Genome-Wide Association Study of Morphological Defects in Nellore Cattle Using a Binary Trait Framework
Abstract
1. Introduction
2. Materials and Methods
2.1. Phenotypic Data
2.2. Genomic Information
2.3. Genome-Wide Association Studies
- is a vector of binary phenotypes;
- is a vector of with representing the probability of an individual being a case given their genotype , fixed effects (contemporary groups) , and the animals were used as a random genetic effect .
- is a vector of genotypes of a variant of interest with its effect ,
- is the incidence of contemporary groups used as a fixed effect with their corresponding coefficients .
- is a vector of effects that captures genetic and common environment effects shared among related individuals, with being the sparse GRM (i.e., GRM with all the small off-diagonal elements set to zero), and being the corresponding variance component.
2.4. Gene Annotation, QTL Identification, and Functional Enrichment Analyses
3. Results
3.1. Genome-Wide Association Studies
3.2. Gene Annotation, QTL Identification, and Functional Enrichment Analyses
4. Discussion
4.1. Methodological Aspects and Statistical Considerations
4.2. Genetic Architecture and Biological Mechanisms of Morphological Defects
4.3. Limitations and Future Research Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMC | A computer program to assess the degree of connectedness among contemporary groups |
| ASIP | Agouti signaling protein |
| BTA | Bos taurus autosome |
| bp | Base pairs |
| CC | Cellular Component (Gene Ontology domain) |
| CG | Contemporary group |
| CCR | C-C motif chemokine receptor |
| CLMP | CXADR-like membrane protein |
| FDR | False discovery rate |
| GALLO | Genomic Annotation in Livestock for positional candidate Loci (R package) |
| GCTA | Genome-wide Complex Trait Analysis |
| GLMM | Generalized linear mixed model |
| GO | Gene Ontology |
| GO:BP | Gene Ontology Biological Process |
| GO:CC | Gene Ontology Cellular Component |
| GO:MF | Gene Ontology Molecular Function |
| GRM | Genomic relationship matrix |
| GWAS | Genome-wide association study |
| HD | High density |
| Kb | Kilobase pairs |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MAF | Minor allele frequency |
| Ne | Effective population size |
| QC | Quality control |
| QTL | Quantitative trait locus |
| SNP | Single-nucleotide polymorphism |
References
- Fernandes Júnior, G.A.; Peripolli, E.; Schmidt, P.I.; Campos, G.S.; Mota, L.F.M.; Mercadante, M.E.Z.; Baldi, F.; Carvalheiro, R.; de Albuquerque, L.G. Current applications and perspectives of genomic selection in Bos indicus (Nellore) cattle. Livest. Sci. 2022, 263, 105001. [Google Scholar] [CrossRef]
- Campos, M.A.F.; de Oliveira, H.R.; de Camargo, G.M.F.; Mulim, H.A.; Cardoso, D.F.; Costa, R.B. Beyond Black and White: Dissecting the Genetic Basis of Skin Depigmentation in Nellore Cattle. Mamm. Genome, 2025; ahead of print. [Google Scholar] [CrossRef]
- Silva, T.L.; Gondro, C.; Fonseca, P.A.S.; Silva, D.A.; Vargas, G.; Neves, H.H.R.; Carvalho Filho, I.; Teixeira, C.S.; Albuquerque, L.G.; Carvalheiro, R. Feet and legs malformation in Nellore cattle: Genetic analysis and prioritization of GWAS results. Front. Genet. 2023, 14, 1118308. [Google Scholar] [CrossRef]
- Campos, M.A.F.; de Oliveira, H.R.; Mulim, H.A.; da Silva Oliveira, E.; Hidalgo, J.; Costa, R.B. Comparison of linear and threshold models for genetic evaluation of morphological defects in Nellore cattle. J. Anim. Sci. 2025; submitted. [Google Scholar]
- Van Eenennaam, A.L.; Drake, D.J. Where in the beef-cattle supply chain might DNA tests generate value? Anim. Prod. Sci. 2012, 52, 185–196. [Google Scholar] [CrossRef]
- Vargas, G.; Neves, H.H.R.; Garzón, N.A.M.; Fonseca, L.F.S.; Fernandes Júnior, G.A.; Albuquerque, L.G.; Carvalheiro, R. Unravelling the effect of structural variants from whole-genome sequence for depigmentation in Nellore cattle. In Proceedings of the World Congress on Genetics Applied to Livestock Production, Rotterdam, The Netherlands, 3–9 July 2022; Wageningen Academic Publishers: Rotterdam, The Netherlands, 2022; pp. 1118–1121. [Google Scholar] [CrossRef]
- Nunes, C.L.C.; Pflanzer, S.B.; Rezende-de-Souza, J.H.; Chizzotti, M.L. Beef production and carcass evaluation in Brazil. Anim. Front. 2024, 14, 15–20. [Google Scholar] [CrossRef]
- ABIEC. Beef Report 2024. Associação Brasileira das Indústrias Exportadoras de Carnes: São Paulo, Brazil. 2024. Available online: https://www.abiec.com.br/wp-content/uploads/beefreport_v2024-ENG.pdf (accessed on 15 September 2025).
- Herrmann, R.; Utz, J.; Rosenberger, E.; Doll, K.; Distl, O. Risk factors for congenital umbilical hernia in German Fleckvieh. Vet. J. 2001, 162, 233–240. [Google Scholar] [CrossRef]
- Machado, P.C.; Brito, L.F.; Martins, R.; Pinto, L.F.B.; Silva, M.R.; Pedrosa, V.B. Genome-wide association analysis reveals novel loci related with visual score traits in Nellore cattle raised in pasture-based systems. Animals 2022, 12, 3526. [Google Scholar] [CrossRef] [PubMed]
- Trigo, B.B.; Alves, N.F.; Milanesi, M.; Garcia, J.F.; Terefe, E.; Hanotte, O.; Tijjani, A.; Utsunomiya, Y.T. A structural variant at ASIP associated with the darkness of hair coat is found in multiple zebu cattle populations. Anim. Genet. 2023, 54, 544–548. [Google Scholar] [CrossRef] [PubMed]
- Ogunbawo, A.R.; Mulim, H.A.; Campos, G.S.; Schinckel, A.P.; Oliveira, H.R. Tailoring genomic selection for Bos taurus indicus: A comprehensive review of SNP arrays and reference genomes. Genes 2024, 15, 1495. [Google Scholar] [CrossRef] [PubMed]
- Widmer, S.; Seefried, F.R.; Häfliger, I.M.; Signer-Hasler, H.; Flury, C.; Drögemüller, C. WNT10B: A locus increasing risk of brachygnathia inferior in Brown Swiss cattle. J. Dairy Sci. 2023, 106, 8969–8978. [Google Scholar] [CrossRef]
- Jiang, L.; Zheng, Z.; Fang, H.; Yang, J. A generalized linear mixed model association tool for biobank-scale data. Nat. Genet. 2021, 53, 1616–1621. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Vargas, G.; Neves, H.H.R.; Cardoso, V.; Munari, D.P.; Carvalheiro, R. Genome-wide association study and functional analysis of feet and leg conformation traits in Nellore cattle. J. Anim. Sci. 2018, 96, 1617–1627. [Google Scholar] [CrossRef]
- Roso, V.M.; Schenkel, F.S. AMC: A computer program to assess the degree of connectedness among contemporary groups. J. Anim. Sci. 2006, 84, 274–283. [Google Scholar]
- Neogen Corporation. GGP Indicus Genotyping Platform. 2021. Available online: https://www.neogen.com (accessed on 6 June 2025).
- Illumina Inc. GenomeStudio Genotyping Module v1.0 User Guide; Part #11319113 Rev. A; Illumina, Inc.: San Diego, CA, USA, 2010. [Google Scholar]
- Sargolzaei, M.; Chesnais, J.P.; Schenkel, F.S. A new approach for efficient genotype imputation using information from relatives. BMC Genomics 2014, 15, 478. [Google Scholar] [CrossRef]
- Neves, H.H.R.; Carvalheiro, R.; O’Brien, A.M.P.; Utsunomiya, Y.T.; do Carmo, A.S.; Schenkel, F.S.; Sölkner, J.; McEwan, J.C.; Van Tassell, C.P.; Sonstegard, T.S.; et al. Accuracy of genomic predictions in Bos indicus (Nellore) cattle. Genet. Sel. Evol. 2014, 46, 17. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.A.M.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 7. [Google Scholar] [CrossRef]
- Ogunbawo, A.R.; Hidalgo, J.; Mulim, H.A.; Carrara, E.R.; Ventura, H.T.; Souza, N.O.; Lourenco, D.; Oliveira, H.R. Applying the algorithm for Proven and Young in GWAS reveals high polygenicity for key traits in Nellore cattle. Front. Genet. 2025, 16, 1549284. [Google Scholar] [CrossRef] [PubMed]
- Corbin, L.J.; Liu, A.Y.H.; Bishop, S.C.; Woolliams, J.A. Estimation of historical effective population size using linkage disequilibria with marker data. J. Anim. Breed. Genet. 2012, 129, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Goddard, M.E.; Hayes, B.J.; Meuwissen, T.H.E. Using the genomic relationship matrix to predict the accuracy of genomic selection. J. Anim. Breed. Genet. 2011, 128, 409–421. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, P.A.S.; Suárez-Vega, A.; Marras, G.; Cánovas, Á. GALLO: An R package for genomic annotation and integration of multiple data sources in livestock for positional candidate loci. GigaScience 2020, 9, giaa149. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing. Available online: https://www.R-project.org/ (accessed on 4 February 2024).
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Vilo, J.; Peterson, H. gprofiler2—An R package for gene list functional enrichment analysis and namespace conversion. F1000Research 2020, 9, 709. [Google Scholar] [CrossRef]
- Gurinovich, A.; Li, M.; Leshchyk, A.; Bae, H.; Song, Z.; Arbeev, K.G.; Nygaard, M.; Feitosa, M.F.; Perls, T.T.; Sebastiani, P. Evaluation of GENESIS, SAIGE, REGENIE and fastGWA-GLMM for genome-wide association studies of binary traits in correlated data. Front. Genet. 2022, 13, 897210. [Google Scholar] [CrossRef]
- Rosendahl, S.; Sulniute, R.; Persson, J.; Forsberg, S.; Häggvik, R.; Drewsen, V.; Koskinen Holm, C.; Kindstedt, E.; Lundberg, P. Lack of CCR3 leads to a skeletal phenotype only in male mice. Biochem. Biophys. Res. Commun. 2022, 620, 98–104. [Google Scholar] [CrossRef]
- Kim, B.J.; Lee, Y.S.; Lee, S.Y.; Baek, W.Y.; Choi, Y.J.; Moon, S.A.; Lee, S.H.; Kim, J.E.; Chang, E.J.; Kim, E.Y.; et al. Osteoclast-secreted SLIT3 coordinates bone resorption and formation. J. Clin. Investig. 2018, 128, 1429–1441. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Mehdi, A.; Srivastava, R.; Verma, N. Immunoregulation of bone remodelling. Int. J. Crit. Illn. Inj. Sci. 2012, 2, 75–81. [Google Scholar] [CrossRef]
- Ng, M.Y.W.; Charsou, C.; Lapao, A.; Singh, S.; Trachsel-Moncho, L.; Schultz, S.W.; Nakken, S.; Munson, M.J.; Simonsen, A. The cholesterol transport protein GRAMD1C regulates autophagy initiation and mitochondrial bioenergetics. Nat. Commun. 2022, 13, 6211. [Google Scholar] [CrossRef]
- Sandhu, J.; Li, S.; Fairall, L.; Pfisterer, S.G.; Gurnett, J.E.; Xiao, X.; Weston, T.A.; Vashi, D.; Ferrari, A.; Ochoa-Callejero, L.; et al. Aster proteins facilitate nonvesicular plasma membrane to ER cholesterol transport in mammalian cells. Cell 2018, 175, 514–529.e20. [Google Scholar] [CrossRef]
- Naito, T.; Yang, H.; Zheng Koh, D.H.; Mahajan, D.; Lu, L.; Saheki, Y. Regulation of cellular cholesterol distribution via non-vesicular lipid transport at ER–Golgi contact sites. Nat. Commun. 2023, 14, 6210. [Google Scholar] [CrossRef]
- Kunej, T.; Šimon, M.; Luštrek, B.; Horvat, S.; Potočnik, K. Examining genotype–phenotype associations of GRAM domain proteins using GWAS, PheWAS and literature review in cattle, human, pig, mouse and chicken. Sci. Rep. 2024, 14, 1177. [Google Scholar] [CrossRef] [PubMed]
- Raschperger, E.; Engstrom, U.; Pettersson, R.F.; Fuxe, J. CLMP, a novel member of the CTX family and a new component of epithelial tight junctions. J. Biol. Chem. 2004, 279, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Lou, B.; Fu, Z.; Mohsen, A.W.; Shen, A.L.; Vockley, J.; Kim, J.J.P. Molecular mechanism of interactions between ACAD9 and binding partners in mitochondrial respiratory complex I assembly. iScience 2021, 24, 103153. [Google Scholar] [CrossRef] [PubMed]
- Sinsheimer, A.; Mohsen, A.W.; Bloom, K.; Karunanidhi, A.; Bharathi, S.; Wu, Y.L.; Schiff, M.; Wang, Y.; Goetzman, E.S.; Ghaloul-Gonzalez, L.; et al. Development and characterization of a mouse model for Acad9 deficiency. Mol. Genet. Metab. 2021, 134, 156–163. [Google Scholar] [CrossRef]
- Repp, B.M.; Wortmann, S.B.; Smeitink, J.A.M.; Rodenburg, R.J.T. Clinical, biochemical and genetic spectrum of 70 patients with ACAD9 deficiency: Is riboflavin supplementation effective? Orphanet J. Rare Dis. 2018, 13, 110. [Google Scholar] [CrossRef]
- Xiong, L.; Pei, J.; Wu, X.; Kalwar, Q.; Liang, C.; Guo, X.; Chu, M.; Bao, P.; Yao, X.; Yan, P. The study of the response of fat metabolism to long-term energy stress based on serum, fatty acid and transcriptome profiles in yaks. Animals 2020, 10, 1150. [Google Scholar] [CrossRef]
- Pulit, S.L.; Stoneman, C.; Morris, A.P.; Wood, A.R.; Glastonbury, C.A.; Tyrrell, J.; Yengo, L.; Ferreira, T.; Marouli, E.; Ji, Y.; et al. Meta-analysis of genome-wide association studies for body fat distribution in 694,649 individuals of European ancestry. Hum. Mol. Genet. 2019, 28, 166–174. [Google Scholar] [CrossRef]
- Dong, K.; Bai, Z.; He, X.; Zhang, L.; Hu, G.; Yao, Y.; Cai, C.L.; Zhou, J. Generation of a novel constitutive smooth muscle cell-specific Myh11-driven Cre mouse model. J. Mol. Cell. Cardiol. 2025, 202, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Alkuraya, F.S.; Cai, X.; Emery, C.; Mochida, G.H.; Al-Dosari, M.S.; Felie, J.M.; Hill, R.S.; Barry, B.J.; Partlow, J.N.; Gascon, G.G.; et al. Human mutations in NDE1 cause extreme microcephaly with lissencephaly. Am. J. Hum. Genet. 2011, 88, 536–547. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Long, B.; Jiao, J.Q.; Wang, J.X.; Liu, J.P.; Li, Q.; Li, P.F. miR-484 regulates mitochondrial network through targeting Fis1. Nat. Commun. 2012, 3, 781. [Google Scholar] [CrossRef]
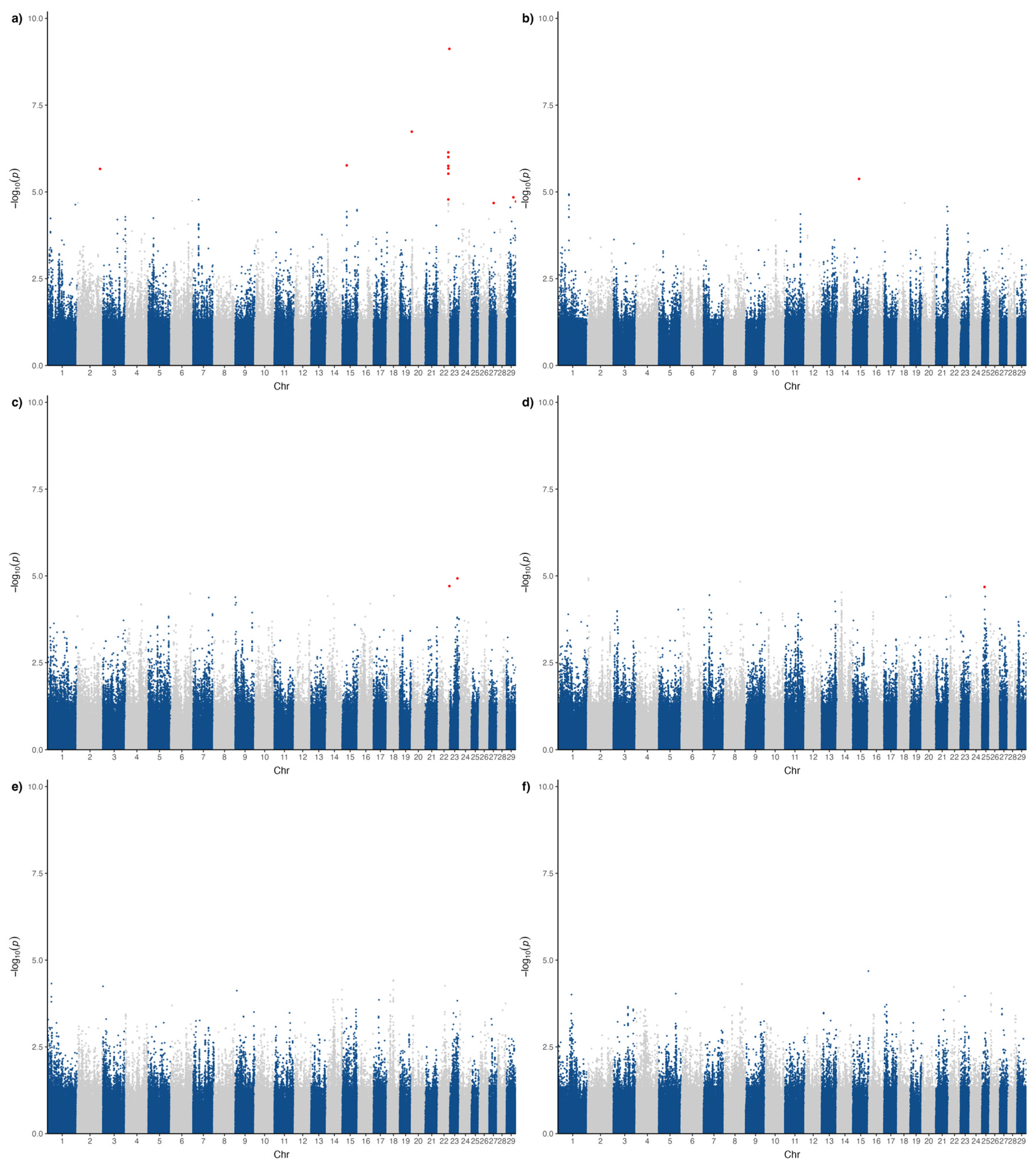
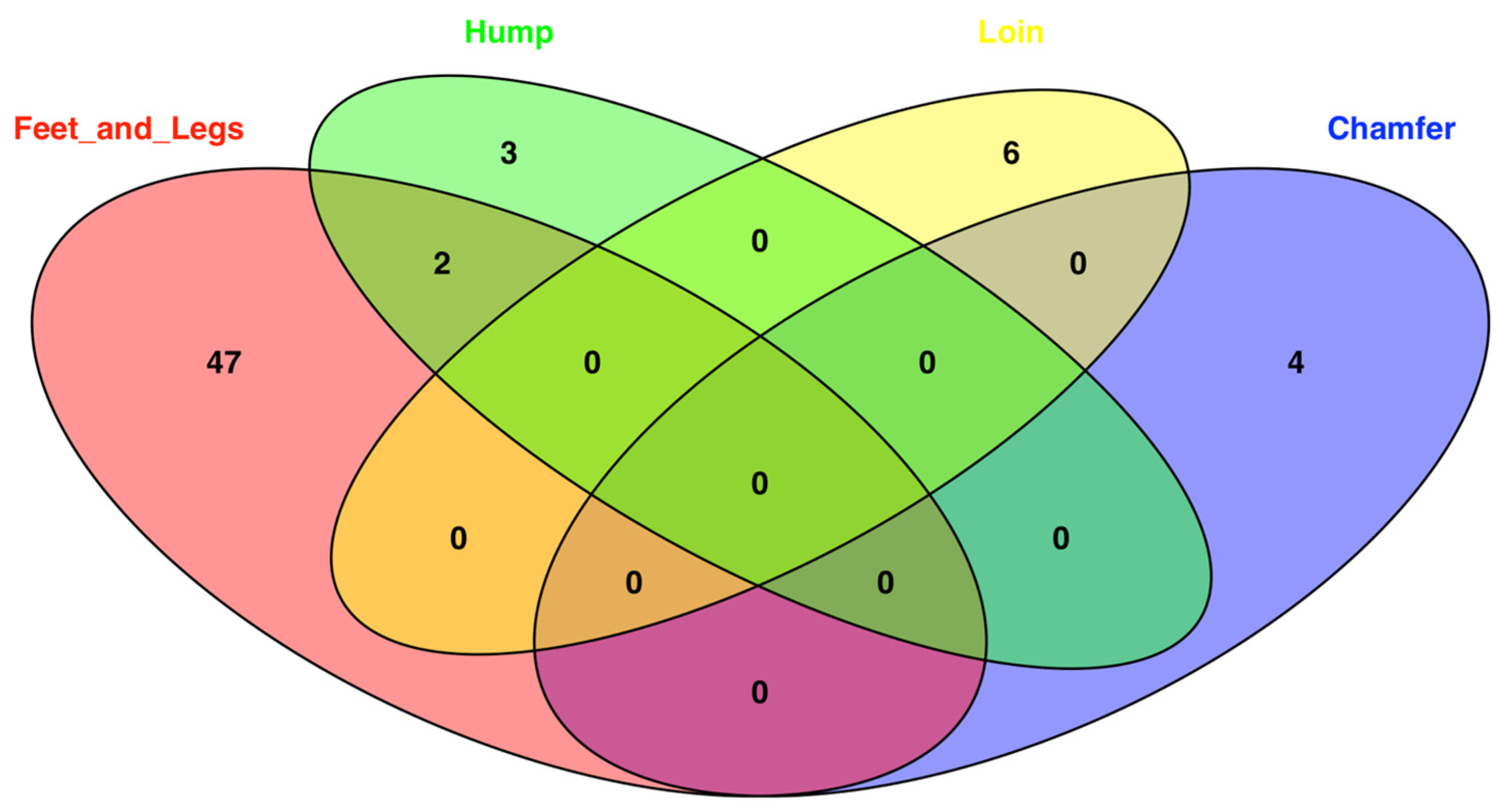
| Trait | Number of Animals | Number of Animals with Defects | Incidence (%) |
|---|---|---|---|
| Feet and legs | 22,493 | 1954 | 8.69 |
| Chamfer | 23,206 | 1053 | 4.54 |
| Hump | 9779 | 439 | 4.49 |
| Loin | 15,225 | 566 | 3.72 |
| Jaw | 9077 | 312 | 3.44 |
| Navel | 3369 | 155 | 4.60 |
| Gene Name | Chromosome | Genomic Region (bp) | Ensembl Gene ID | |
|---|---|---|---|---|
| Start | End | |||
| Feet and legs | ||||
| LCK | 2 | 121,262,594 | 121,283,528 | ENSBTAG00000012695 |
| USP28 | 15 | 24,336,893 | 24,400,181 | ENSBTAG00000002323 |
| SLIT3 | 20 | 447,017 | 1,163,732 | ENSBTAG00000017746 |
| CCR1 | 22 | 53,199,437 | 53,237,483 | ENSBTAG00000019428 |
| CCRL2 | 22 | 52,998,319 | 53,000,315 | ENSBTAG00000006155 |
| CCR5 | 22 | 53,024,929 | 53,032,609 | ENSBTAG00000067584 |
| CCR2 | 22 | 53,041,056 | 53,057,320 | ENSBTAG00000056962 |
| CCR3 | 22 | 53,134,643 | 53,166,540 | ENSBTAG00000001338 |
| CD6 | 29 | 37,311,550 | 37,357,284 | ENSBTAG00000018367 |
| CD5 | 29 | 37,422,886 | 37,444,466 | ENSBTAG00000013730 |
| Chamfer | ||||
| GRAMD1B | 15 | 33,964,574 | 34,225,319 | ENSBTAG00000001410 |
| CLMP | 15 | 33,689,099 | 33,794,265 | ENSBTAG00000020046 |
| Hump | ||||
| IQSEC1 | 22 | 58,592,571 | 58,878,769 | ENSBTAG00000003237 |
| ACAD9 | 22 | 58,892,389 | 58,940,024 | ENSBTAG00000003242 |
| ATXN1 | 23 | 40,688,296 | 40,830,587 | ENSBTAG00000019675 |
| GMPR | 23 | 40,838,557 | 40,899,426 | ENSBTAG00000015743 |
| Loin | ||||
| MARF1 | 25 | 14,044,148 | 14,083,629 | ENSBTAG00000020387 |
| NDE1 | 25 | 14,084,964 | 14,144,211 | ENSBTAG00000015986 |
| MYH11 | 25 | 14,124,964 | 14,277,425 | ENSBTAG00000015988 |
| BMERB1 | 25 | 13,881,271 | 14,026,628 | ENSBTAG00000011692 |
| GO Identification | Category | p-Value | Term | Ensembl Gene ID |
|---|---|---|---|---|
| Feet and legs | ||||
| GO:0070098 | GO:BP | 3.89 × 10−6 | chemokine-mediated signaling pathway | ENSBTAG00000017746, ENSBTAG00000019428, ENSBTAG00000031355, ENSBTAG00000056962, ENSBTAG00000001338 |
| GO:0006935 | GO:BP | 1.74 × 10−3 | chemotaxis | ENSBTAG00000017746, ENSBTAG00000019428, ENSBTAG00000031355, ENSBTAG00000056962, ENSBTAG00000001338 |
| GO:0006955 | GO:BP | 1.93 × 10−3 | immune response | ENSBTAG00000001292, ENSBTAG00000019428, ENSBTAG00000031355, ENSBTAG00000056962, ENSBTAG00000001338, ENSBTAG00000018367, ENSBTAG00000019015, ENSBTAG00000048470 |
| KEGG:04514 | KEGG | 8.39 × 10−3 | Cell adhesion molecules | ENSBTAG00000019486, ENSBTAG00000039149, ENSBTAG00000018367 |
| Chamfer | ||||
| GO:0015485 | GO:MF | 0.009 | cholesterol binding | ENSBTAG00000001410 |
| GO:0071397 | GO:BP | 0.024 | cellular response to cholesterol | ENSBTAG00000001410 |
| Hump | ||||
| GO:0032011 | GO:BP | 0.049 | ARF protein signal transduction | ENSBTAG00000003237 |
| GO:0051791 | GO:BP | 0.049 | medium-chain fatty acid metabolic process | ENSBTAG00000003242 |
| GO:0003920 | GO:MF | 0.014 | GMP reductase activity | ENSBTAG00000015743 |
| Loin | ||||
| GO:0097435 | GO:BP | 0.018 | supramolecular fiber organization | ENSBTAG00000015986, ENSBTAG00000015988, ENSBTAG00000011692 |
| GO:0031109 | GO:BP | 0.018 | microtubule polymerization or depolymerization | ENSBTAG00000015986, ENSBTAG00000011692 |
| GO:0021822 | GO:BP | 0.018 | negative regulation of cell motility involved in cerebral cortex radial glia guided migration | ENSBTAG00000011692 |
| Chromosome | SNP ID | Position (bp) | QTL Type | Name |
|---|---|---|---|---|
| Feet and legs | ||||
| 15 | Meat and Carcass | Shear force; Marbling score | ||
| rs41746697 | 24,243,265 | Production | Body weight | |
| Reproduction | Pregnancy rate; Conception rate | |||
| 20 | rs133818511 | 1,067,779 | Production | Body depth |
| Reproduction | Calving ease; Pregnancy rate | |||
| Exterior | Foot angle; Feet and leg conformation; Udder attachment; Stature; Strength | |||
| Production | Length of productive life; Methane production | |||
| Health | Somatic cell score | |||
| 22 | rs137317872 | 52,912,130 | Meat and Carcass | Connective tissue amount |
| Health | Bovine respiratory disease susceptibility; Clinical mastitis | |||
| 22 | rs136044991 | 58,804,102 | Meat and Carcass | Muscle taurine content |
| Production | Body depth; Body weight | |||
| 27 | rs135251990 | 24,315,347 | Meat and Carcass | Marbling score |
| 29 | rs42183554 | 37,423,894 | Meat and Carcass | Tenderness score |
| Chamfer | ||||
| 15 | rs136075448 | 33,880,728 | Production | Dry matter intake |
| Meat and Carcass | Marbling score | |||
| Hump | ||||
| 22 | rs136044991 | 58,804,102 | Meat and Carcass | Muscle taurine content |
| Production | Body depth; Body weight | |||
| Health | Bovine respiratory disease susceptibility | |||
| 23 | rs134164538 | 40,775,703 | Meat and Carcass | Marbling score; Shear force |
| Reproduction | Inseminations per conception; Interval to first estrus after calving | |||
| Loin | ||||
| 25 | rs137228331 | 14,067,719 | Production | Dry matter intake; Residual feed intake |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campos, M.A.F.; Rojas de Oliveira, H.; Mulim, H.A.; Oliveira, E.d.S.; Fonseca, P.A.d.S.; Camargo, G.M.F.d.; Costa, R.B. Genome-Wide Association Study of Morphological Defects in Nellore Cattle Using a Binary Trait Framework. Genes 2025, 16, 1204. https://doi.org/10.3390/genes16101204
Campos MAF, Rojas de Oliveira H, Mulim HA, Oliveira EdS, Fonseca PAdS, Camargo GMFd, Costa RB. Genome-Wide Association Study of Morphological Defects in Nellore Cattle Using a Binary Trait Framework. Genes. 2025; 16(10):1204. https://doi.org/10.3390/genes16101204
Chicago/Turabian StyleCampos, Milena A. F., Hinayah Rojas de Oliveira, Henrique A. Mulim, Eduarda da Silva Oliveira, Pablo Augusto de Souza Fonseca, Gregorio M. F. de Camargo, and Raphael Bermal Costa. 2025. "Genome-Wide Association Study of Morphological Defects in Nellore Cattle Using a Binary Trait Framework" Genes 16, no. 10: 1204. https://doi.org/10.3390/genes16101204
APA StyleCampos, M. A. F., Rojas de Oliveira, H., Mulim, H. A., Oliveira, E. d. S., Fonseca, P. A. d. S., Camargo, G. M. F. d., & Costa, R. B. (2025). Genome-Wide Association Study of Morphological Defects in Nellore Cattle Using a Binary Trait Framework. Genes, 16(10), 1204. https://doi.org/10.3390/genes16101204







