DNA Barcoding Identification of Angelicae Sinensis Radix and Its Adulterants Based on Internal Transcribed Spacer 2 Region and Secondary Structure Prediction
Abstract
1. Introduction
2. Materials and Methods
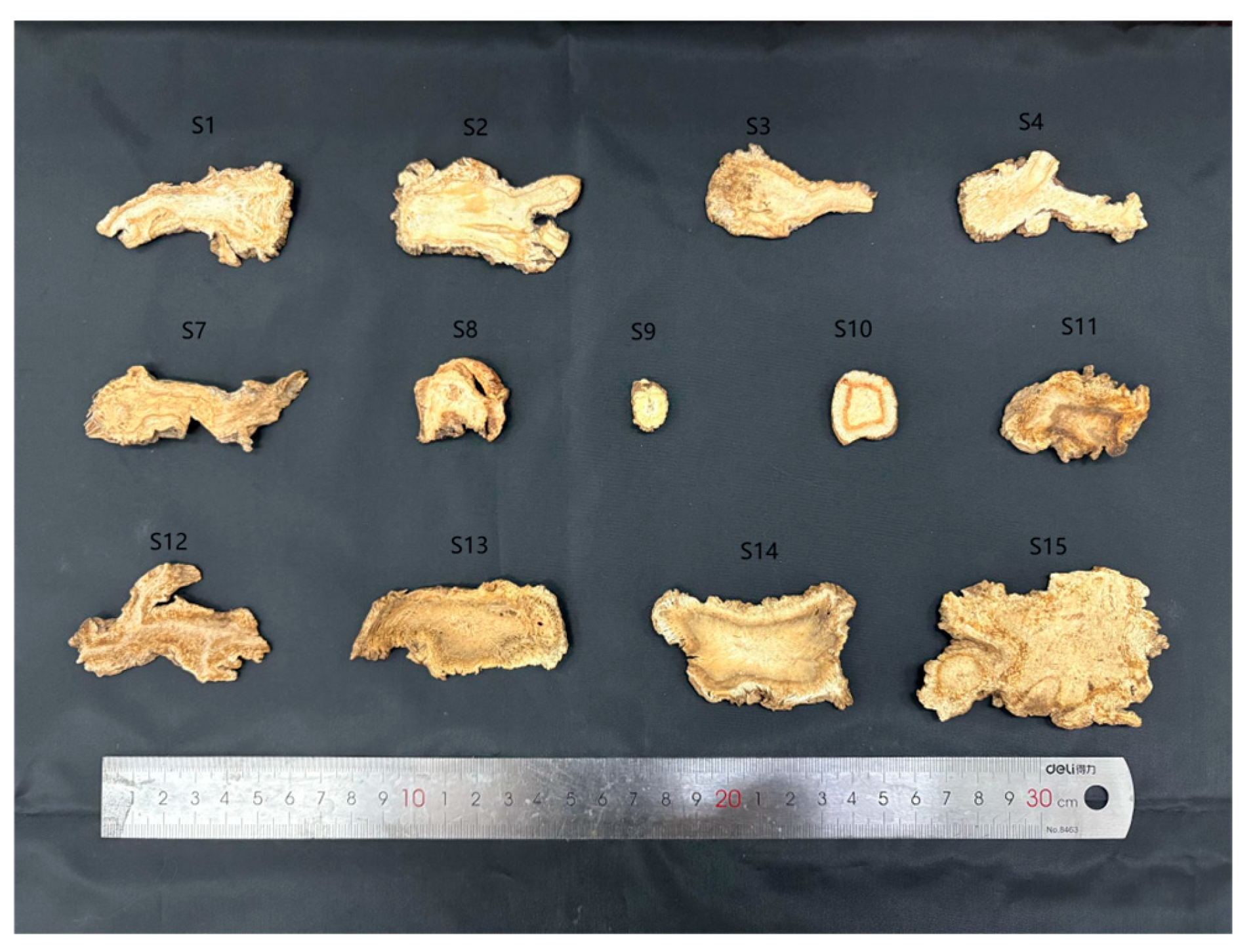
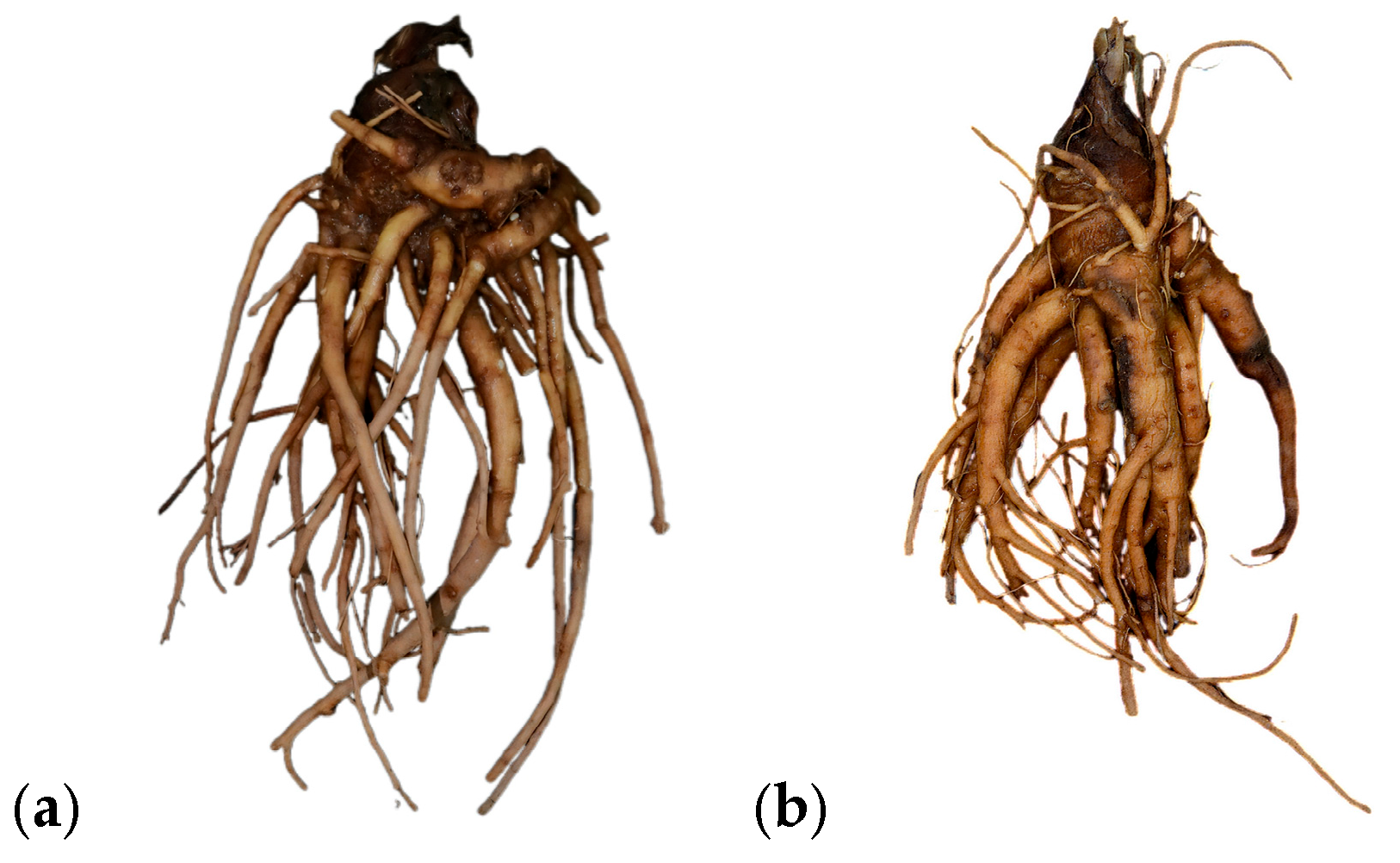
2.1. Materials
2.2. Methods
3. Results
3.1. Total DNA Extraction
3.2. PCR Results and Sequence Characteristics
3.2.1. PCR Results
3.2.2. Sequence Characteristics
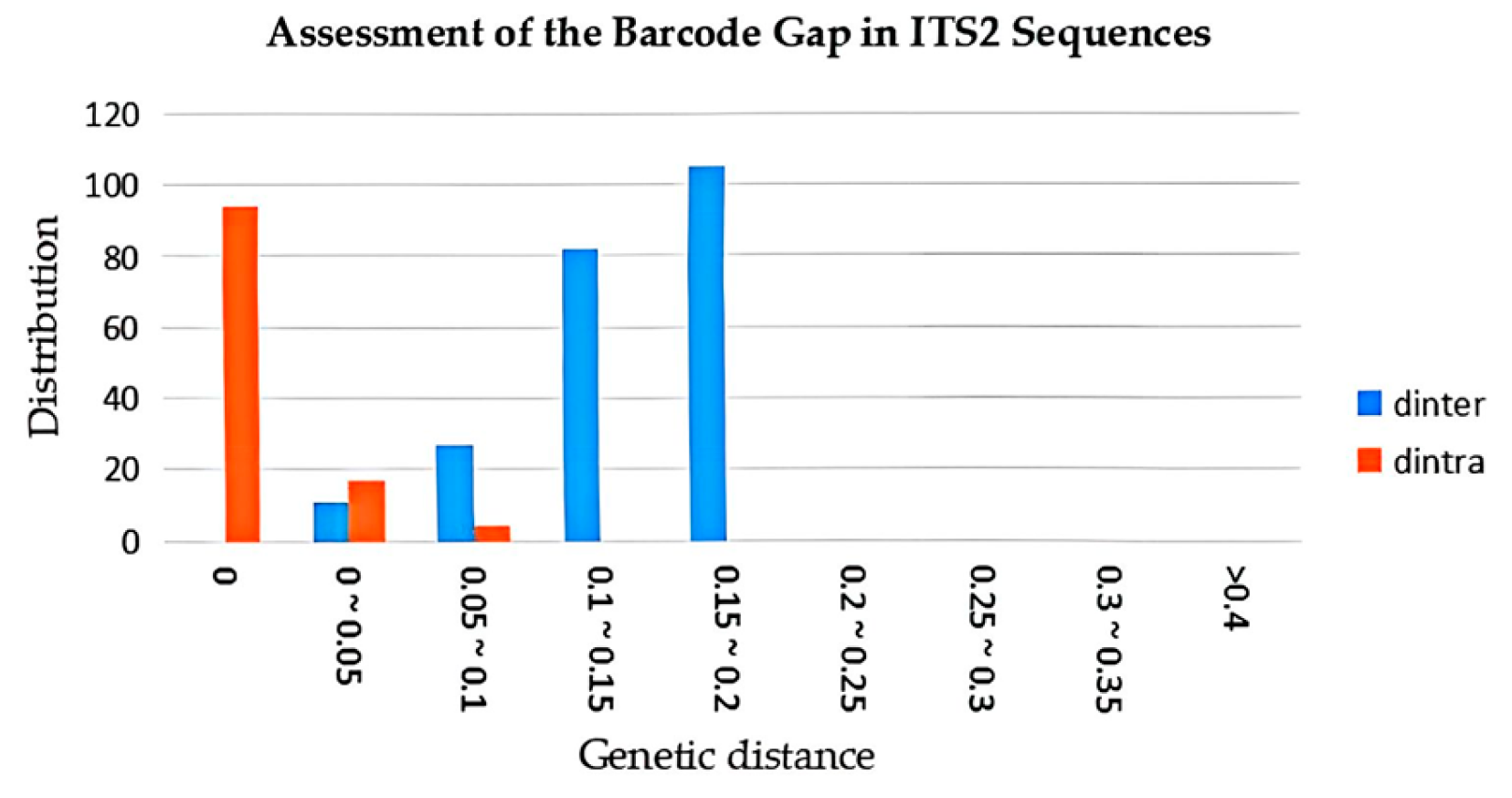
3.3. Intraspecific and Interspecific Variation Comparison
3.4. ITS2 Secondary Structure Prediction and Phylogenetic Tree Construction
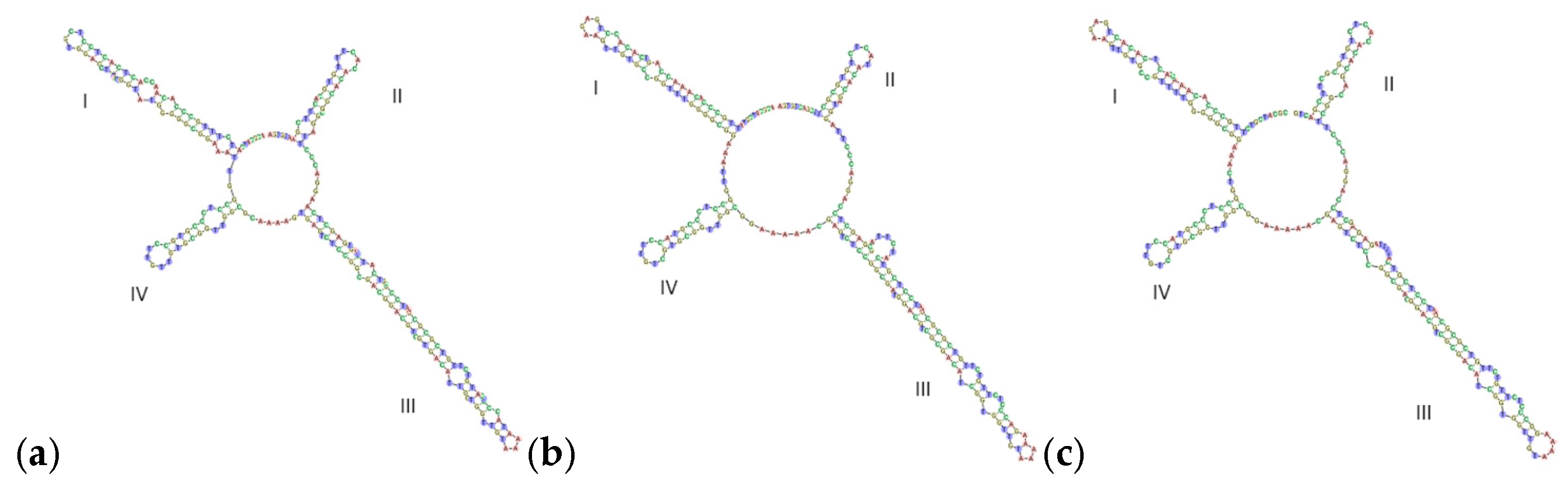
3.4.1. ITS2 Secondary Structure Prediction
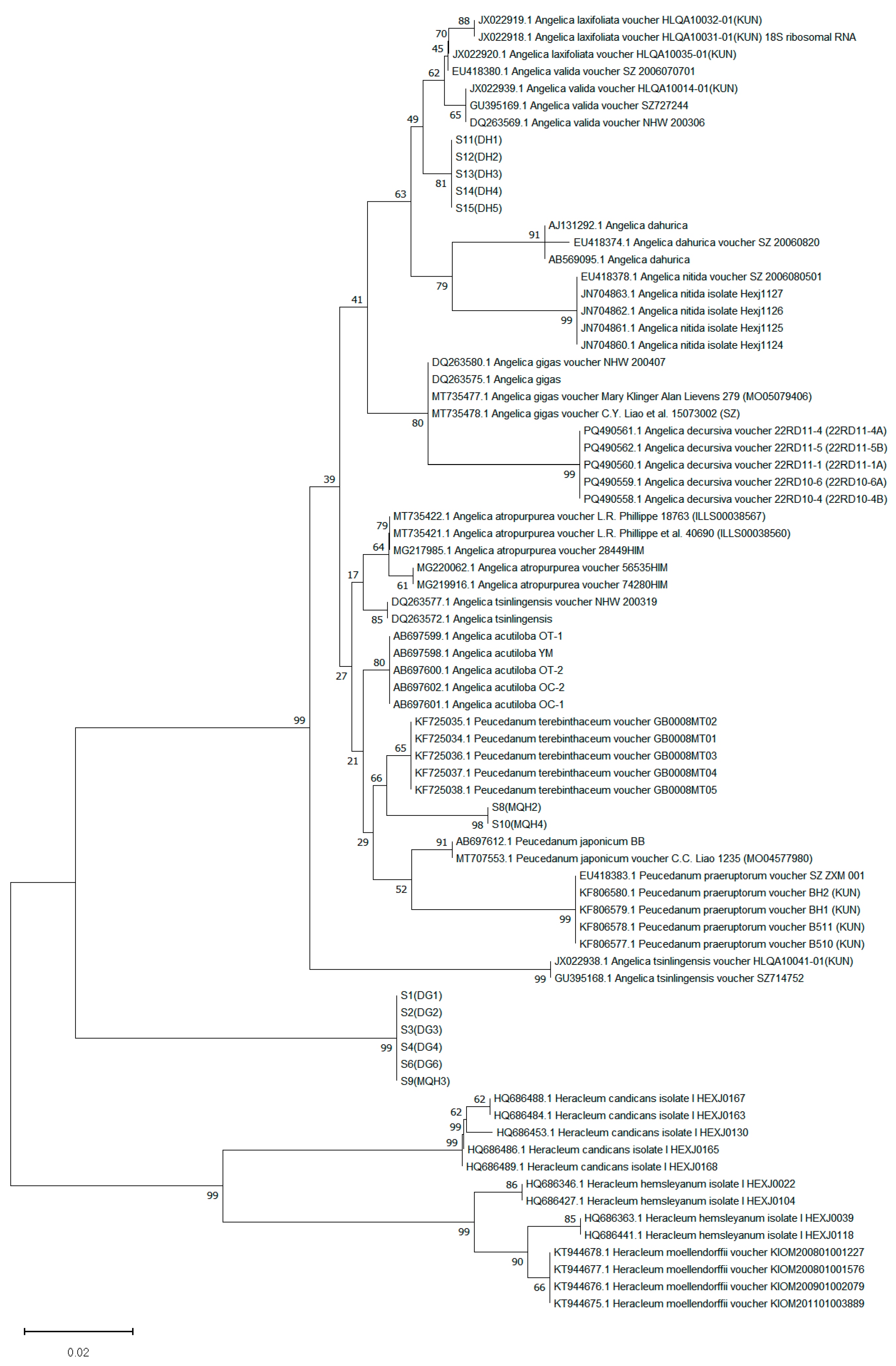
3.4.2. Phylogenetic Tree Construction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ITS2 | Internal Transcribed Spacer 2 |
| DNA | Deoxyribo Nucleic Acid |
| RNA | Ribonucleic Acid |
| PCR | Polymerase Chain Reaction |
| dintra | Intraspecific Distance |
| dinter | Interspecific Distance |
| K2P | Kimura 2-Parameter |
| NJ | Neighbor-Joining |
| NCBI | National Center for Biotechnology Information |
| CTAB | Cetyltrime Thylammonium Bromide |
| IUB | International Union of Biochemistry |
References
- Zhou, L.; Zeng, J. Research Advances on Chemical Constituents and Pharmacological Effects of Angelica pubescens. Mod. Chin. Med. 2019, 21, 1739–1748. [Google Scholar]
- Ni, L.; Yao, Y.; Zhang, X.; Sun, L.; Liu, J.; Kuang, X.; Zhao, J.; Gan, X.; Yi, J.; Zhou, L. Classification, Identification and Research Progress of Commonly Used Varieties of Qianhu (Peucedanum). Guid. J. Tradit. Chin. Med. Pharm. 2024, 30, 77–82. [Google Scholar]
- Zheng, S.; Zhao, S.; Zeng, Y.; Liu, M.; Shang, X.; Wang, J. Research Status and Thinking on Identification of Variety and Origin of Chinese Medicinal Materials. Mod. Chin. Med. 2021, 23, 2037–2045. [Google Scholar]
- Cheng, L.; Lin, Z.; Xin, C.; Sun, H.; Li, X. Molecular Identification and Phylogenetic Analysis of Papaver Based on ITS2 Barcoding. J. Forensic Sci. 2022, 67, 712–719. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, S.; Wu, W.; Xie, J.; Cheng, X.; Ye, Z.; Yin, X.; Liu, Y.; Huang, Z. Molecular Identification and Phylogenetic Analysis of the Traditional Chinese Medicinal Plant Kochia Scoparia Using ITS2 Barcoding. Interdiscip. Sci. Comput. Life Sci. 2021, 13, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-W.; Zhou, J. DNA Barcoding of Notopterygii Rhizoma et Radix (Qiang-Huo) and Identification of Adulteration in Its Medicinal Services. Sci. Rep. 2024, 14, 2879. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Xie, Z.; Wu, J.; Tao, J.; Xu, X. DNA Barcoding Identification of Kadsurae Caulis and Spatholobi Caulis Based on Internal Transcribed Spacer 2 Region and Secondary Structure Prediction. Pharmacogn. Mag. 2016, 12, S165–S169. [Google Scholar] [CrossRef] [PubMed]
- Pere, K.; Mburu, K.; Muge, E.K.; Wagacha, J.M.; Nyaboga, E.N. Molecular Identification, Genetic Diversity, and Secondary Structure Predictions of Physalis Species Using ITS2 DNA Barcoding. BMC Plant Biol. 2024, 24, 1178. [Google Scholar] [CrossRef] [PubMed]
- Devi, M.P.; Dasgupta, M.; Mohanty, S.; Sharma, S.K.; Hegde, V.; Roy, S.S.; Renadevan, R.; Kumar, K.B.; Patel, H.K.; Sahoo, M.R. DNA Barcoding and ITS2 Secondary Structure Predictions in Taro (Colocasia esculenta L. Schott) from the North Eastern Hill Region of India. Genes 2022, 13, 2294. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhao, W.; Zhao, Y.; Li, M. Differences in Morphological Characteristics, Photosynthetic Capacity and Chloroplast Genomes Reveal Molecular Markers to Distinguish Angelica sinensis, A. acutiloba and A. gigas. BMC Plant Biol. 2025, 25, 961. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, K.; Shindo, S.; Watanabe, H.; Ikegami, F. Identification of Angelica acutiloba and Related Species by Analysis of Inter- and Intra-Specific Sequence Variations in Chloroplast and Nuclear DNA Sequences. Am. J. Plant Sci. 2012, 3, 1260–1265. [Google Scholar] [CrossRef]
- Liu, X.; Ou, Q.; Shi, Y.; Xin, T.; Song, J.; Luo, J.; Cui, W. Identification of Angelica sinensis and its adulterants based on ITS2 sequence. Chin. Herb. Med. 2018, 49, 4877–4883. [Google Scholar]
- Schultz, J.; Müller, T.; Achtziger, M.; Seibel, P.N.; Dandekar, T.; Wolf, M. The Internal Transcribed Spacer 2 Database–A Web Server for (Not Only) Low Level Phylogenetic Analyses. Nucleic Acids Res. 2006, 34, W704–W707. [Google Scholar] [CrossRef] [PubMed]
- Selig, C.; Wolf, M.; Müller, T.; Dandekar, T.; Schultz, J. The ITS2 Database II: Homology Modelling RNA Structure for Molecular Systematics. Nucleic Acids Res. 2008, 36, D377–D380. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Molecular Biology and Evolution. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
| Sample | Concentration (ng/μL) | A260/A280 |
|---|---|---|
| S1 | 482.8 | 2.07 |
| S2 | 651.5 | 1.93 |
| S3 | 480.0 | 2.07 |
| S4 | 615.7 | 1.96 |
| S5 | 33.0 | 1.97 |
| S6 | 541.1 | 1.98 |
| S7 | 19.2 | 1.78 |
| S8 | 96.3 | 2.09 |
| S9 | 97.8 | 1.97 |
| S10 | 149.2 | 2.13 |
| S11 | 260.2 | 2.13 |
| S12 | 288.1 | 2.15 |
| S13 | 62.3 | 2.12 |
| S14 | 160.6 | 2.01 |
| S15 | 249.4 | 2.16 |
| Samples | Length (bp) | GC Content (%) | Variable Sites |
|---|---|---|---|
| 1 1 1 1 1 2 2 3 3 3 3 3 3 3 3 3 4 4 4 4 4 4 4 4 4 5 5 5 5 5 5 6 6 6 6 6 6 7 9 2 3 6 7 8 4 9 1 2 3 4 5 6 7 8 9 0 1 2 3 4 5 6 7 8 0 1 5 7 8 9 1 2 3 4 5 6 | |||
| S1, S2, S3, S4, S6 | 229 | 54.15 | ACTGCACCTCTCGTGGAGCTGTACTGGATGCGGAATTGGC |
| S8, S10 | 227 | 55.51 | GCGCACACATGAGAAGTTGTGCCGTTTGGCGAACTGGCCT |
| S11, S12, S13, S14, S15 | 227 | 55.51 | GAGCACACACTGAGAAGTTGTGCCGGTTGCGAATTGGCC |
| S9 | 227 | 54.62 | ACTGCACCTCTCGTGGAGCTGTACTGGATGCGGAATTGGC |
| 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 6 6 7 7 7 7 7 7 7 7 8 8 8 8 8 8 8 9 9 9 9 9 9 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 2 2 2 2 2 2 2 2 3 3 3 3 3 3 3 3 4 4 4 4 4 4 4 5 5 5 5 5 5 5 5 5 5 6 6 6 7 9 1 2 3 4 5 6 7 8 0 1 4 6 7 8 9 0 1 3 4 5 1 3 4 7 8 9 0 2 3 4 5 6 7 8 9 0 1 2 4 5 7 8 9 0 1 3 4 5 6 8 2 3 4 5 7 8 9 0 1 2 3 4 5 6 7 8 9 0 1 4 TCCGTGCCTGTTCGGTTGCGCATAGTCCGCGACGGACGTTGCATTGTGGTGTAATACCTCATGTCTTGTCGCGC CGTACCTTGCGCGTTGGCGAACATCCGGCACGGACGTCGACTCGGTGTTGAAGGCCTCTTGTCTTGTCGCGCG TCGTACCTTGCGCGTTGGCGAACATCCGGCATGGACGTCGACTCGGTGTTGAAAGACCTCTTGTCTTGTCGCGC TCCGTGCCTGTTCGGTTGCGCATAGTCCGCGACGGACGTTGCATTGTGGTGTAATACCTCATGTCTTGTCGCGCG 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 6 6 6 6 6 7 7 7 7 7 7 7 7 7 7 8 8 8 8 9 9 9 9 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 2 2 2 2 2 2 2 2 2 2 5 6 7 8 9 0 1 2 3 4 5 6 7 8 9 0 1 2 4 6 7 8 9 0 2 3 4 5 7 8 9 0 1 2 3 4 5 6 7 8 9 0 1 2 3 4 5 6 7 8 9 GAATCCGCGTCATCTTAGTGAGCTCAAGGACCTTAGGCGGCACACACTTTGTGCACTTTCAATG AATCCTCGTCATTTTAGAGAGCTCCAGGACCTTCGGCAGCACACACTCTGTGCGCTTCGACTG GAATCCTCGTCATCTTAGCGAGCTCCAGGACCTTAGGTAGCACATACTCTGTGCGCTTCGACTG AATCCGCGTCATCTTAGTGAGCTCAAGGACCTTAGGCGGCACACACTTTGTGCACTTCGAATG | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.; Zeng, Q.; Gong, M.; Wu, H.; Chen, W.; Xu, X. DNA Barcoding Identification of Angelicae Sinensis Radix and Its Adulterants Based on Internal Transcribed Spacer 2 Region and Secondary Structure Prediction. Genes 2025, 16, 1333. https://doi.org/10.3390/genes16111333
Chen Z, Zeng Q, Gong M, Wu H, Chen W, Xu X. DNA Barcoding Identification of Angelicae Sinensis Radix and Its Adulterants Based on Internal Transcribed Spacer 2 Region and Secondary Structure Prediction. Genes. 2025; 16(11):1333. https://doi.org/10.3390/genes16111333
Chicago/Turabian StyleChen, Zifeng, Qiman Zeng, Meihui Gong, Huimin Wu, Wenli Chen, and Xinjun Xu. 2025. "DNA Barcoding Identification of Angelicae Sinensis Radix and Its Adulterants Based on Internal Transcribed Spacer 2 Region and Secondary Structure Prediction" Genes 16, no. 11: 1333. https://doi.org/10.3390/genes16111333
APA StyleChen, Z., Zeng, Q., Gong, M., Wu, H., Chen, W., & Xu, X. (2025). DNA Barcoding Identification of Angelicae Sinensis Radix and Its Adulterants Based on Internal Transcribed Spacer 2 Region and Secondary Structure Prediction. Genes, 16(11), 1333. https://doi.org/10.3390/genes16111333





