Genomic Relationship Between Brochothrix campestris and Its Phages: A Cross-Replicon and Interspecies Perspective
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ATCC | American Type Culture Collection |
| BLASTN | Basic Local Alignment Search Tool using a Nucleotide query |
| BLASTP | Basic Local Alignment Search Tool using a Protein query |
| BP | Base pairs |
| CIP | Collection of the Institut Pasteur |
| CRISPR-CAS | Clustered Regularly Interspaced Short Palindromic Repeats–CRISPR-associated protein |
| DDBJ | DNA DataBank of Japan |
| DSMZ (DSM) | Deutsche Sammlung von Mikroorganismen und Zellkulturen |
| ENA | European Nucleotide Archive |
| GEIs | Genomic islands |
| Kbp | Kilo-base pairs |
| Mbp | Mega-base pairs |
| NCBI | National Center for Biotechnology Information |
| PP | Phage-plasmid |
| TBLASTN | Translated Basic Local Alignment Search Tool using a Nucleotide query |
| TMW | Treated Municipal Wastewater |
References
- Illikoud, N.; Rossero, A.; Chauvet, R.; Courcoux, P.; Pilet, M.-F.; Charrier, T.; Jaffrès, E.; Zagorec, M. Genotypic and phenotypic characterization of the food spoilage bacterium Brochothrix thermosphacta. Food Microbiol. 2019, 81, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Talon, R.; Grimont, P.A.D.; Grimont, F.; Gasser, F.; Boeufgras, J.M. Brochothrix campestris sp. nov. Int. J. Syst. Evol. Microbiol. 1988, 38, 99–102. [Google Scholar] [CrossRef]
- Gribble, A.; Brightwell, G. Spoilage characteristics of Brochothrix thermosphacta and campestris in chilled vacuum packaged lamb, and their detection and identification by real time PCR. Meat Sci. 2013, 94, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Siragusa, G.R.; Cutter, C.N. Brochocin-C, a new bacteriocin produced by Brochothrix campestris. Appl. Environ. Microbiol. 1993, 59, 2326–2328. [Google Scholar] [CrossRef]
- McCormick, J.K.; Poon, A.; Sailer, M.; Gao, Y.; Roy, K.L.; McMullen, L.M.; Vederas, J.C.; Stiles, M.E.; Van Belkum, M.J. Genetic characterization and heterologous expression of brochocin-C, an antibotulinal, two-peptide bacteriocin produced by Brochothrix campestris ATCC 43754. Appl. Environ. Microbiol. 1998, 64, 4757–4766. [Google Scholar] [CrossRef]
- Greer, G.G.; Dilts, B.D. Control of meatborne Listeria monocytogenes and Brochothrix thermosphacta by a bacteriocinogenic Brochothrix campestris ATCC 43754. Food Microbiol. 2006, 23, 785–790. [Google Scholar] [CrossRef]
- Gingras, L.; Piché, L.C.; Saucier, L.; Vincent, A.T. The complete genomic sequence of the type strain Brochothrix thermosphacta DSM 20171 highlights a diversity of prophages in this species. Microbiol. Resour. Announc. 2024, 13, e00239-24. [Google Scholar] [CrossRef]
- Pasechnek, A.; Rabinovich, L.; Stadnyuk, O.; Azulay, G.; Mioduser, J.; Argov, T.; Borovok, I.; Sigal, N.; Herskovits, A.A. Active lysogeny in Listeria monocytogenes is a bacteria-phage adaptive response in the mammalian environment. Cell Rep. 2020, 32, 107956. [Google Scholar] [CrossRef]
- Vu, H.T.K.; Benjakul, S.; Vongkamjan, K. Characterization of Listeria prophages in lysogenic isolates from foods and food processing environments. PLoS ONE 2019, 14, e0214641. [Google Scholar] [CrossRef]
- Vu, H.T.K.; Stasiewicz, M.J.; Benjakul, S.; Vongkamjan, K. Genomic analysis of prophages recovered from Listeria monocytogenes lysogens found in seafood and seafood-related environment. Microorganisms 2021, 9, 1354. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Jones, D. The genus Brochothrix. In The Prokaryotes: Volume 4: Bacteria: Firmicutes, Cyanobacteria; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 477–491. [Google Scholar]
- Brito, I.L. Examining horizontal gene transfer in microbial communities. Nat. Rev. Microbiol. 2021, 19, 442–453. [Google Scholar] [CrossRef]
- Rodríguez-Beltrán, J.; DelaFuente, J.; León-Sampedro, R.; MacLean, R.C.; San Millán, Á. Beyond horizontal gene transfer: The role of plasmids in bacterial evolution. Nat. Rev. Microbiol. 2021, 19, 347–359. [Google Scholar] [CrossRef]
- Bertani, G. Studies on lysogenesis I. J. Bacteriol. 1951, 62, 293–300. [Google Scholar] [CrossRef]
- Ravin, V.K.; Shulga, M.G. Evidence for extrachromosomal location of prophage N15. Virology 1970, 40, 800–807. [Google Scholar] [CrossRef]
- Pfeifer, E.; Moura de Sousa, J.A.; Touchon, M.; Rocha, E.P.C. Bacteria have numerous distinctive groups of phage–plasmids with conserved phage and variable plasmid gene repertoires. Nucleic Acids Res. 2021, 49, 2655–2673. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genom. 2021, 7, 000685. [Google Scholar] [CrossRef]
- Bouras, G.; Nepal, R.; Houtak, G.; Psaltis, A.J.; Wormald, P.-J.; Vreugde, S. Pharokka: A fast scalable bacteriophage annotation tool. Bioinformatics 2023, 39, btac776. [Google Scholar] [CrossRef]
- Terzian, P.; Olo Ndela, E.; Galiez, C.; Lossouarn, J.; Pérez Bucio, R.E.; Mom, R.; Toussaint, A.; Petit, M.-A.; Enault, F. PHROG: Families of prokaryotic virus proteins clustered using remote homology. NAR Genom. Bioinform. 2021, 3, lqab067. [Google Scholar] [CrossRef]
- Dhillon, B.K.; Laird, M.R.; Shay, J.A.; Winsor, G.L.; Lo, R.; Nizam, F.; Pereira, S.K.; Waglechner, N.; McArthur, A.G.; Langille, M.G.I.; et al. IslandViewer 3: More flexible, interactive genomic island discovery, visualization and analysis. Nucleic Acids Res. 2015, 43, W104–W108. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- García-Alcalde, F.; Okonechnikov, K.; Carbonell, J.; Cruz, L.M.; Götz, S.; Tarazona, S.; Dopazo, J.; Meyer, T.F.; Conesa, A. Qualimap: Evaluating next-generation sequencing alignment data. Bioinformatics 2012, 28, 2678–2679. [Google Scholar] [CrossRef] [PubMed]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef] [PubMed]
- Höll, L.; Hilgarth, M.; Geissler, A.J.; Behr, J.; Vogel, R.F. Prediction of in situ metabolism of photobacteria in modified atmosphere packaged poultry meat using metatranscriptomic data. Microbiol. Res. 2019, 222, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Kilcher, S.; Loessner, M.J.; Klumpp, J. Brochothrix thermosphacta bacteriophages feature heterogeneous and highly mosaic genomes and utilize unique prophage insertion sites. J. Bacteriol. 2010, 192, 5441–5453. [Google Scholar] [CrossRef]
- Rocha, E.P.C.; Bikard, D. Microbial defenses against mobile genetic elements and viruses: Who defends whom from what? PLoS Biol. 2022, 20, e3001514. [Google Scholar] [CrossRef]
- Stanborough, T.; Fegan, N.; Powell, S.M.; Tamplin, M.; Chandry, P.S. Insight into the genome of Brochothrix thermosphacta, a problematic meat spoilage bacterium. Appl. Environ. Microbiol. 2017, 83, e02786-16. [Google Scholar] [CrossRef]
- Pfeifer, E.; Rocha, E.P.C. Phage-plasmids promote recombination and emergence of phages and plasmids. Nat. Commun. 2024, 15, 1545. [Google Scholar] [CrossRef]
- Pfeifer, E.; Bonnin, R.A.; Rocha, E.P.C. Phage-plasmids spread antibiotic resistance genes through infection and lysogenic conversion. mBio 2022, 13, e01851-22. [Google Scholar] [CrossRef]
- Girons, I.S.; Bourhy, P.; Ottone, C.; Picardeau, M.; Yelton, D.; Hendrix, R.W.; Glaser, P.; Charon, N. The LE1 bacteriophage replicates as a plasmid within Leptospira biflexa: Construction of an L. biflexa-Escherichia coli shuttle vector. J. Bacteriol. 2000, 182, 5700–5705. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Wang, J.; Zhu, Y.; Tang, B.; Zhang, Y.; He, P.; Zhang, Y.; Liu, B.; Guo, X.; Zhao, G.; et al. Identification of three extra-chromosomal replicons in Leptospira pathogenic strain and development of new shuttle vectors. BMC Genom. 2015, 16, 90. [Google Scholar] [CrossRef] [PubMed]
- Kommineni, S.; Bretl, D.J.; Lam, V.; Chakraborty, R.; Hayward, M.; Simpson, P.; Cao, Y.; Bousounis, P.; Kristich, C.J.; Salzman, N.H. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 2015, 526, 719–722. [Google Scholar] [CrossRef] [PubMed]
- Solis-Balandra, M.A.; Sanchez-Salas, J.L. Classification and multi-functional use of bacteriocins in health, biotechnology, and food industry. Antibiotics 2024, 13, 666. [Google Scholar] [CrossRef]
- Sugrue, I.; Ross, R.P.; Hill, C. Bacteriocin diversity, function, discovery and application as antimicrobials. Nat. Rev. Microbiol. 2024, 22, 556–571. [Google Scholar] [CrossRef]
- Darbandi, A.; Asadi, A.; Mahdizade Ari, M.; Ohadi, E.; Talebi, M.; Halaj Zadeh, M.; Darb Emamie, A.; Ghanavati, R.; Kakanj, M. Bacteriocins: Properties and potential use as antimicrobials. J. Clin. Lab. Anal. 2022, 36, e24093. [Google Scholar] [CrossRef]
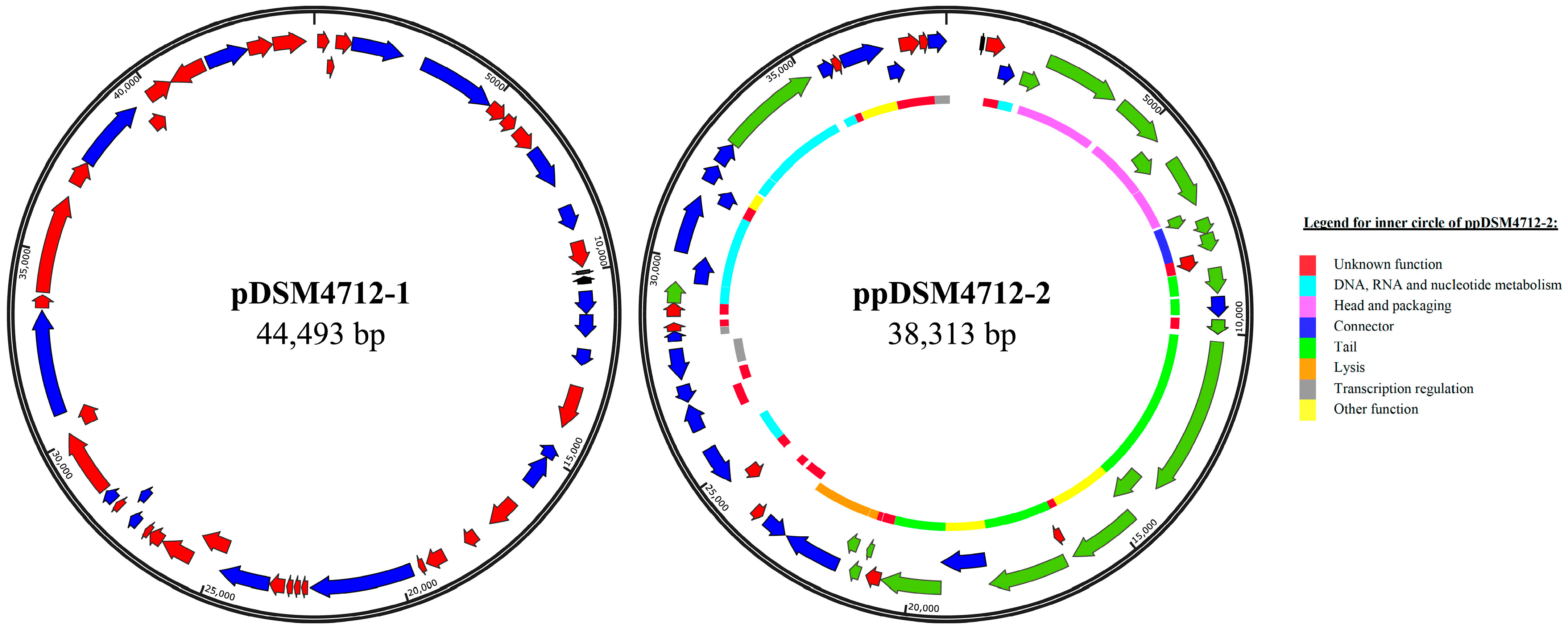
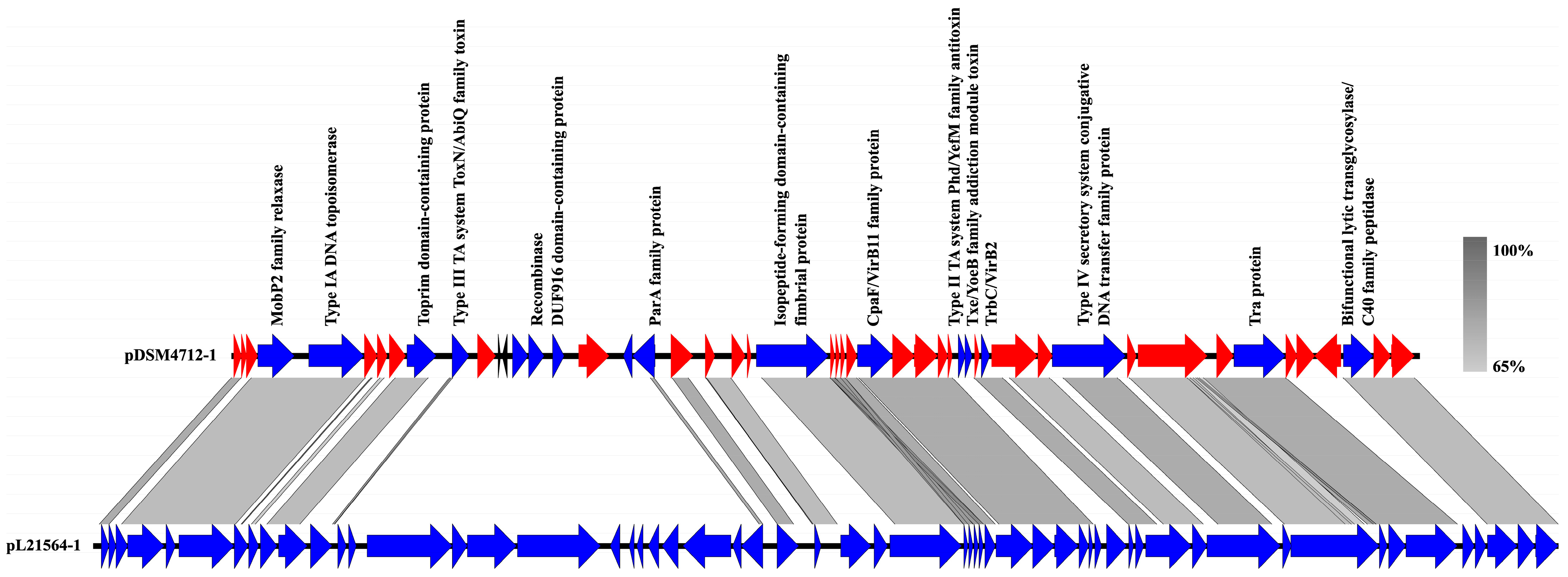
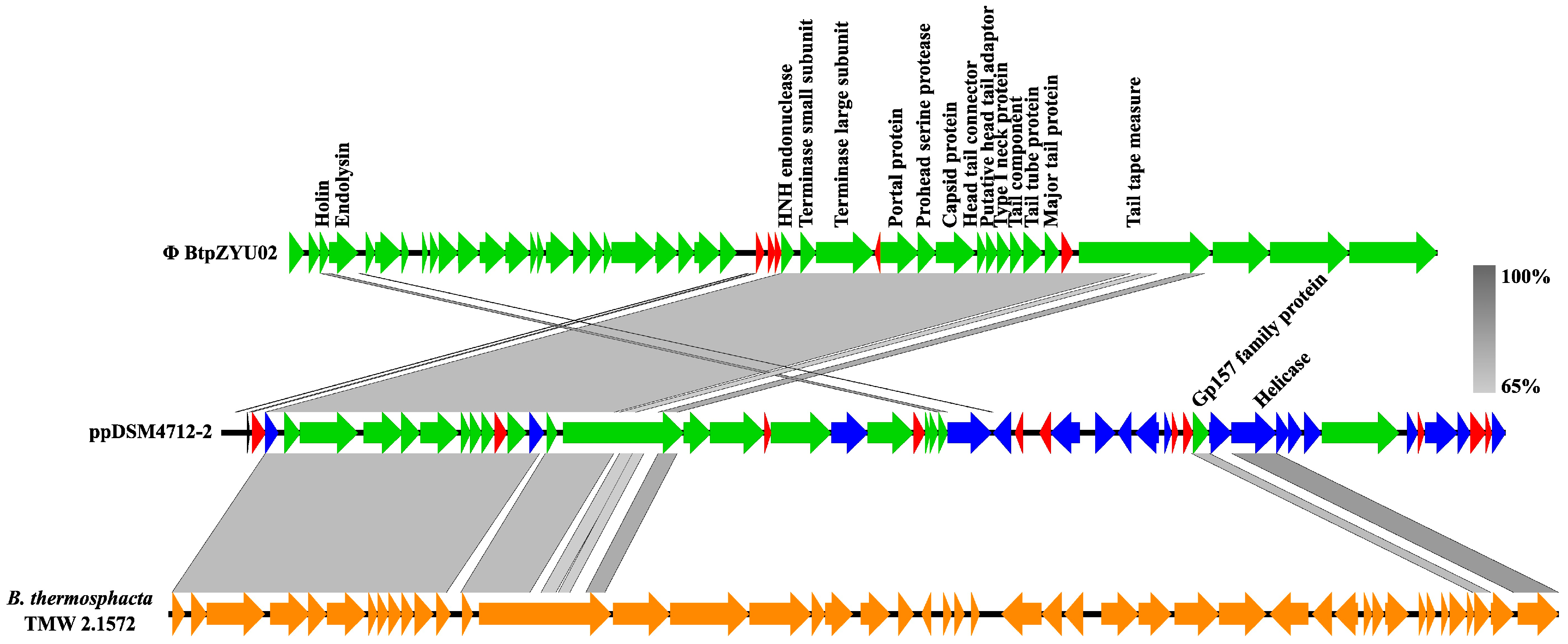
| Features | Chromosome | pDSM4712-1 | ppDSM4712-2 |
|---|---|---|---|
| Length (bp) | 2,324,176 | 44,493 | 38,313 |
| GC (%) | 40.54 | 35.19 | 37.85 |
| Number of genes | 2145 | 48 | 50 |
| tRNA | 85 | 0 | 1 |
| GenBank | CP175511 | PQ657675 | PQ657676 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Attéré, S.A.; Piché, L.C.; Vincent, A.T. Genomic Relationship Between Brochothrix campestris and Its Phages: A Cross-Replicon and Interspecies Perspective. Genes 2025, 16, 1218. https://doi.org/10.3390/genes16101218
Attéré SA, Piché LC, Vincent AT. Genomic Relationship Between Brochothrix campestris and Its Phages: A Cross-Replicon and Interspecies Perspective. Genes. 2025; 16(10):1218. https://doi.org/10.3390/genes16101218
Chicago/Turabian StyleAttéré, Sabrina A., Laurie C. Piché, and Antony T. Vincent. 2025. "Genomic Relationship Between Brochothrix campestris and Its Phages: A Cross-Replicon and Interspecies Perspective" Genes 16, no. 10: 1218. https://doi.org/10.3390/genes16101218
APA StyleAttéré, S. A., Piché, L. C., & Vincent, A. T. (2025). Genomic Relationship Between Brochothrix campestris and Its Phages: A Cross-Replicon and Interspecies Perspective. Genes, 16(10), 1218. https://doi.org/10.3390/genes16101218







