li, Y.; Xiang, T.; Ma, Y.; Liu, X.; Li, X.; Zhao, S.; Zhu, M. Characterization of genetic fundamentals for piglet mortality at birth in Yorkshire, Landrace, and Duroc sows. Anim. Genet. 2022, 53, 142–145. [Google Scholar] [CrossRef]
- Bharati, J.; Kumar, S.; Buragohain, B.M.; Das, D.; Devi, S.J.; Mohan, N.H.; Gupta, V.K. Identification of differentially expressed genes and pathways in the post-ovulatory ampulla of cyclic pigs through a transcriptomics approach. Mol. Biol. Rep. 2025, 52, 481. [Google Scholar] [CrossRef]




| Item | AFS | AFF | cEBV | GP | CNB | TNB |
|---|---|---|---|---|---|---|
| Average | 345.36 | 459.17 | 100.43 | 113.8 | 9.34 | 9.79 |
| Standard deviation | 151.34 | 151.45 | 21.31 | 2.05 | 2.14 | 2.24 |
| Item | NBA | NNB | NH | NW | NS | NM |
| Average | 8.9 | 0.77 | 8.43 | 0.48 | 0.41 | 0.22 |
| Standard deviation | 2.26 | 1.19 | 2.08 | 0.93 | 0.85 | 0.62 |
| Item | ND | mNBA | fNBA | LBW | IBW | NLT |
| Average | 0.14 | 4.69 | 4.41 | 10.8 | 1.21 | 7.24 |
| Standard deviation | 0.53 | 1.87 | 1.55 | 3.03 | 0.26 | 0.44 |
| Item | NRT | TNT | NP | PB | WA | NW |
| Average | 7.26 | 14.51 | 3.92 | 76.24 | 28.32 | 8.73 |
| Standard deviation | 0.45 | 0.89 | 2.07 | 25.01 | 1.73 | 2.17 |
| Item | IWW | LWW | DGBW | AN | BWN | BLN |
| Average | 6.55 | 56.34 | 207.19 | 34.86 | 8.61 | 48.32 |
| Standard deviation | 1.21 | 17.39 | 37.68 | 12.97 | 6.05 | 4.84 |
| Item | CCN | ACN | LCN | |||
| Average | 43.33 | 39.62 | 10.78 | |||
| Standard deviation | 3.03 | 3.55 | 2.36 |
| Trait | SNP | chr | pos | maf | Effect | p.Value | expl.var |
|---|---|---|---|---|---|---|---|
| age at first service (AFS) | CNC10022746 | 2 | 132,234,238 | 0.080607 | 92.95919 | 5.17 × 10−8 | 6.088653 |
| CNCB10003227 | 4 | 42,176,304 | 0.492991 | 64.73012 | 6.69 × 10−7 | 9.956982 | |
| CNCB10001752 | 2 | 43,415,752 | 0.063084 | 91.56389 | 9.13 × 10−7 | 4.711177 | |
| CNC10081297 | 8 | 64,117,040 | 0.133178 | 65.51755 | 1.07 × 10−6 | 4.711258 | |
| CNC10230240 | 23 | 7304,150 | 0.432243 | −62.55 | 1.14 × 10−6 | 9.128621 | |
| CNCB10001743 | 2 | 41,306,798 | 0.364486 | −63.3215 | 1.71 × 10−6 | 8.830169 | |
| CNCB10002698 | 3 | 81,907,335 | 0.060748 | 90.11435 | 3.96 × 10−6 | 4.405144 | |
| CNC10140681 | 14 | 34,388,947 | 0.496495 | 56.95693 | 4.18 × 10−6 | 7.710316 | |
| CNC10022920 | 2 | 142,568,860 | 0.140187 | −65.1442 | 4.80 × 10−6 | 4.863218 | |
| CNC10032654 | 3 | 135,578,375 | 0.35514 | −60.7004 | 7.48 × 10−6 | 8.022486 | |
| age at first farrowing (AFF) | CNC10022746 | 2 | 132,234,238 | 0.080607 | 93.22857 | 4.89 × 10−8 | 6.113294 |
| CNCB10003227 | 4 | 42,176,304 | 0.492991 | 64.87131 | 6.49 × 10−7 | 9.982999 | |
| CNC10081297 | 8 | 64,117,040 | 0.133178 | 65.6649 | 1.03 × 10−6 | 4.724206 | |
| CNCB10001752 | 2 | 43,415,752 | 0.063084 | 91.12987 | 1.05 × 10−6 | 4.658467 | |
| CNC10230240 | 23 | 7,304,150 | 0.432243 | −62.7986 | 1.06 × 10−6 | 9.185247 | |
| CNCB10001743 | 2 | 41,306,798 | 0.364486 | −63.0733 | 1.92 × 10−6 | 8.745775 | |
| CNC10140681 | 14 | 34,388,947 | 0.496495 | 57.01878 | 4.17 × 10−6 | 7.713572 | |
| CNCB10002698 | 3 | 81,907,335 | 0.060748 | 89.98522 | 4.18 × 10−6 | 4.384855 | |
| CNC10022920 | 2 | 142,568,860 | 0.140187 | −65.5027 | 4.36 × 10−6 | 4.908299 | |
| number of non-viable piglets born (NNB) | 12_57934316 | 12 | 57,934,316 | 0.425754 | −0.84146 | 3.17 × 10−7 | 23.52132 |
| X_3172285 | 23 | 3,172,285 | 0.441995 | −0.83655 | 8.39 × 10−7 | 23.45181 | |
| 12_56124655 | 12 | 56,124,655 | 0.472158 | 0.748347 | 2.29 × 10−6 | 18.96438 | |
| X_3176453 | 23 | 3,176,453 | 0.439675 | −0.79842 | 2.41 × 10−6 | 21.33899 | |
| 12_56141636 | 12 | 56,141,636 | 0.470998 | 0.744105 | 3.16 × 10−6 | 18.74499 | |
| Y_2815432 | 24 | 2,815,432 | 0.408353 | −0.83261 | 3.64 × 10−6 | 22.75738 | |
| 12_56118044 | 12 | 56,118,044 | 0.472158 | 0.736089 | 4.69 × 10−6 | 18.34819 | |
| number of healthy offspring (NH) | CNC10131876 | 13 | 95,848,356 | 0.456977 | 1.018798 | 7.56 × 10−6 | 12.84277 |
| number of weak offspring (NW) | CNC10021700 | 2 | 79,598,020 | 0.037123 | 0.843657 | 5.93 × 10−7 | 7.213283 |
| CNC10021702 | 2 | 79,632,416 | 0.037123 | 0.843657 | 5.93 × 10−7 | 7.213283 | |
| CNC10021703 | 2 | 79,654,987 | 0.037123 | 0.843657 | 5.93 × 10−7 | 7.213283 | |
| CNC10021544 | 2 | 73,221,843 | 0.035963 | 0.801703 | 2.75 × 10−6 | 6.317745 | |
| CNC10021546 | 2 | 73,295,370 | 0.035963 | 0.801703 | 2.75 × 10−6 | 6.317745 | |
| CNCB10007357 | 10 | 15,723,170 | 0.12181 | 0.434066 | 5.57 × 10−6 | 5.714374 | |
| number of stillborn piglets (NS) | CNC10060156 | 6 | 7,647,082 | 0.091647 | 0.534188 | 1.93 × 10−6 | 5.945918 |
| 4_60598574 | 4 | 60,598,574 | 0.091647 | 0.543139 | 2.68 × 10−6 | 6.146847 | |
| 4_60633363 | 4 | 60,633,363 | 0.091647 | 0.543139 | 2.68 × 10−6 | 6.146847 | |
| 4_60143356 | 4 | 60,143,356 | 0.054524 | 0.636295 | 2.83 × 10−6 | 5.224121 | |
| CNC10060142 | 6 | 7,132,347 | 0.096288 | 0.516031 | 3.71 × 10−6 | 5.79974 | |
| CNC10152171 | 15 | 109,828,092 | 0.063805 | 0.628791 | 7.54 × 10−6 | 5.911394 | |
| CNCB10004308 | 6 | 6,241,186 | 0.430394 | 0.401679 | 7.76 × 10−6 | 9.900437 | |
| CNCB10004309 | 6 | 6,317,896 | 0.430394 | 0.401679 | 7.76 × 10−6 | 9.900437 | |
| number of mummified piglets (NM) | 2_35090057 | 2 | 35,090,057 | 0.022042 | 0.856174 | 6.46 × 10−7 | 7.81605 |
| 2_35184348 | 2 | 35,184,348 | 0.022042 | 0.856174 | 6.46 × 10−7 | 7.81605 | |
| 2_35304516 | 2 | 35,304,516 | 0.022042 | 0.856174 | 6.46 × 10−7 | 7.81605 | |
| 10_1094605 | 10 | 1,094,605 | 0.082367 | 0.493245 | 7.79 × 10−7 | 9.095816 | |
| 5_18526734 | 5 | 18,526,734 | 0.026682 | 0.836982 | 1.23 × 10−6 | 8.999204 | |
| 7_87309677 | 7 | 87,309,677 | 0.075406 | 0.508977 | 1.41 × 10−6 | 8.934071 | |
| 2_32394198 | 2 | 32,394,198 | 0.465197 | 0.402239 | 1.63 × 10−6 | 19.91114 | |
| 11_55264554 | 11 | 55,264,554 | 0.058005 | 0.576168 | 1.73 × 10−6 | 8.972348 | |
| 1_608863 | 26 | 608,863 | 0.056845 | 0.555878 | 2.17 × 10−6 | 8.194593 | |
| 12_55351712 | 12 | 55,351,712 | 0.023202 | 0.80172 | 3.02 × 10−6 | 7.205587 | |
| 12_55045321 | 12 | 55,045,321 | 0.023202 | 0.775936 | 3.86 × 10−6 | 6.749571 | |
| 12_55094012 | 12 | 55,094,012 | 0.023202 | 0.775936 | 3.86 × 10−6 | 6.749571 | |
| 5_18448981 | 5 | 18,448,981 | 0.054524 | 0.544356 | 3.91 × 10−6 | 7.556202 | |
| 7_88172698 | 7 | 88,172,698 | 0.053364 | 0.561383 | 4.93 × 10−6 | 7.874957 | |
| 11_56289609 | 11 | 56,289,609 | 0.038283 | 0.609406 | 5.37 × 10−6 | 6.763383 | |
| 2_34903944 | 2 | 34,903,944 | 0.423434 | −0.44095 | 6.96 × 10−6 | 23.48065 | |
| 5_18115698 | 5 | 18,115,698 | 0.037123 | 0.582897 | 7.68 × 10−6 | 6.007498 | |
| number of deformed piglets (ND) | 8_129315758 | 8 | 129,315,758 | 0.020882 | 0.666854 | 1.25 × 10−8 | 8.789016 |
| 3_110221403 | 3 | 110,221,403 | 0.051044 | 0.394235 | 6.00 × 10−7 | 7.277446 | |
| CNC10072333 | 7 | 116,430,733 | 0.093968 | 0.279291 | 3.65 × 10−6 | 6.419684 | |
| CNC10070593 | 7 | 27,389,510 | 0.055684 | 0.358172 | 4.58 × 10−6 | 6.520967 | |
| 9_5732524 | 9 | 5,732,524 | 0.047564 | 0.370071 | 4.92 × 10−6 | 5.997376 | |
| percentage of pure black piglets (PB) | CNC10020510 | 2 | 23,172,010 | 0.487952 | −11.0069 | 1.5 × 10−6 | 10.35707 |
| CNCB10010204 | 14 | 131,325,841 | 0.46988 | −10.773 | 5.25 × 10−6 | 9.891355 | |
| CNC10130116 | 13 | 6,000,732 | 0.457831 | 9.161748 | 6.9 × 10−6 | 7.128846 | |
| CNCB10010264 | 14 | 140,199,680 | 0.445783 | 10.00824 | 7.02 × 10−6 | 8.467228 | |
| CNCB10011174 | 16 | 233,939 | 0.451807 | −8.96875 | 7.05 × 10−6 | 6.816688 | |
| chest circumference at transfer to nursery (CCN) | CNC10051999 | 5 | 105,361,021 | 0.091667 | −3.20897 | 1.96 × 10−5 | 18.98266 |
| Number | Trait | vc-Heritability | pev-Heritability |
|---|---|---|---|
| 1 | age at first service (AFS) | <0.01 | <0.01 |
| 2 | age at first farrowing (AFF) | <0.01 | <0.01 |
| 3 | comprehensive estimated breeding value (cEBV) | 0.21 ± 0.07 | 0.53 |
| 4 | gestation period (GP) | 0.1 ± 0.07 | 0.49 |
| 5 | comprehensive number of piglets born (CNB) | <0.01 | <0.01 |
| 6 | total number of piglets born (TNB) | <0.01 | <0.01 |
| 7 | number of piglets born alive at birth (NBA) | 0.05 ± 0.06 | 0.48 |
| 8 | number of non-viable piglets born (NNB) | 0.21 ± 0.07 | 0.53 |
| 9 | number of healthy offspring (NH) | <0.01 | <0.01 |
| 10 | number of weak offspring (NW) | 0.06 ± 0.06 | 0.48 |
| 11 | number of stillborn piglets (NS) | 0.01 ± 0.07 | 0.45 |
| 12 | number of mummified piglets (NM) | 0.65 ± 0.1 | 0.7 |
| 13 | number of deformed piglets (ND) | 0.19 ± 0.07 | 0.53 |
| 14 | number of male piglets born alive at birth (mNBA) | <0.01 | <0.01 |
| 15 | number of female piglets born alive at birth (fNBA) | 0.03 ± 0.06 | 0.05 |
| 16 | litter birth weight (LBW) | 0.01 ± 0.09 | 0.4 |
| 17 | individual birth weight (IBW) | 0.01 ± 0.08 | 0.41 |
| 18 | number of left teats in piglets (NLT) | <0.01 | 0.47 |
| 19 | number of right teats in piglets (NRT) | <0.01 | 0.47 |
| 20 | total number of teats in piglets (TNT) | <0.01 | <0.01 |
| 21 | number of spotted piglets (NP) | 0.33 ± 0.12 | 0.27 |
| 22 | percentage of pure black piglets (PB) | <0.01 | <0.01 |
| 23 | weaning age (WA) | <0.01 | <0.01 |
| 24 | number of weaned piglets (NW) | <0.01 | <0.01 |
| 25 | individual weaning weight (IWW) | <0.01 | <0.01 |
| 26 | litter weaning weight (LWW) | <0.01 | 0.03 |
| 27 | daily gain from birth to weaning (DGBW) | 0.7 ± 0.34 | 0.45 |
| 28 | age at transfer to nursery (AN) | 0.12 ± 0.12 | 0.12 |
| 29 | body weight at transfer to nursery (BWN) | <0.01 | 0.03 |
| 30 | body length at transfer to nursery (BLN) | 0.3 ± 0.14 | 0.24 |
| 31 | chest circumference at transfer to nursery (CCN) | <0.01 | <0.01 |
| 32 | abdominal circumference at transfer to nursery (ACN) | 0.59 ± 0.13 | 0.43 |
| 33 | leg circumference at transfer to nursery (LCN) | 0.59 ± 0.13 | 0.43 |
| Trait | SNP | GenBank | SNP_Ensembl |
|---|---|---|---|
| age at first service (AFS) | CNC10022746 | ADAMTS19 | intron_variant |
| CNCB10003227 | VIRMA | intron_variant | |
| CNC10081297 | EPHA5 | intergenic_region | |
| CNCB10001743 | SERGEF | intron_variant | |
| CNC10022920 | PCDHAC2 | intron_variant | |
| age at first farrowing (AFF) | CNC10022746 | ADAMTS19 | intron_variant |
| CNCB10003227 | VIRMA | intron_variant | |
| CNC10081297 | EPHA5 | intergenic_region | |
| CNCB10001743 | SERGEF | intron_variant | |
| CNC10022920 | PCDHAC2 | intron_variant | |
| number of weak offspring (NW) | CNC10021700 | ZNF354B | intron_variant |
| CNC10021702 | PROP1 | upstream_gene_variant | |
| CNC10021544 | CATSPERD | intron_variant | |
| CNC10021546 | HSD11B1L | upstream_gene_variant | |
| number of stillborn piglets (NS) | CNC10060156 | DYNLRB2 | downstream_gene_variant |
| 4_60598574 | HNF4G | intron_variant | |
| 4_60633363 | HNF4G | intron_variant | |
| CNCB10004309 | HSD17B2 | intron_variant | |
| number of mummified piglets (NM) | 2_35090057 | LUZP2 | intron_variant |
| 2_35184348 | LUZP2 | intron_variant | |
| 2_35304516 | LUZP2 | intron_variant | |
| 5_18526734 | AAAS | synonymous_variant | |
| 7_87309677 | SLCO3A1-SV2B | intergenic_region | |
| 2_32394198 | KIF18A-BDNF | intergenic_region | |
| 5_18448981 | RARG | upstream_gene_variant | |
| 5_18115698 | KRT79 | intron_variant | |
| number of deformed piglets (ND) | 3_110221403 | PCARE | downstream_gene_variant |
| CNC10072333 | DICER1 | intron_variant |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, Y.; Li, W.; Dai, C.; Liao, C.; Cheng, J.; Li, H.; Zhao, W. Genome-Wide Association Study of First-Parity Reproductive Traits in Suzi Pig. Genes 2025, 16, 1335. https://doi.org/10.3390/genes16111335
Fu Y, Li W, Dai C, Liao C, Cheng J, Li H, Zhao W. Genome-Wide Association Study of First-Parity Reproductive Traits in Suzi Pig. Genes. 2025; 16(11):1335. https://doi.org/10.3390/genes16111335
Chicago/Turabian StyleFu, Yanfeng, Weining Li, Chaohui Dai, Chao Liao, Jinhua Cheng, Hui Li, and Weimin Zhao. 2025. "Genome-Wide Association Study of First-Parity Reproductive Traits in Suzi Pig" Genes 16, no. 11: 1335. https://doi.org/10.3390/genes16111335
APA StyleFu, Y., Li, W., Dai, C., Liao, C., Cheng, J., Li, H., & Zhao, W. (2025). Genome-Wide Association Study of First-Parity Reproductive Traits in Suzi Pig. Genes, 16(11), 1335. https://doi.org/10.3390/genes16111335






