Characterization of LTR Retrotransposon Reverse Transcriptase in Tamarix chinensis L. and Activity Analysis Under Salt and Alkali Stresses
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Salt/Alkali Stress Treatments
2.2. Amplification and Sequencing of LTR Retrotransposons in T. chinensis
2.3. Classification and Activity Screening of T. chinensis LTR Retrotransposons
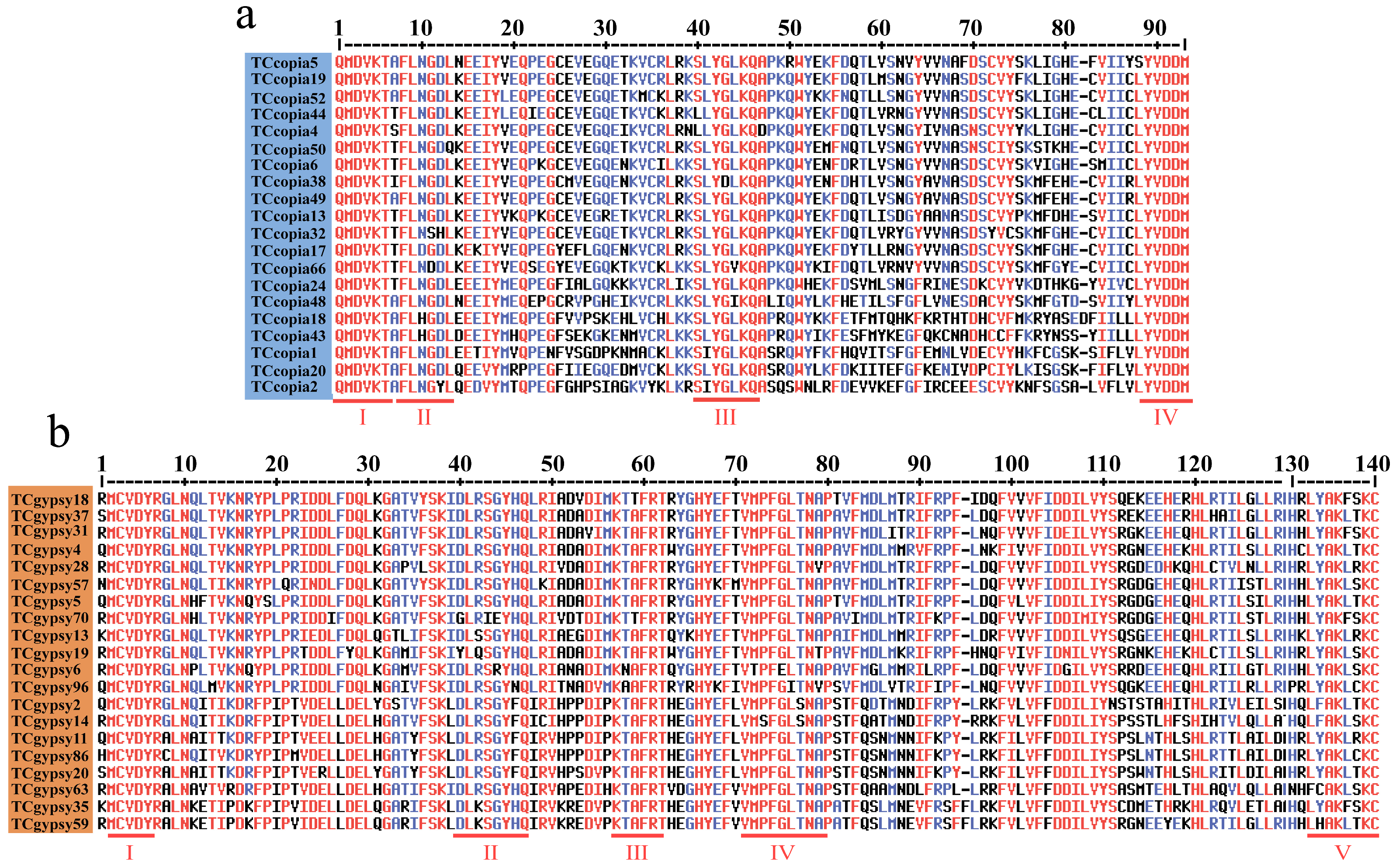
2.4. Sequence Characterization of Ty1-copia and Ty3-gypsy RT Domains in T. chinensis
2.5. Phylogenetic Analysis of Ty1-copia and Ty3-gypsy in T. chinensis
2.6. Analysis of Ty1-copia and Ty3-gypsy Dominant Abundance Under Salt and Alkali Stresses
3. Results
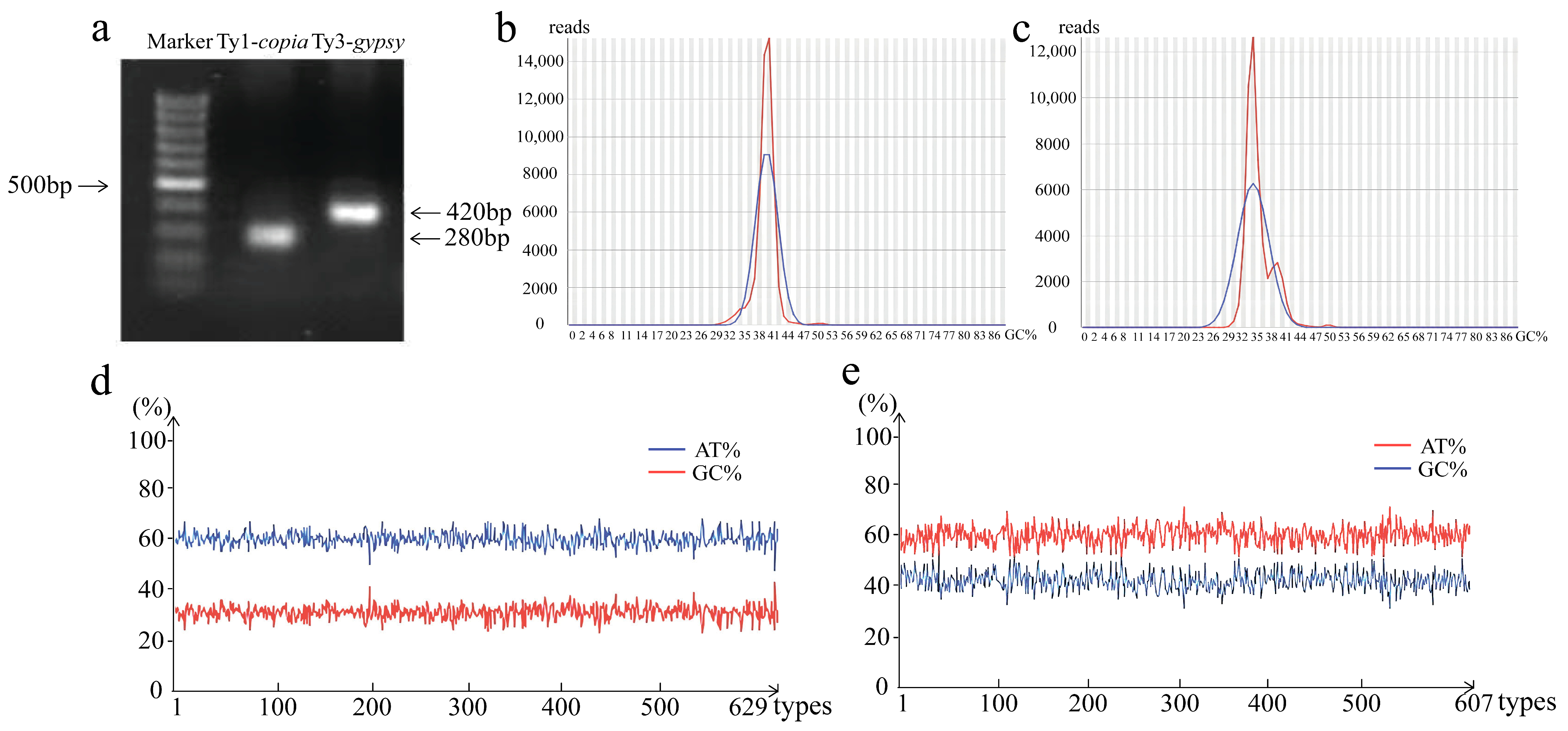
3.1. Clustering and Characterization of LTR Retrotransposon RT Domains in T. chinensis
3.2. Activity and Conserved Domain Analysis of LTR Retrotransposons
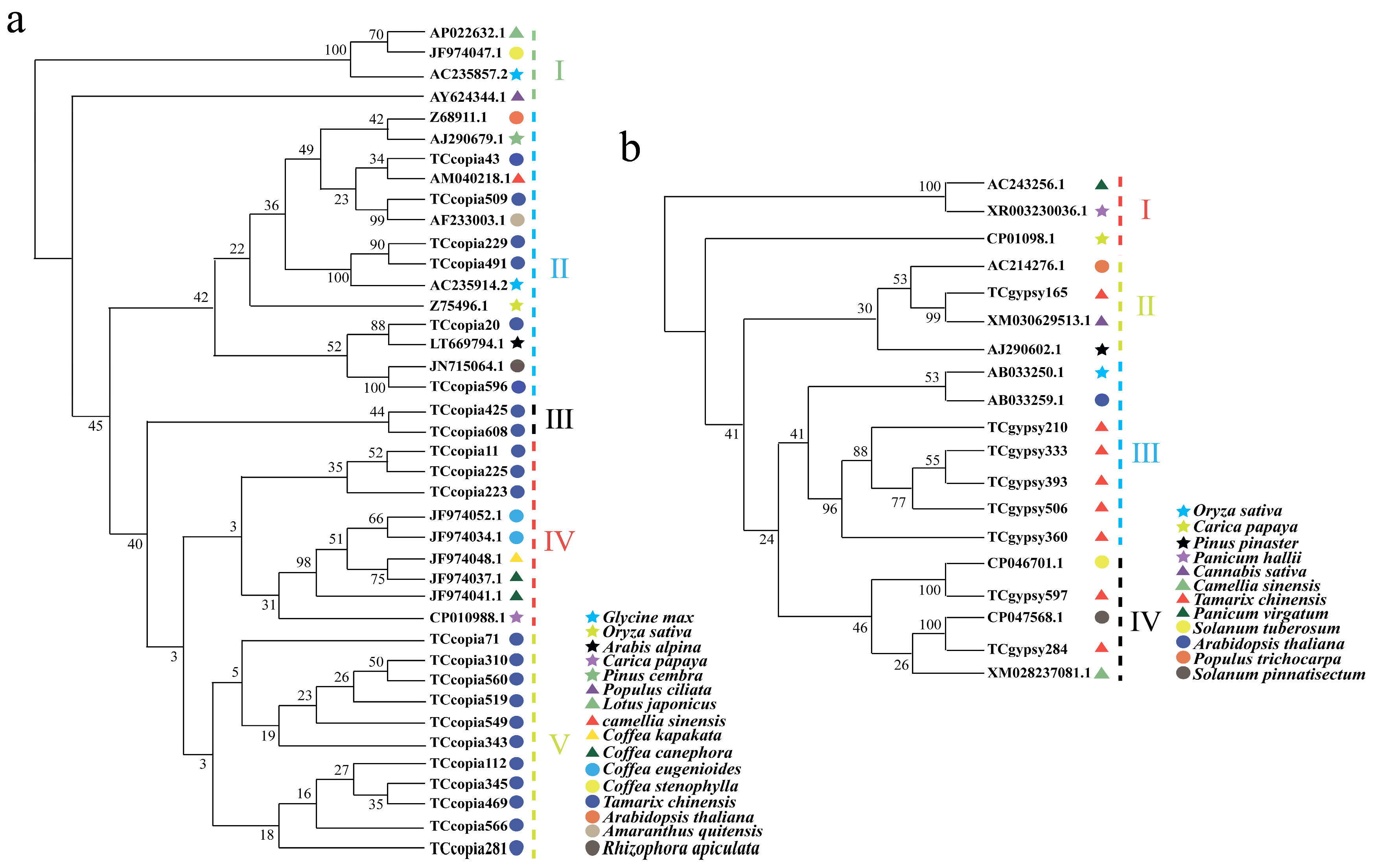
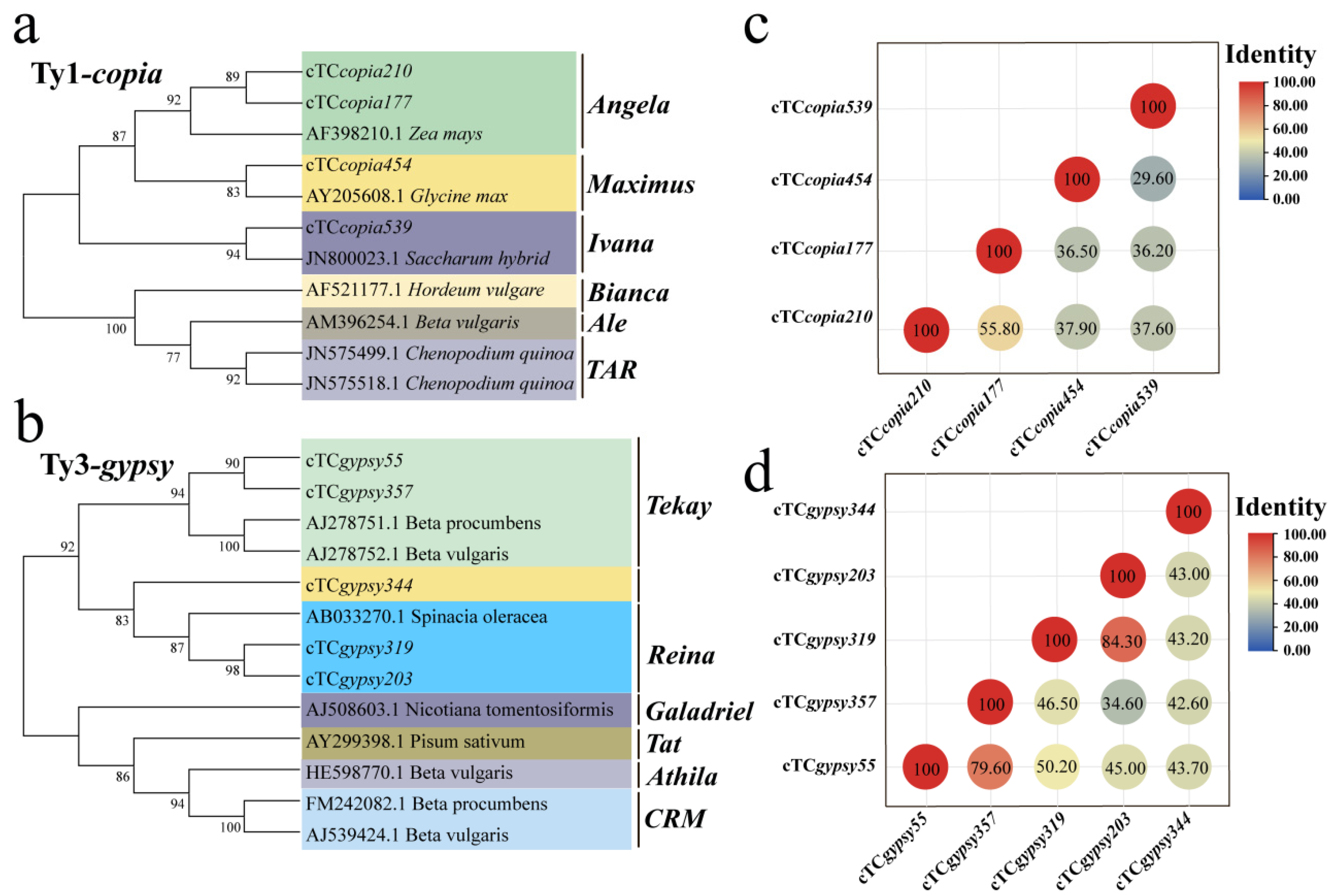
3.3. Phylogenetic Analysis of LTR Retrotransposons Across Species
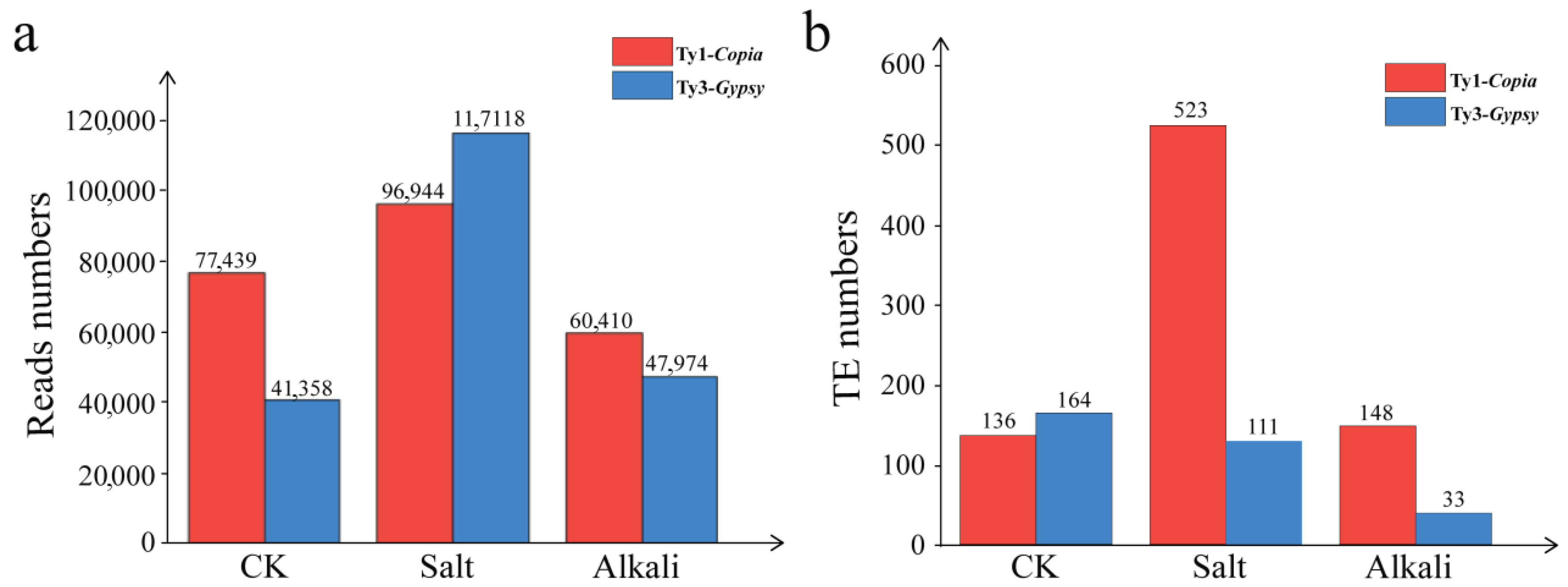
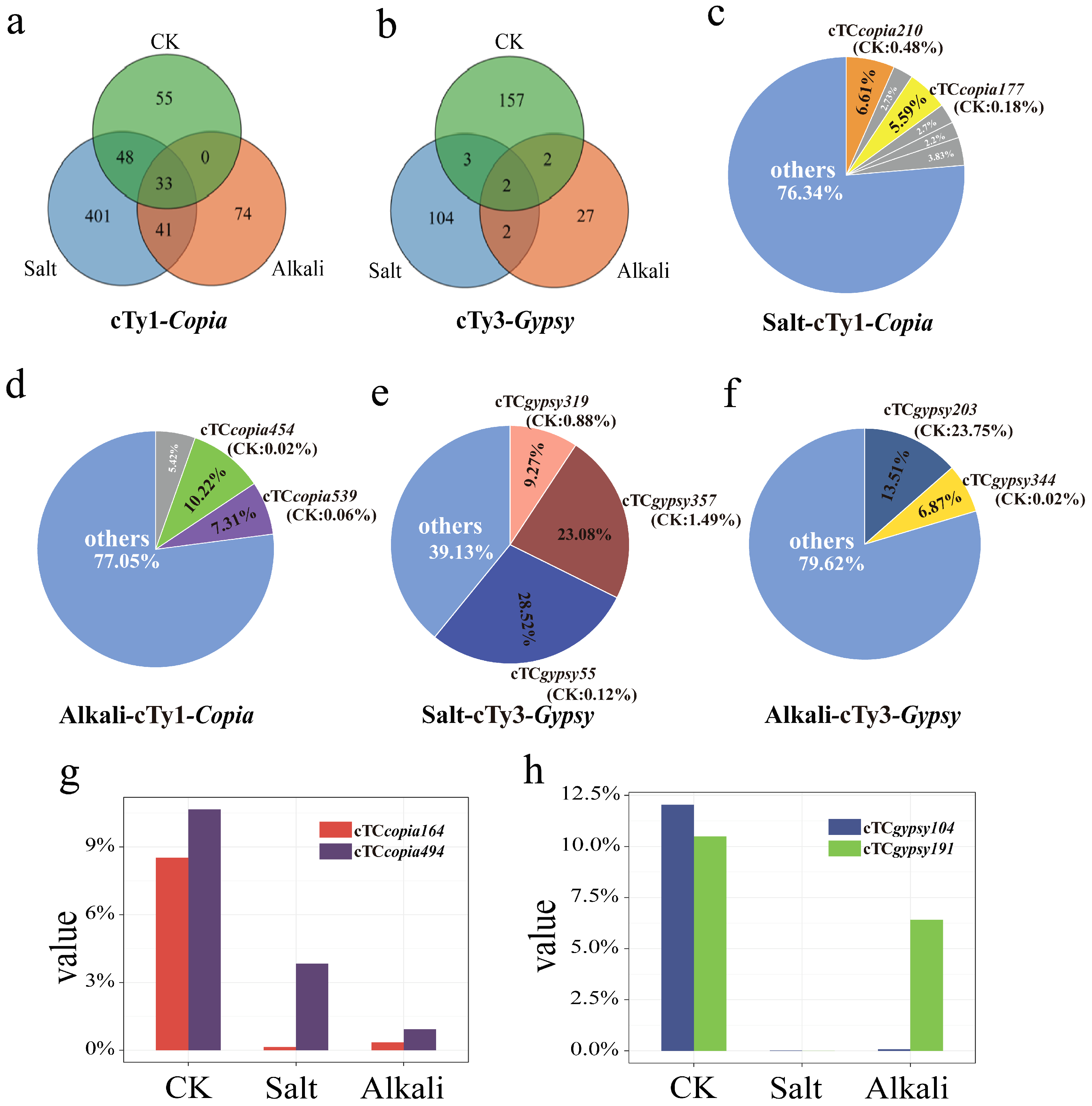
3.4. Sequencing and Clustering Analysis of LTR Retrotransposons in T. chinensis Under Salt and Alkali Stresses
3.5. Expression Specificity of LTR Retrotransposons in T. chinensis Under Salt and Alkali Stresses
3.6. Phylogenetic Analysis of LTR Retrotransposons in T. chinensis Under Salt and Alkali Stresses
4. Discussion
4.1. Sequence Characteristics of LTR Retrotransposons in the T. chinensis Genome
4.2. Transpositional Activity and Phylogenetic Relationships of Ty1-copia and Ty3-gypsy in T. chinensis
4.3. LTR Retrotransposon Activity and Dominance in T. chinensis Under Salt and Alkali Stresses
4.4. Characterization Analysis of LTR Retrotransposons Under Salt and Alkali Stresses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nascimento, J.; Sader, M.; Ribeiro, T.; Pedrosa-Harand, A. Influence of Ty3/gypsy and Ty1/copia LTR-retrotransposons on the large genomes of Alstroemeriaceae: Genome landscape of Bomarea edulis (Tussac) Herb. Protoplasma 2025, 262, 881–894. [Google Scholar] [CrossRef] [PubMed]
- Chu, J.; Newman, J.; Cho, J. Molecular Mimicry of Transposable Elements in Plants. Plant Cell Physiol. 2025, 66, 490–495. [Google Scholar] [CrossRef]
- Xiao, Y.; Wang, J. Understanding the Regulation Activities of Transposons in Driving the Variation and Evolution of Polyploid Plant Genome. Plants 2025, 14, 1160. [Google Scholar] [CrossRef]
- Gao, D.; Li, Y.; Kim, K.D.; Abernathy, B.; Jackson, S.A. Landscape and evolutionary dynamics of terminal repeat retrotransposons in miniature in plant genomes. Genome Biol. 2016, 17, 7. [Google Scholar] [CrossRef] [PubMed]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef]
- Xiong, Y.; Eickbush, T.H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990, 9, 3353–3362. [Google Scholar] [CrossRef]
- Peterson-Burch, B.D.; Voytas, D.F. Genes of the Pseudoviridae (Ty1/copia retrotransposons). Mol. Biol. Evol. 2002, 19, 1832–1845. [Google Scholar] [CrossRef]
- Huang, C.R.; Burns, K.H.; Boeke, J.D. Active transposition in genomes. Annu. Rev. Genet. 2012, 46, 651–675. [Google Scholar] [CrossRef]
- Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef]
- Paterson, A.H.; Bowers, J.E.; Bruggmann, R.; Dubchak, I.; Grimwood, J.; Gundlach, H.; Haberer, G.; Hellsten, U.; Mitros, T.; Poliakov, A.; et al. The Sorghum bicolor genome and the diversification of grasses. Nature 2009, 457, 551–556. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Zhang, Z.; Sun, H.; Li, H.; Dai, H. Isolation of Ty1-copia-like retrotransposon sequences from the apple genome by chromosome walking based on modified SiteFinding-polymerase chain reaction. Acta Biochim. Biophys. Sin. 2007, 39, 675–683. [Google Scholar] [CrossRef]
- Rajput, M.K.; Upadhyaya, K.C. Characterization of heterogeneity in Ty1-copia GROUP retrotransposons in chickpea (Cicer arietinum L.). Mol. Biol. (Mosk) 2010, 44, 601–607. [Google Scholar] [CrossRef]
- Della Valle, F.; Reddy, P.; Aguirre Vazquez, A.; Izpisua Belmonte, J.C. Reactivation of retrotransposable elements is associated with environmental stress and ageing. Nat. Rev. Genet. 2025, 26, 547–558. [Google Scholar] [CrossRef]
- Nguyen, V.H.; Mittelsten Scheid, O.; Gutzat, R. Heat stress response and transposon control in plant shoot stem cells. Plant Physiol. 2025, 197, kiaf110. [Google Scholar] [CrossRef]
- Zedek, F.; Smerda, J.; Smarda, P.; Bureš, P. Correlated evolution of LTR retrotransposons and genome size in the genus Eleocharis. BMC Plant Biol. 2010, 10, 265. [Google Scholar] [CrossRef] [PubMed]
- Qiu, F.; Ungerer, M.C. Genomic abundance and transcriptional activity of diverse gypsy and copia long terminal repeat retrotransposons in three wild sunflower species. BMC Plant Biol. 2018, 18, 6. [Google Scholar] [CrossRef] [PubMed]
- Butelli, E.; Licciardello, C.; Zhang, Y.; Liu, J.; Mackay, S.; Bailey, P.; Reforgiato-Recupero, G.; Martin, C. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 2012, 24, 1242–1255. [Google Scholar] [CrossRef]
- Kobayashi, S.; Goto-Yamamoto, N.; Hirochika, H. Retrotransposon-induced mutations in grape skin color. Science 2004, 304, 982. [Google Scholar] [CrossRef] [PubMed]
- Ito, H.; Kim, J.M.; Matsunaga, W.; Saze, H.; Matsui, A.; Endo, T.A.; Harukawa, Y.; Takagi, H.; Yaegashi, H.; Masuta, Y.; et al. A Stress-Activated Transposon in Arabidopsis Induces Transgenerational Abscisic Acid Insensitivity. Sci. Rep. 2016, 6, 23181. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Jiang, Y.; Ding, M.; He, S.; Zhang, H.; Lin, L.; Rong, J. Molecular characterization of a transcriptionally active Ty1/copia-like retrotransposon in Gossypium. Plant Cell Rep. 2015, 34, 1037–1047. [Google Scholar] [CrossRef]
- He, P.; Ma, Y.; Zhao, G.; Dai, H.; Li, H.; Chang, L.; Zhang, Z. FaRE1: A transcriptionally active Ty1-copia retrotransposon in strawberry. J. Plant Res. 2010, 123, 707–714. [Google Scholar] [CrossRef]
- Ujino-Ihara, T. Transcriptome analysis of heat stressed seedlings with or without pre-heat treatment in Cryptomeria japonica. Mol. Genet. Genom. 2020, 295, 1163–1172. [Google Scholar] [CrossRef]
- Rama Rao, Y.; Yadav, P.; Rani, V.; Muley, D.; Sahoo, R.K.; Brajendra; Bhushan Kumar, S.; Gill, R.; Singh Gill, S.; Ansari, M.W.; et al. Putrescine mitigates NaCl-induced stress by modulating gene expression, antioxidants, and ethylene level in tomato. Plant Signal. Behav. 2025, 20, 2515431. [Google Scholar] [CrossRef]
- Jiang, D.; Huang, W.; Liu, J. Phylogenetic relationship of WRKY transcription factors in Solanum and potato genes in response to hormonal and biotic stresses. Plant Signal. Behav. 2025, 20, 2491465. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, Y.; Song, X.; Wang, S.; Zhang, M.; Lu, J.; Xu, S.; Wang, H. Ozone stress-induced DNA methylation variations and their transgenerational inheritance in foxtail millet. Front. Plant Sci. 2024, 15, 1463584. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, R.; Ito, H. Transposition of the heat-activated retrotransposon ONSEN results in enhanced hypocotyl elongation. Genes Genet. Syst. 2025, 100, 24-00110. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Z.; Wang, P.; Jiang, B.; Lei, X.; Wu, J.; Dong, W.; Gao, C. A 2-Cys peroxiredoxin gene from Tamarix hispida improved salt stress tolerance in plants. BMC Plant Biol. 2020, 20, 360. [Google Scholar] [CrossRef]
- Cui, Y.; Ning, Z.; Li, M.; Qin, X.; Yue, X.; Chen, X.; Zhu, C.; Sun, H.; Huang, Y. Microbial network-driven remediation of saline-alkali soils by salt-tolerant plants. Front. Microbiol. 2025, 16, 1565399. [Google Scholar] [CrossRef]
- Kidwell, K.K.; Osborn, T.C. Simple plant DNA isolation procedures. In Plant Genomes: Methods for Genetic and Physical Mapping; Beckmann, J.S., Osborn, T.C., Eds.; Springer: Dordrecht, The Netherlands, 1992; pp. 1–13. [Google Scholar]
- Hirochika, H.; Hirochika, R. Ty1-copia group retrotransposons as ubiquitous components of plant genomes. Jpn. J. Genet. 1993, 68, 35–46. [Google Scholar] [CrossRef]
- Dixit, A.; Ma, K.H.; Yu, J.W.; Cho, E.G.; Park, Y.J. Reverse transcriptase domain sequences from Mungbean (Vigna radiata) LTR retrotransposons: Sequence characterization and phylogenetic analysis. Plant Cell Rep. 2006, 25, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Huang, Y.; Luo, L.; Hu, X.; Yu, F.; Yang, Y.; Deng, Z.; Wu, J.; Chen, R.; Zhang, M. Characterization, Genomic Organization, Abundance, and Chromosomal Distribution of Ty1-copia Retrotransposons in Erianthus arundinaceus. Front. Plant Sci. 2017, 8, 924. [Google Scholar] [CrossRef]
- Diao, X.; Freeling, M.; Lisch, D. Horizontal transfer of a plant transposon. PLoS Biol. 2006, 4, e5. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Liu, W.; Peng, Q.; He, X.; Xie, D. Characterization and chromosomal organization of Ty1-copia retrotransposons in wax gourd. Gene 2014, 551, 26–32. [Google Scholar] [CrossRef]
- Fan, F.; Wen, X.; Ding, G.; Cui, B. Isolation, identification, and characterization of genomic LTR retrotransposon sequences from masson pine (Pinus massoniana). Tree Genet. Genomes 2013, 9, 1237–1246. [Google Scholar] [CrossRef]
- Monsen, Ø.; Grønvold, L.; Datsomor, A.; Harvey, T.; Kijas, J.; Suh, A.; Hvidsten, T.R.; Sandve, S.R. The role of transposon activity in shaping cis-regulatory element evolution after whole-genome duplication. Genome Res. 2025, 35, 475–488. [Google Scholar] [CrossRef]
- Zhao, J.; Ding, Y.; Ramakrishnan, M.; Zou, L.H.; Chen, Y.; Zhou, M. LTR retrotransposon-derived novel lncRNA2 enhances cold tolerance in Moso bamboo by modulating antioxidant activity and photosynthetic efficiency. PeerJ 2025, 13, e19056. [Google Scholar] [CrossRef] [PubMed]
- Wicker, T.; Keller, B. Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Res. 2007, 17, 1072–1081. [Google Scholar] [CrossRef]
- Llorens, C.; Muñoz-Pomer, A.; Bernad, L.; Botella, H.; Moya, A. Network dynamics of eukaryotic LTR retroelements beyond phylogenetic trees. Biol. Direct 2009, 4, 41. [Google Scholar] [CrossRef]
- Gebrie, A. Transposable elements as essential elements in the control of gene expression. Mob. DNA 2023, 14, 9. [Google Scholar] [CrossRef]
- Qu, X.; Pan, Y.; Wang, P.; Ran, L.; Qin, G.; Li, Q.; Kang, P. Response of Phyllosphere and Rhizosphere Microbial Communities to Salt Stress of Tamarix chinensis. Plants 2024, 13, 1091. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.Y.; Dai, H.Y.; Zhao, G.L.; Ma, Y.; Ou, C.Q.; Li, H.; Li, L.G.; Zhang, Z.H. Genome-wide characterization of long terminal repeat -retrotransposons in apple reveals the differences in heterogeneity and copy number between Ty1-copia and Ty3-gypsy retrotransposons. J. Integr. Plant Biol. 2008, 50, 1130–1139. [Google Scholar] [CrossRef] [PubMed]
- El Baidouri, M.; Carpentier, M.C.; Cooke, R.; Gao, D.; Lasserre, E.; Llauro, C.; Mirouze, M.; Picault, N.; Jackson, S.A.; Panaud, O. Widespread and frequent horizontal transfers of transposable elements in plants. Genome Res. 2014, 24, 831–838. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Li, B.; Wang, Y.; Wang, S.; Zhang, M.; Li, M.; Zheng, T.; Wang, H. Characterization of LTR Retrotransposon Reverse Transcriptase in Tamarix chinensis L. and Activity Analysis Under Salt and Alkali Stresses. Genes 2025, 16, 1262. https://doi.org/10.3390/genes16111262
Wang L, Li B, Wang Y, Wang S, Zhang M, Li M, Zheng T, Wang H. Characterization of LTR Retrotransposon Reverse Transcriptase in Tamarix chinensis L. and Activity Analysis Under Salt and Alkali Stresses. Genes. 2025; 16(11):1262. https://doi.org/10.3390/genes16111262
Chicago/Turabian StyleWang, Long, Bo Li, Yuqian Wang, Shiji Wang, Meichun Zhang, Mengyao Li, Tong Zheng, and Hongyan Wang. 2025. "Characterization of LTR Retrotransposon Reverse Transcriptase in Tamarix chinensis L. and Activity Analysis Under Salt and Alkali Stresses" Genes 16, no. 11: 1262. https://doi.org/10.3390/genes16111262
APA StyleWang, L., Li, B., Wang, Y., Wang, S., Zhang, M., Li, M., Zheng, T., & Wang, H. (2025). Characterization of LTR Retrotransposon Reverse Transcriptase in Tamarix chinensis L. and Activity Analysis Under Salt and Alkali Stresses. Genes, 16(11), 1262. https://doi.org/10.3390/genes16111262





