Genome-Wide Identification and Expression Analysis of the bHLH Transcription Factor Family in Lilium bakerianum var. rubrum
Abstract
1. Introduction
2. Materials and Methods
2.1. Full-Length Transcriptome Sequencing and Annotation
2.2. Identification of bHLH TFs
2.3. Phylogenetic Analysis and Classification of the bHLH TFs
2.4. Transcriptome Sequencing and Expression Level Analysis
3. Results
3.1. Summary of Transcriptome Assembly and Annotation
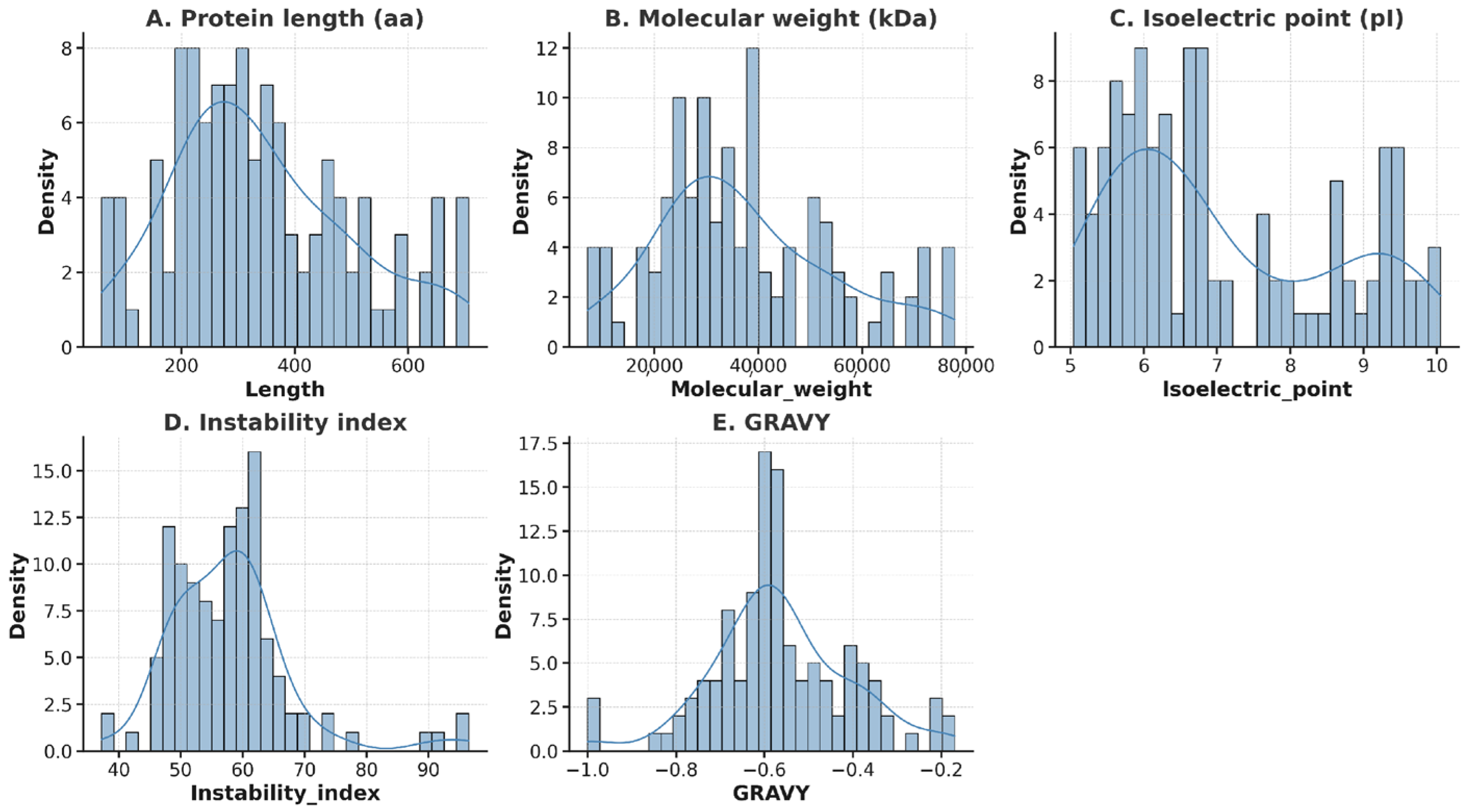
3.2. Identification and Features of bHLH TFs
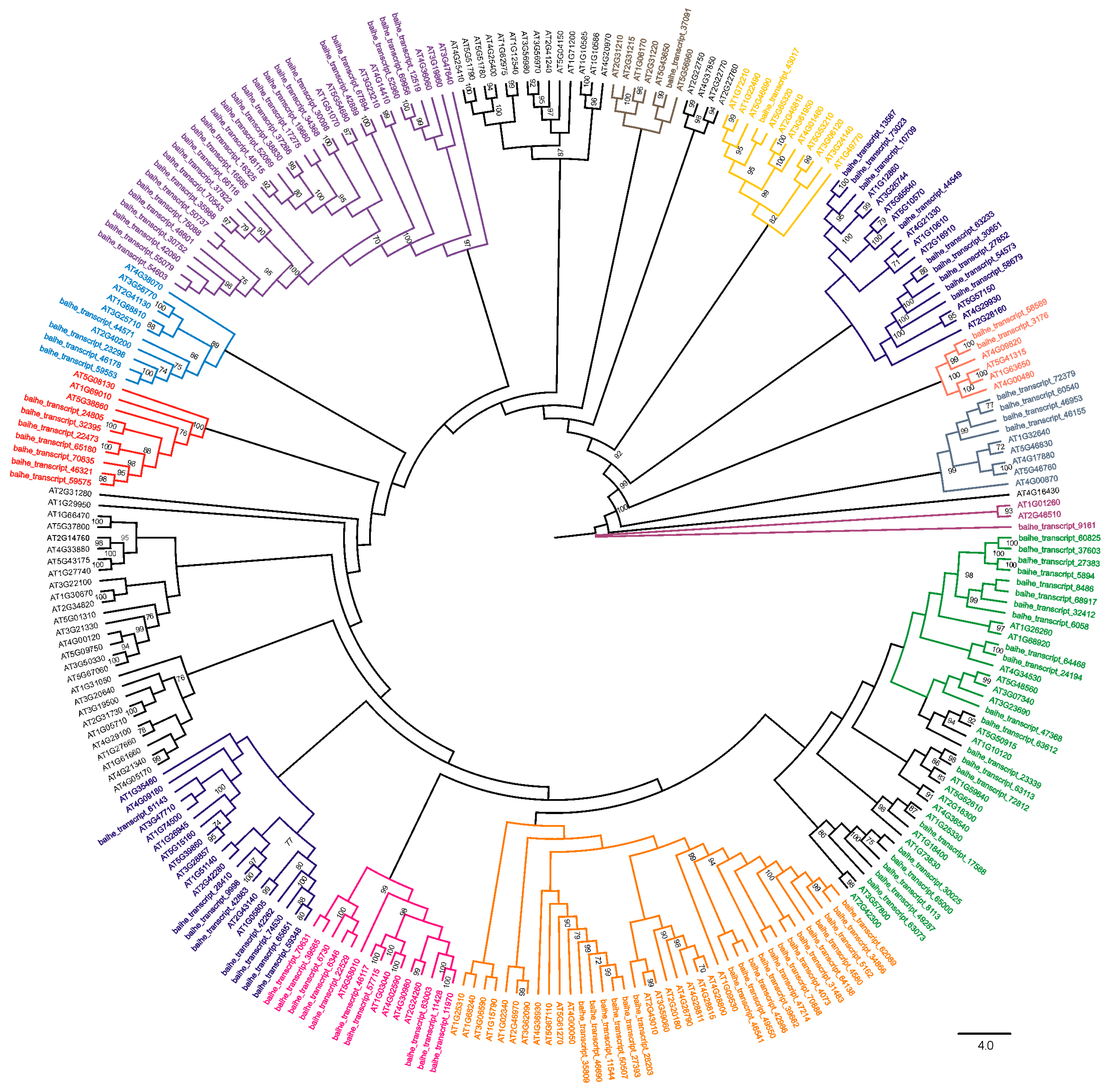
3.3. Subfamily Classification of bHLH Genes
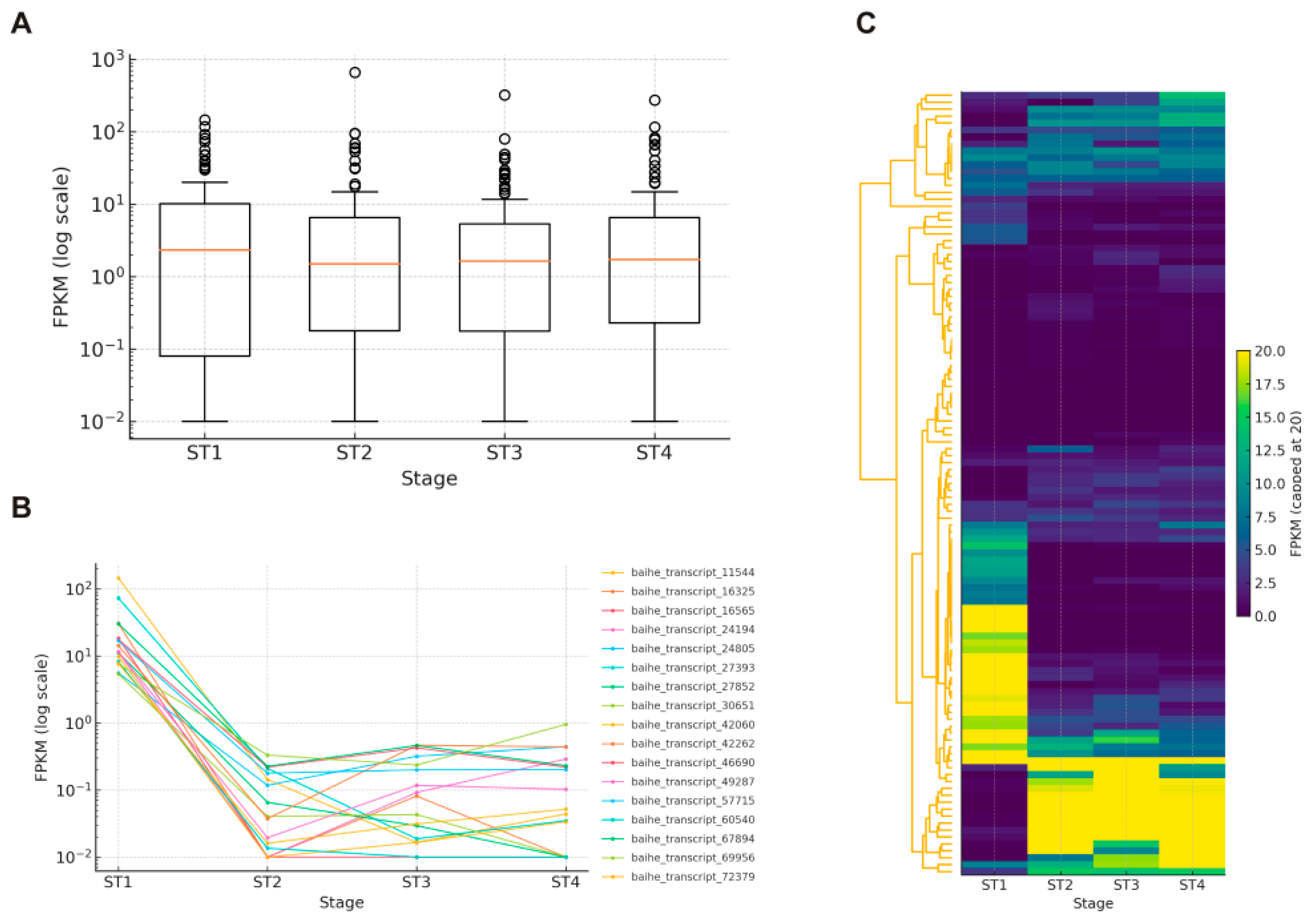
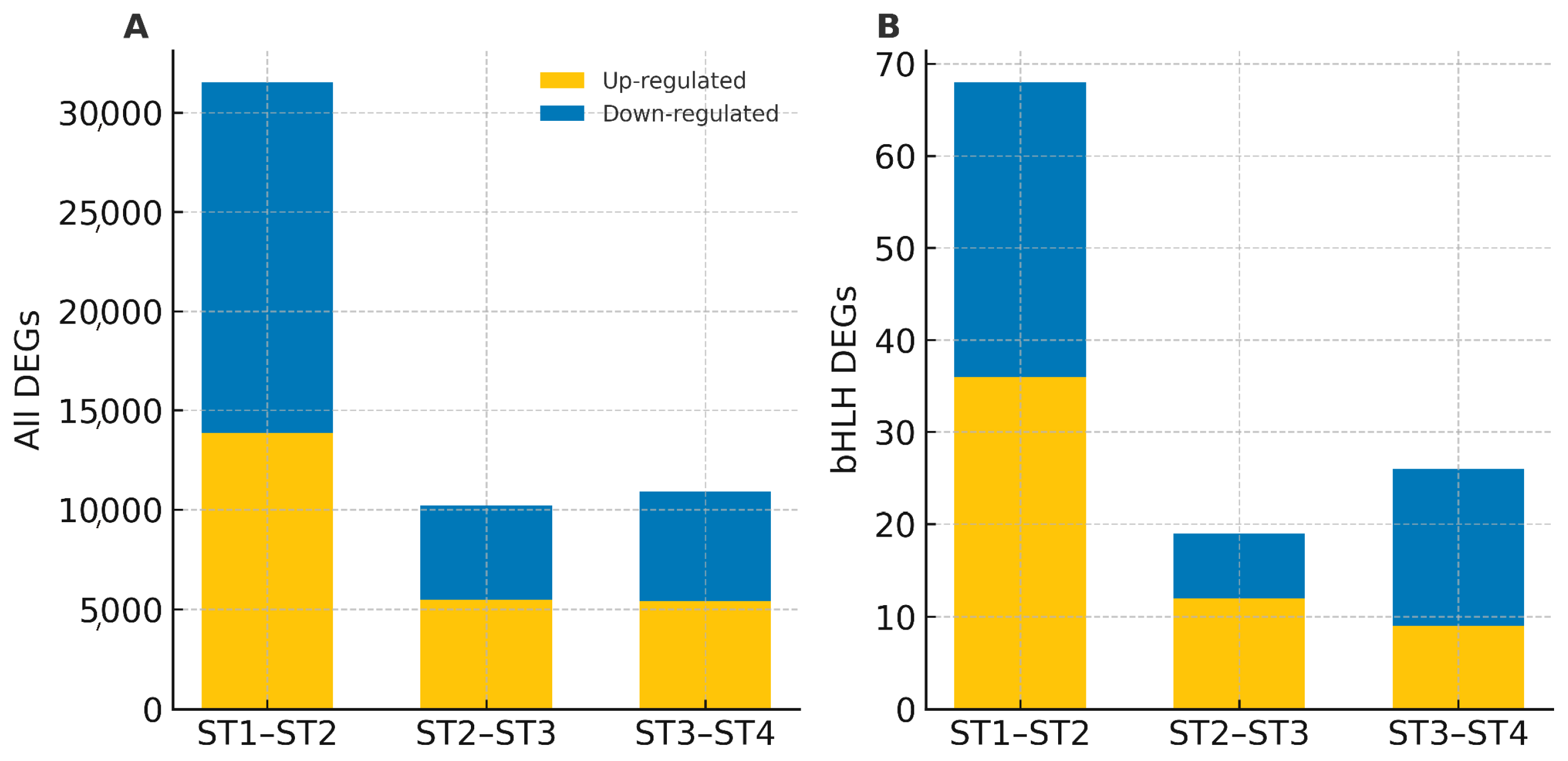
3.4. Expression Patterns of bHLH Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| bHLH | Basic Helix–Loop–Helix |
| LBVR | L. bakerianum var. rubrum |
| TF | Transcription Factor |
| HMM | Hidden Markov Model |
| aa | Amino Acids |
| GRAVY | Grand Average of Hydropathicity |
| ML | Maximum Likelihood |
| FPKM | Fragments per Kilobase of Transcript per Million Mapped Reads |
| DEGs | Differentially Expressed Genes |
| ORFs | Open Reading Frames |
| BUSCO | Benchmarking Universal Single-Copy Orthologs |
References
- Jones, S. An overview of the basic helix-loop-helix proteins. Genome Biol. 2004, 5, 226. [Google Scholar] [CrossRef]
- Atchley, W.R.; Fitch, W.M. A natural classification of the basic helix–loop–helix class of transcription factors. Proc. Natl. Acad. Sci. USA 1997, 94, 5172–5176. [Google Scholar] [CrossRef]
- Pires, N.; Dolan, L. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 2010, 27, 862–874. [Google Scholar] [CrossRef] [PubMed]
- Buck, M.J.; Atchley, W.R. Phylogenetic analysis of plant basic helix-loop-helix proteins. J. Mol. Evol. 2003, 56, 742–750. [Google Scholar] [CrossRef] [PubMed]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.; Chen, H. Comparative functional genomics analysis of bHLH gene family in rice, maize and wheat. BMC Plant Biol. 2018, 18, 309. [Google Scholar] [CrossRef]
- Rensing, S.A.; Lang, D.; Zimmer, A.D.; Terry, A.; Salamov, A.; Shapiro, H.; Nishiyama, T.; Perroud, P.F.; Lindquist, E.A.; Kamisugi, Y.; et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 2008, 319, 64–69. [Google Scholar] [CrossRef]
- Heim, M.A.; Jakoby, M.; Werber, M.; Martin, C.; Weisshaar, B.; Bailey, P.C. The basic helix–loop–helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 2003, 20, 735–747. [Google Scholar] [CrossRef]
- MacAlister, C.A.; Ohashi-Ito, K.; Bergmann, D.C. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 2007, 445, 537–540. [Google Scholar] [CrossRef]
- Dombrecht, B.; Xue, G.P.; Sprague, S.J.; Kirkegaard, J.A.; Ross, J.J.; Reid, J.B.; Fitt, G.P.; Sewelam, N.; Schenk, P.M.; Manners, J.M.; et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 2007, 19, 2225–2245. [Google Scholar] [CrossRef]
- Qi, Y.; Zhou, L.; Han, L.; Zou, H.; Miao, K.; Wang, Y. PsbHLH1, a novel transcription factor involved in regulating anthocyanin biosynthesis in tree peony (Paeonia suffruticosa). Plant Physiol. Biochem. 2020, 154, 396–408. [Google Scholar] [CrossRef]
- Ma, Y.; Yang, G.; Duan, R.; Li, X.; Zeng, S.; Yan, Y.; Zheng, C.; Hu, Y. Transcriptome analysis of alfalfa (Medicago sativa L.) roots reveals overwintering changes in different varieties. Czech J. Genet. Plant Breed. 2024, 60, 97–104. [Google Scholar] [CrossRef]
- Li, H.; Yao, Y.; An, L.; Li, X.; Cui, Y.; Bai, Y.; Yao, X.; Wu, K. Isolation and expression analysis of the HvnAnt2 gene in qingke barley (Hordeum vulgare L. var. nudum Hook. f.) varieties with different grain colours. Czech J. Genet. Plant Breed. 2024, 60, 107–118. [Google Scholar] [CrossRef]
- So, K.; Ri, U.; Sun, S.; Che, H.; He, L.; Ri, H.; Zhang, Y. bHLH transcription factor from Lilium pumilum, LpbHLH144 confers the salt and alkali stress tolerance of tobacco. Plant Physiol. Biochem. 2025, 226, 110076. [Google Scholar] [CrossRef] [PubMed]
- Yuan, G.; Wu, Z.; Liu, X.; Li, T.; Teng, N. Characterization and functional analysis of LoUDT1, a bHLH transcription factor related to anther development in the lily oriental hybrid Siberia (Lilium spp.). Plant Physiol. Biochem. 2021, 166, 1087–1095. [Google Scholar] [CrossRef]
- An, W.; Sun, Y.; Gao, Z.; Liu, X.; Guo, Q.; Sun, S.; Zhang, M.; Han, Y.; Irfan, M.; Chen, L.; et al. LvbHLH13 regulates anthocyanin biosynthesis by activating the LvMYB5 promoter in lily (Lilium ‘Viviana’). Horticulturae 2024, 10, 926. [Google Scholar] [CrossRef]
- Feng, Y.; Guo, Z.; Zhong, J.; Liang, Y.; Zhang, P.; Sun, M. The LibHLH22 and LibHLH63 from Lilium ‘Siberia’ can positively regulate volatile terpenoid biosynthesis. Horticulturae 2023, 9, 459. [Google Scholar] [CrossRef]
- Fang, S.; Lin, M.; Ali, M.M.; Zheng, Y.; Yi, X.; Wang, S.; Chen, F.; Lin, Z. LhANS-rr1, LhDFR, and LhMYB114 regulate anthocyanin biosynthesis in flower buds of Lilium ‘Siberia’. Genes 2023, 14, 559. [Google Scholar] [CrossRef]
- Zhang, K.; Lyu, T.; Lyu, Y. Transcriptional insights into lily stem bulblet formation: Hormonal regulation, sugar metabolism, and transcriptional networks in LA Lily ‘Aladdin’. Horticulturae 2024, 10, 171. [Google Scholar] [CrossRef]
- Chinese Flora Editorial Committee. Flora Reipublicae Popularis Sinicae, Vol. 14: Liliaceae; Science Press: Beijing, China, 1980; p. 138. [Google Scholar]
- Wang, M.; Zhang, S.; Li, R.; Zhao, Q. Unraveling the specialized metabolic pathways in medicinal plant genomes: A review. Front. Plant Sci. 2024, 15, 1459533. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Haas, B.J. TransDecoder v5.0.0; Github Repository. Available online: https://github.com/TransDecoder/TransDecoder (accessed on 22 January 2025).
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007, 35, D61–D65. [Google Scholar] [CrossRef]
- Bairoch, A.; Apweiler, R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000, 28, 45–48. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Krylov, D.M.; Makarova, K.S.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; Rao, B.S.; et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004, 5, R7. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-Scale Protein Function Classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Zheng, Y.; Jiao, C.; Sun, H.; Rosli, H.G.; Pombo, M.A.; Zhang, P.; Banf, M.; Dai, X.; Martin, G.B.; Giovannoni, J.J.; et al. iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [PubMed]
- Thumuluri, V.; Almagro Armenteros, J.J.; Johansen, A.R.; Nielsen, H.; Winther, O. DeepLoc 2.0: Multi-label subcellular localization prediction using protein language models. Nucleic Acids Res. 2022, 50, W228–W234. [Google Scholar] [CrossRef]
- Høie, M.H.; Kiehl, E.N.; Petersen, B.; Nielsen, M.; Winther, O.; Nielsen, H.; Hallgren, J.; Marcatili, P. NetSurfP-3.0: Accurate and fast prediction of protein structural features by protein language models and deep learning. Nucleic Acids Res. 2022, 50, W510–W515. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Kondou, Y.; Nakazawa, M.; Kawashima, M.; Ichikawa, T.; Yoshizumi, T.; Suzuki, K.; Ishikawa, A.; Koshi, T.; Matsui, R.; Muto, S.; et al. Retarded growth of embryo1, a new basic helix-loop-helix protein, expresses in endosperm to control embryo growth. Plant Physiol. 2008, 147, 1924–1935. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, X.; Li, K.; Liu, H.; Lin, C. Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genet. 2013, 9, e1003861. [Google Scholar] [CrossRef] [PubMed]
- Di Marzo, M.; Roig-Villanova, I.; Zanchetti, E.; Caselli, F.; Gregis, V.; Bardetti, P.; Chiara, M.; Guazzotti, A.; Caporali, E.; Mendes, M.A.; et al. MADS-box and bHLH transcription factors coordinate transmitting tract development in Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 526. [Google Scholar] [CrossRef]
- Cifuentes-Esquivel, N.; Bou-Torrent, J.; Galstyan, A.; Gallemí, M.; Sessa, G.; Salla Martret, M.; Roig-Villanova, I.; Ruberti, I.; Martínez-García, J.F. The bHLH proteins BEE and BIM positively modulate the shade avoidance syndrome in Arabidopsis seedlings. Plant J. 2013, 75, 989–1002. [Google Scholar] [CrossRef]
- Zhang, T.; Lv, W.; Zhang, H.; Ma, L.; Li, P.; Ge, L.; Li, G. Genome-wide analysis of the basic Helix-Loop-Helix (bHLH) transcription factor family in maize. BMC Plant Biol. 2018, 18, 235. [Google Scholar] [CrossRef]
- Xin, Z.; Huang, H.; Li, T.; Liu, L.; Du, X.; Li, G.; Zhang, K.; Wang, D.; Yang, Y. Comprehensive analysis of bHLH genes in wheat and functional characterization of TabHLH319 in salt tolerance. Plant Cell Rep. 2025, 44, 199. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, A. Genomic Characterization and expression analysis of Basic Helix-Loop-Helix (bHLH) family genes in traditional Chinese herb Dendrobium officinale. Plants 2020, 9, 1044. [Google Scholar] [CrossRef]
- Wang, M.J.; Ou, Y.; Li, Z.; Zheng, Q.D.; Ke, Y.J.; Lai, H.P.; Lan, S.R.; Peng, D.H.; Liu, Z.J.; Ai, Y. Genome-wide identification and analysis of bHLH transcription factors related to anthocyanin biosynthesis in Cymbidium ensifolium. Int. J. Mol. Sci. 2023, 24, 3825. [Google Scholar] [CrossRef]
- Liu, R.; Wang, Y.; Tang, S.; Cai, J.; Liu, S.; Zheng, P.; Sun, B. Genome-wide identification of the tea plant bHLH transcription factor family and discovery of candidate regulators of trichome formation. Sci. Rep. 2021, 11, 10764. [Google Scholar] [CrossRef]
- Qin, S.; Liang, Y.; Ye, Y.; Wei, G.; Lin, Q.; Qin, W.; Wei, F. Genome-wide analysis of the bHLH gene family in Spatholobus suberectus identifies SsbHLH112 as a regulator of flavonoid biosynthesis. BMC Plant Biol. 2025, 25, 594. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, B.; Yuan, F. Genome-wide identification of bHLH transcription factors and functional analysis in salt gland development of the recretohalophyte sea lavender (Limonium bicolor). Hortic. Res. 2024, 11, uhae036. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Duan, X.; Jiang, H.; Sun, Y.; Tang, Y.; Yuan, Z.; Guo, J.; Liang, W.; Chen, L.; Yin, J.; et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006, 141, 1167–1184. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, R.; Khurana, R.; Malik, N.; Badoni, S.; Parida, S.K.; Kapoor, S.; Tyagi, A.K. bHLH142 regulates various metabolic pathway-related genes to affect pollen development and anther dehiscence in rice. Sci. Rep. 2017, 7, 43397. [Google Scholar] [CrossRef]
- Ortolan, F.; Fonini, L.S.; Pastori, T.; Mariath, J.E.; Saibo, N.J.; Margis-Pinheiro, M.; Lazzarotto, F. Tightly controlled expression of OsbHLH35 is critical for anther development in rice. Plant Sci. 2021, 302, 110716. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, Z.; Wang, M.; Zhang, M.; Chen, J.; Wu, H. Genome-Wide Identification and Expression Analysis of the bHLH Transcription Factor Family in Lilium bakerianum var. rubrum. Genes 2025, 16, 1153. https://doi.org/10.3390/genes16101153
Gu Z, Wang M, Zhang M, Chen J, Wu H. Genome-Wide Identification and Expression Analysis of the bHLH Transcription Factor Family in Lilium bakerianum var. rubrum. Genes. 2025; 16(10):1153. https://doi.org/10.3390/genes16101153
Chicago/Turabian StyleGu, Zhijia, Mingcheng Wang, Minhui Zhang, Junji Chen, and Hongzhi Wu. 2025. "Genome-Wide Identification and Expression Analysis of the bHLH Transcription Factor Family in Lilium bakerianum var. rubrum" Genes 16, no. 10: 1153. https://doi.org/10.3390/genes16101153
APA StyleGu, Z., Wang, M., Zhang, M., Chen, J., & Wu, H. (2025). Genome-Wide Identification and Expression Analysis of the bHLH Transcription Factor Family in Lilium bakerianum var. rubrum. Genes, 16(10), 1153. https://doi.org/10.3390/genes16101153






