Genome-Wide Patterns of Homozygosity and Heterozygosity and Candidate Genes in Greek Insular and Mainland Native Goats
Abstract
1. Introduction
2. Materials and Methods
2.1. Goat Samples, SNP Genotyping, and Data Editing
2.2. Genome-Wide Diversity and Principal Component Analysis
2.3. Detection of Runs of Homozygosity and Heterozygosity
2.4. Gene and Functional Annotation
3. Results
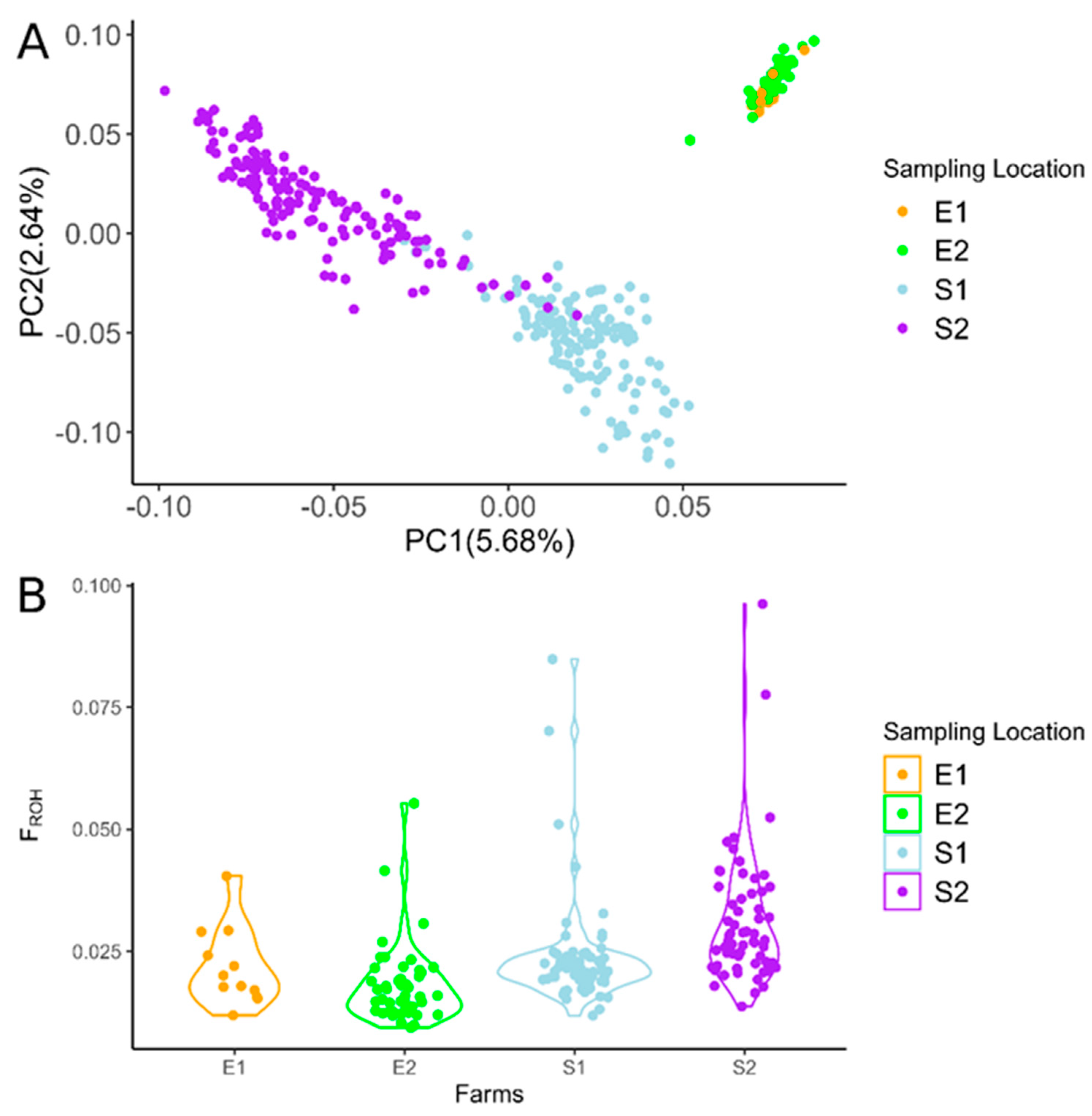
3.1. Genome-Wide Diversity and Principal Component Analysis
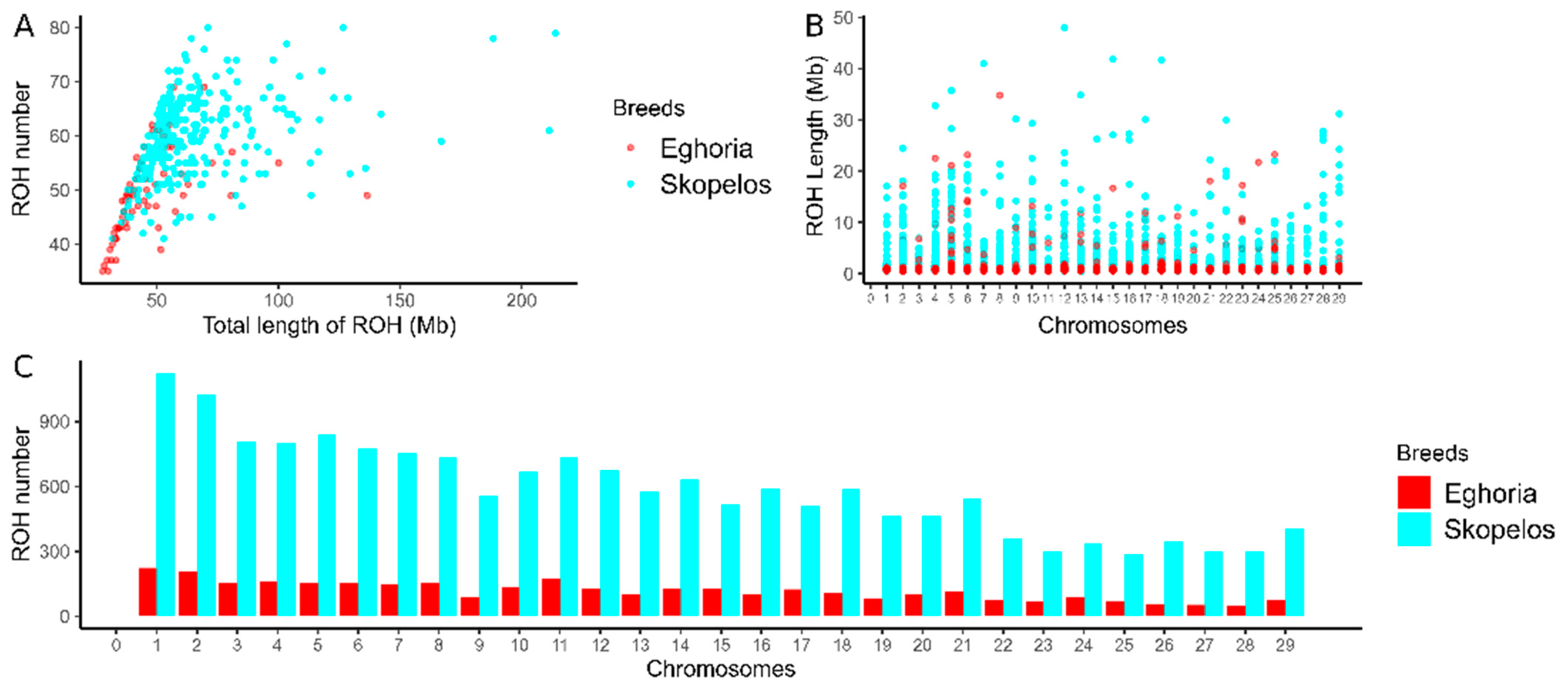
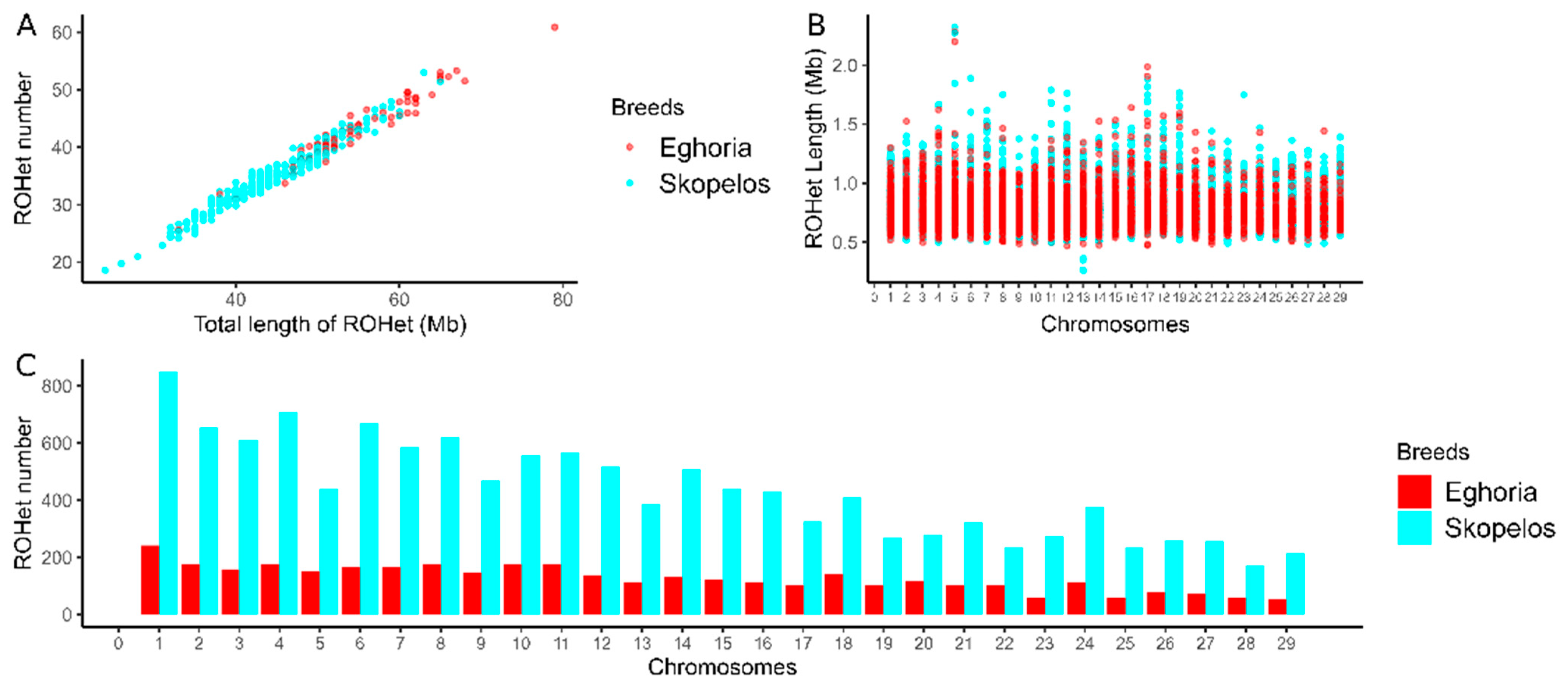
3.2. ROH and ROHet Patterns
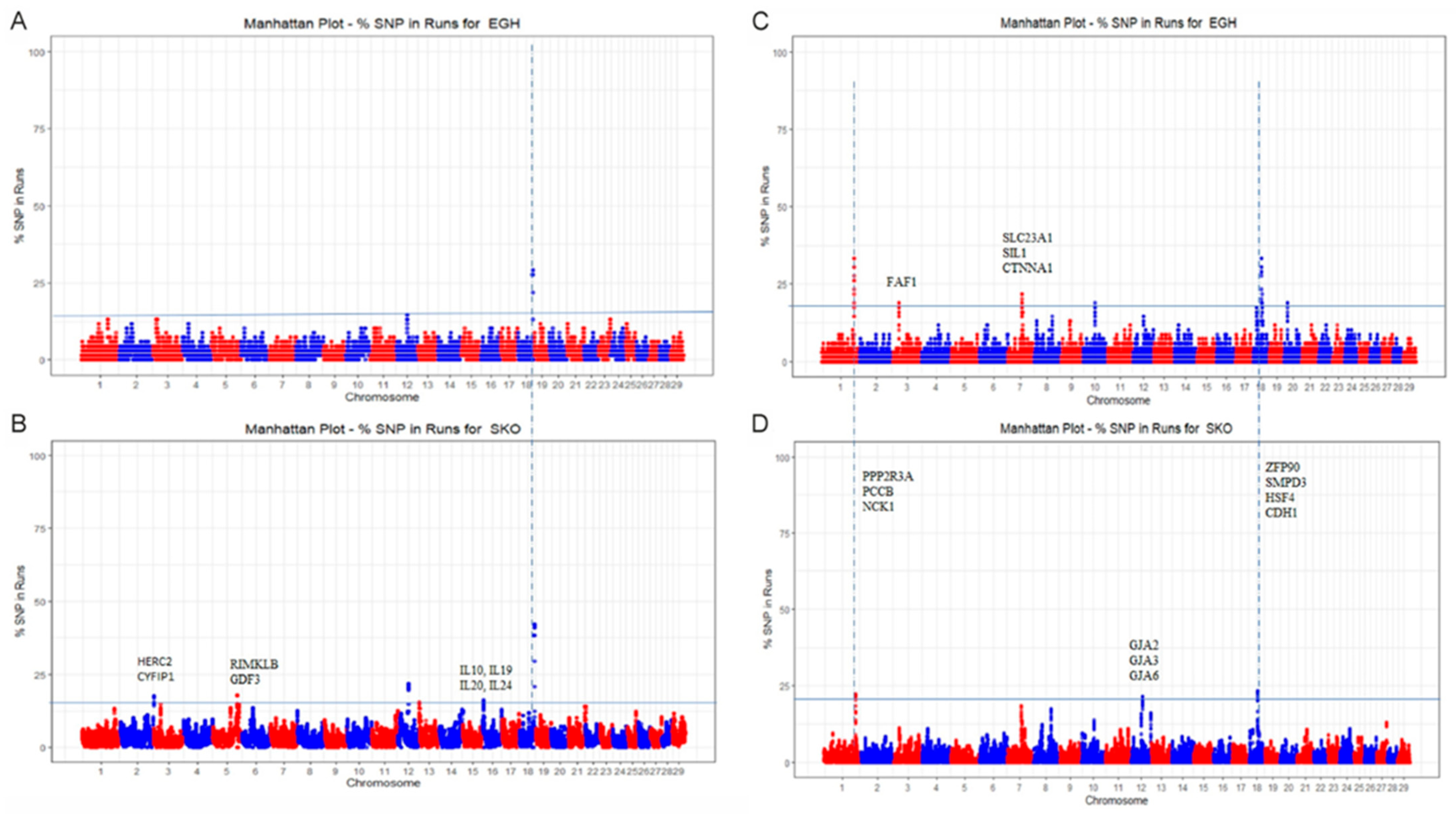
3.3. Gene Identification and Functional Annotation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ceballos, F.C.; Joshi, P.K.; Clark, D.W.; Ramsay, M.; Wilson, J.F. Runs of Homozygosity: Windows into Population History and Trait Architecture. Nat. Rev. Genet. 2018, 19, 220–234. [Google Scholar] [CrossRef] [PubMed]
- Broman, K.W.; Weber, J.L. Long homozygous chromosomal segments in reference families from the centre d’étude du Polymorphisme Humain. Am. J. Hum. Genet. 1999, 65, 1493–1500. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, T.F.; Amills, M.; Bertolini, F.; Rothschild, M.; Marras, G.; Boink, G.; Jordana, J.; Capote, J.; Carolan, S.; Hallsson, J.H.; et al. Patterns of homozygosity in insular and continental goat breeds. Genet. Sel. Evol. 2018, 50, 56. [Google Scholar] [CrossRef] [PubMed]
- Biscarini, F.; Mastrangelo, S.; Catillo, G.; Senczuk, G.; Ciampolini, R. Insights into genetic diversity, runs of homozygosity and heterozygosity-rich regions in Maremmana semi-feral cattle using pedigree and genomic data. Animals 2020, 10, 2285. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Mei, S.; Zhou, J.; Zhang, Y.; Qiao, M.; Sun, H.; Li, Z.; Li, L.; Dong, B.; Oyelami, F.O.; et al. Genome-wide assessment of runs of homozygosity and estimates of genomic inbreeding in a Chinese composite pig breed. Front. Genet. 2021, 12, 720081. [Google Scholar] [CrossRef]
- Selli, A.; Ventura, R.V.; Fonseca, P.A.S.; Buzanskas, M.E.; Andrietta, L.T.; Balieiro, J.C.C.; Brito, L.F. Detection and visualization of heterozygosity-rich regions and runs of homozygosity in worldwide sheep populations. Animals 2021, 11, 2696. [Google Scholar] [CrossRef]
- Bertolini, F.; Cardoso, T.F.; Marras, G.; Nicolazzi, E.L.; Rothschild, M.F.; Amills, M. Genome-wide patterns of homozygosity provide clues about the population history and adaptation of goats. Genet. Sel. Evol. 2018, 50, 59. [Google Scholar] [CrossRef]
- Signer-Hasler, H.; Henkel, J.; Bangerter, E.; Bulut, Z.; The VarGoats Consortium; Drögemüller, C.; Leeb, T.; Flury, C. Runs of Homozygosity in Swiss Goats Reveal Genetic Changes Associated with Domestication and Modern Selection. Genet. Sel. Evol. 2022, 54, 6. [Google Scholar] [CrossRef]
- Manunza, A.; Diaz, J.R.; Sayre, B.L.; Cozzi, P.; Bobbo, T.; Deniskova, T.; Dotsev, A.; Zinovieva, N.; Stella, A. Discovering Novel Clues of Natural Selection on Four Worldwide Goat Breeds. Sci. Rep. 2023, 13, 2110. [Google Scholar] [CrossRef]
- Zhao, Q.; Huang, C.; Chen, Q.; Su, Y.; Zhang, Y.; Wang, R.; Su, R.; Xu, H.; Liu, S.; Ma, Y.; et al. Genomic Inbreeding and Runs of Homozygosity Analysis of Cashmere Goat. Animals 2024, 14, 1246. [Google Scholar] [CrossRef]
- Dixit, S.P.; Singh, S.; Ganguly, I.; Bhatia, A.K.; Sharma, A.; Kumar, N.A.; Dang, A.K.; Jayakumar, S. Genome-wide runs of homozygosity revealed selection signatures in Bos indicus. Front. Genet. 2020, 11, 92. [Google Scholar] [CrossRef] [PubMed]
- Zavarez, L.B.; Utsunomiya, Y.T.; Carmo, A.S.; Neves, H.H.; Carvalheiro, R.; Ferenčaković, M.; Pérez O’Brien, A.M.; Curik, I.; Cole, J.B.; Van Tassell, C.P.; et al. Assessment of Autozygosity in Nellore Cows (Bos indicus) through High-Density SNP Genotypes. Front. Genet. 2015, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Guldbrandtsen, B.; Bosse, M.; Lund, M.S.; Sahana, G. Runs of Homozygosity and Distribution of Functional Variants in the Cattle Genome. BMC Genom. 2015, 16, 542. [Google Scholar] [CrossRef]
- Szmatoła, T.; Gurgul, A.; Jasielczuk, I.; Ząbek, T.; Ropka-Molik, K.; Litwińczuk, Z.; Bugno-Poniewierska, M. A Comprehensive Analysis of Runs of Homozygosity of Eleven Cattle Breeds Representing Different Production Types. Animals 2019, 9, 1024. [Google Scholar] [CrossRef]
- Ghoreishifar, S.M.; Moradi-Shahrbabak, H.; Fallahi, M.H.; Jalil Sarghale, A.; Moradi-Shahrbabak, M.; Abdollahi-Arpanahi, R.; Khansefid, M. Correction to: Genomic Measures of Inbreeding Coefficients and Genome-Wide Scan for Runs of Homozygosity Islands in Iranian River Buffalo, Bubalus bubalis. BMC Genet. 2020, 21, 42, Erratum in BMC Genet. 2020, 21, 16. [Google Scholar] [CrossRef]
- Mulim, H.A.; Brito, L.F.; Pinto, L.F.; Ferraz, J.B.; Grigoletto, L.; Silva, M.R.; Pedrosa, V.B. Characterization of Runs of Homozygosity, Heterozygosity-Enriched Regions, and Population Structure in Cattle Populations Selected for Different Breeding Goals. BMC Genom. 2021, 23, 209. [Google Scholar] [CrossRef]
- Tsartsianidou, V.; Sánchez-Molano, E.; Kapsona, V.V.; Basdagianni, Z.; Chatziplis, D.; Arsenos, G.; Triantafyllidis, A.; Banos, G. A comprehensive genome-wide scan detects genomic regions related to local adaptation and climate resilience in Mediterranean domestic sheep. Genet. Sel. Evol. 2021, 53, 90. [Google Scholar] [CrossRef]
- Mastrangelo, S.; Ciani, E.; Sardina, M.T.; Sottile, G.; Pilla, F.; Portolano, B.; The Bi. Ov. Ita Consortium. Runs of Homozygosity Reveal Genome-Wide Autozygosity in Italian Sheep Breeds. Anim. Genet. 2018, 49, 71–81. [Google Scholar] [CrossRef]
- Mastrangelo, S.; Tolone, M.; Sardina, M.T.; Sottile, G.; Sutera, A.M.; Di Gerlando, R.; Portolano, B. Genome-Wide Scan for Runs of Homozygosity Identifies Potential Candidate Genes Associated with Local Adaptation in Valle del Belice Sheep. Genet. Sel. Evol. 2017, 49, 84. [Google Scholar] [CrossRef]
- Talebi, R.; Szmatoła, T.; Mészáros, G.; Qanbari, S. Runs of homozygosity in modern chicken revealed by sequence data. G3 Genes Genomes Genet. 2020, 10, 4615–4623. [Google Scholar] [CrossRef]
- Keller, M.C.; Visscher, P.M.; Goddard, M.E. Quantification of inbreeding due to distant ancestors and its detection using dense single nucleotide polymorphism data. Genetics 2011, 189, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Nagy, I.; Nguyen, T.A. Characterizing and Eliminating the Inbreeding Load. Vet. Sci. 2024, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.L.; Hall, S.J.; Del Corvo, M.; Ballingall, K.T.; Colli, L.; Ajmone Marsan, P.; Biscarini, F. Inbreeding and Purging at the Genomic Level: The Chillingham Cattle Reveal Extensive, Non-Random SNP Heterozygosity. Anim. Genet. 2016, 47, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Arias, K.D.; Fernandez, I.; Traoré, A.; Goyache, F. West African Cattle Share Non-Random Heterozygosity-Rich Region Islands Enriched on Adaptation-Related Genes Despite Their Different Origins. Front. Anim. Sci. 2024, 5, 1468575. [Google Scholar] [CrossRef]
- Falchi, L.; Cesarani, A.; Criscione, A.; Hidalgo, J.; Garcia, A.; Mastrangelo, S.; Macciotta, N.P.P. Effect of Genotyping Density on the Detection of Runs of Homozygosity and Heterozygosity in Cattle. J. Anim. Sci. 2024, 102, skae147. [Google Scholar] [CrossRef]
- Li, G.; Tang, J.; Huang, J.; Jiang, Y.; Fan, Y.; Wang, X.; Ren, J. Genome-wide estimates of runs of homozygosity, heterozygosity, and genetic load in two Chinese indigenous goat breeds. Front. Genet. 2022, 13, 774196. [Google Scholar] [CrossRef]
- Somenzi, E.; Senczuk, G.; Ciampolini, R.; Cortellari, M.; Vajana, E.; Tosser-Klopp, G.; Pilla, F.; Ajmone-Marsan, P.; Crepaldi, P.; Colli, L. The SNP-Based Profiling of Montecristo Feral Goat Populations Reveals a History of Isolation, Bottlenecks, and the Effects of Management. Genes 2022, 13, 213. [Google Scholar] [CrossRef]
- Chessari, G.; Criscione, A.; Marletta, D.; Crepaldi, P.; Portolano, B.; Manunza, A.; Cesarani, A.; Biscarini, F.; Mastrangelo, S. Characterization of heterozygosity-rich regions in Italian and worldwide goat breeds. Sci. Rep. 2024, 14, 3. [Google Scholar] [CrossRef]
- Ruan, D.; Yang, J.; Zhuang, Z.; Ding, R.; Huang, J.; Quan, J.; Gu, T.; Hong, L.; Zheng, E.; Li, Z.; et al. Assessment of heterozygosity and genome-wide analysis of heterozygosity regions in two Duroc pig populations. Front. Genet. 2022, 12, 812456. [Google Scholar] [CrossRef]
- Bordonaro, S.; Chessari, G.; Mastrangelo, S.; Senczuk, G.; Chessa, S.; Castiglioni, B.; Tumino, S.; Marletta, D.; Criscione, A. Genome-wide population structure, homozygosity, and heterozygosity patterns of Nero Siciliano pig in the framework of Italian and cosmopolitan breeds. Anim. Genet. 2023, 54, 591–605. [Google Scholar] [CrossRef]
- Bizarria dos Santos, W.; Pimenta Schettini, G.; Fonseca, M.G.; Pereira, G.L.; Loyola Chardulo, L.A.; Rodrigues Machado Neto, O.; Baldassini, W.A.; Nunes de Oliveira, H.; Abdallah Curi, R. Fine-Scale Estimation of Inbreeding Rates, Runs of Homozygosity and Genome-Wide Heterozygosity Levels in the Mangalarga Marchador Horse Breed. J. Anim. Breed. Genet. 2021, 138, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Amills, M.; Capote, J.; Tosser-Klopp, G. Goat domestication and breeding: A jigsaw of historical, biological and molecular data with missing pieces. Anim. Genet. 2017, 48, 631–644. [Google Scholar] [CrossRef] [PubMed]
- EUROSTAT. Goats Population—Annual Data. 2023. Available online: https://ec.europa.eu/eurostat/databrowser/view/apro_mt_lsgoat/default/table?lang=en (accessed on 3 May 2023).
- Gelasakis, A.I.; Rose, G.; Giannakou, R.; Valergakis, G.E.; Theodoridis, A.; Fortomaris, P.; Arsenos, G. Typology and characteristics of dairy goat production systems in Greece. Livest. Sci. 2017, 197, 22–29. [Google Scholar] [CrossRef]
- FAO. Domestic Animal Diversity Information System (DAD-IS). 2022. Available online: https://www.fao.org/dad-is/browse-by-country-and-species/en/ (accessed on 3 May 2023).
- Krina, A.; Koutsias, N.; Pleniou, M.; Xystrakis, F. Climatic classification of Greece: Update—Future estimation—Relation with forest vegetation. In Proceedings of the 18th Congress of the Hellenic Forestry Society & International Workshop, Edessa, Greece, 2 October 2017; pp. 1088–1095. [Google Scholar]
- Pappas, B.G.; Boyazoglu, J.; Vasiloudis, C. The Skopelos Goat Breed of Greece. Anim. Genet. Resour. Inf. 1992, 9, 69–76. [Google Scholar] [CrossRef]
- Nardone, A.; Ronchi, B.; Lacetera, N.; Ranieri, M.S.; Bernabucci, U. Effects of Climate Changes on Animal Production and Sustainability of Livestock Systems. Livest. Sci. 2010, 130, 57–69. [Google Scholar] [CrossRef]
- Cozzi, P.; Manunza, A.; Ramirez-Diaz, J.; Tsartsianidou, V.; Gkagkavouzis, K.; Peraza, P.; Johansson, A.M.; Arranz, J.J.; Freire, F.; Kusza, S.; et al. SMARTER-Database: A Tool to Integrate SNP Array Datasets for Sheep and Goat Breeds. Gigabyte 2024, 2024, gigabyte139. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.A.M.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 1–16. [Google Scholar] [CrossRef]
- Meyermans, R.; Gorssen, W.; Buys, N.; Janssens, S. How to study runs of homozygosity using plink? A guide for analyzing medium density SNP data in livestock and pet species. BMC Genom. 2020, 21, 94. [Google Scholar] [CrossRef]
- Manichaikul, A.; Mychaleckyj, J.C.; Rich, S.S.; Daly, K.; Sale, M.; Chen, W.M. Robust relationship inference in genome-wide association studies. Bioinformatics 2010, 26, 2867–2873. [Google Scholar] [CrossRef]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef]
- Berg, P.; Groeneveld, L.F.; Brekke, C.; Våge, D.I.; Sørheim, K.M.; Grøva, L. Genetic characterization of a small closed island population of Norwegian coastal goat. Acta Agric. Scand. 2020, 69, 47–52. [Google Scholar] [CrossRef]
- McQuillan, R.; Leutenegger, A.L.; Abdel-Rahman, R.; Franklin, C.S.; Pericic, M.; Barac-Lauc, L.; Smolej-Narancic, N.; Janicijevic, B.; Polasek, O.; Tenesa, A.; et al. Runs of Homozygosity in European populations. Am. J. Hum. Genet. 2008, 83, 359–372. [Google Scholar] [CrossRef]
- Biscarini, F.; Manunza, A.; Cozzi, P.; Stella, A. Common heterozygosity-rich regions across the genomes of commercial and local goat breeds. In Proceedings of the 12th World Congress on Genetics Applied to Livestock Production, Rotterdam, The Netherlands, 3–8 July 2022; pp. 798–801. [Google Scholar]
- Pemberton, T.J.; Absher, D.; Feldman, M.W.; Myers, R.M.; Rosenberg, N.A.; Li, J.Z. Genomic patterns of homozygosity in worldwide human populations. Am. J. Hum. Genet. 2012, 91, 275–292. [Google Scholar] [CrossRef] [PubMed]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Martin, F.J.; Gall, A.; Szpak, M.; Flicek, P. Accessing livestock resources in Ensembl. Front. Genet. 2021, 12, 650228. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. G:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, 191–198. [Google Scholar] [CrossRef]
- Wanjala, G.; Kusuma Astuti, P.; Bagi, Z.; Kichamu, N.; Strausz, P.; Kusza, S. A review on the potential effects of environmental and economic factors on sheep genetic diversity: Consequences of climate change. Saudi J. Biol. Sci. 2023, 30, 103505. [Google Scholar] [CrossRef]
- Abioja, M.O.; Logunleko, M.O.; Majekodunmi, B.C.; Adekunle, E.O.; Shittu, O.O.; Odeyemi, A.J.; Nwosu, E.U.; Oke, O.E.; Iyasere, O.S.; Abiona, J.A.; et al. Roles of candidate genes in the adaptation of goats to heat stress: A review. Small Rumin. Res. 2023, 218, 106878. [Google Scholar] [CrossRef]
- Rangkasenee, N.; Murani, E.; Brunner, R.; Schellander, K.; Cinar, M.U.; Scholz, A.M.; Luther, H.; Hofer, A.; Ponsuksili, S.; Wimmers, K. KRT8, FAF1 and PTH1R gene polymorphisms are associated with leg weakness traits in pigs. Mol. Biol. Rep. 2013, 40, 2859–2866. [Google Scholar] [CrossRef]
- Guang-Xin, E.; Duan, X.H.; Zhang, J.H.; Huang, Y.F.; Zhao, Y.J.; Na, R.S.; Zhao, Z.Q.; Ma, Y.H.; Chu, M.X.; Basang, W.D.; et al. Genome-wide selection signatures analysis of litter size in Dazu black goats using single-nucleotide polymorphism. 3 Biotech 2019, 9, 336. [Google Scholar] [CrossRef]
- Caroprese, M.; Ciliberti, M.G.; Albenzio, M.; Sevi, A. Climate change impact on immune response in sheep. In Sheep Production Adapting to Climate Change; Springer: Singapore, 2017; pp. 95–116. [Google Scholar] [CrossRef]
- Naveed, M.; Kazmi, S.K.; Amin, M.; Asif, Z.; Islam, U.; Shahid, K.; Tehreem, S. Comprehensive review on the molecular genetics of autosomal recessive primary microcephaly (MCPH). Genet. Res. 2018, 100, e7. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Wu, Q.; Sun, L.; Pan, N.C.; Wang, X. Cenpj regulates cilia disassembly and neurogenesis in the developing mouse cortex. J. Neurosci. 2019, 39, 1994–2010. [Google Scholar] [CrossRef] [PubMed]
- Michailidou, S.; Tsangaris, G.T.; Tzora, A.; Skoufos, I.; Banos, G.; Argiriou, A.; Arsenos, G. Analysis of genome-wide DNA arrays reveals the genomic population structure and diversity in autochthonous Greek goat breeds. PLoS ONE 2019, 14, e0226179. [Google Scholar] [CrossRef] [PubMed]
- Colli, L.; Milanesi, M.; Talenti, A.; Bertolini, F.; Chen, M.; Crisà, A.; Daly, K.G.; Del Corvo, M.; Guldbrandtsen, B.; Lenstra, J.A.; et al. Genome-wide SNP profiling of worldwide goat populations reveals strong partitioning of diversity and highlights post-domestication migration routes. Genet. Sel. Evol. 2018, 50, 58. [Google Scholar] [CrossRef]
- Monau, P.I.; Visser, C.; Muchadeyi, F.C.; Okpeku, M.; Nsoso, S.J.; Van Marle-Köster, E. Population structure of indigenous southern African goats based on the Illumina Goat50K SNP panel. Trop. Anim. Health Prod. 2020, 52, 1795–1802. [Google Scholar] [CrossRef]
- Browning, S.R.; Browning, B.L. Identity by descent between distant relatives: Detection and applications. Annu. Rev. Genet. 2012, 46, 617–633. [Google Scholar] [CrossRef]
- Bertolini, F.; Servin, B.; Talenti, A.; Rochat, E.; Kim, E.S.; Oget, C.; Palhière, I.; Crisà, A.; Catillo, G.; Steri, R.; et al. Signatures of selection and environmental adaptation across the goat genome post-domestication. Genet. Sel. Evol. 2018, 50, 57. [Google Scholar] [CrossRef]
- Kim, E.S.; Elbeltagy, A.R.; Aboul-Naga, A.M.; Rischkowsky, B.; Sayre, B.; Mwacharo, J.M.; Rothschild, M.F. Multiple genomic signatures of selection in goats and sheep indigenous to a hot arid environment. Heredity 2016, 116, 255–264. [Google Scholar] [CrossRef]
- Mdladla, K.; Dzomba, E.F.; Muchadeyi, F.C. Landscape genomics and pathway analysis to understand genetic adaptation of South African indigenous goat populations. Heredity 2018, 120, 369–378. [Google Scholar] [CrossRef]
- Onzima, R.B.; Upadhyay, M.R.; Doekes, H.P.; Brito, L.F.; Bosse, M.; Kanis, E.; Groenen, M.A.M.; Crooijmans, R.P.M.A. Genome-wide characterization of selection signatures and runs of homozygosity in Ugandan goat breeds. Front. Genet. 2018, 9, 318. [Google Scholar] [CrossRef] [PubMed]
- Teubner, B.; Michel, V.; Pesch, J.; Lautermann, J.; Cohen-Salmon, M.; Söhl, G.; Jahnke, K.; Winterhager, E.; Herberhold, C.; Hardelin, J.P.; et al. Connexin30 (Gjb6)-deficiency causes severe hearing impairment and lack of endocochlear potential. Hum. Mol. Genet. 2003, 12, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Lamartine, J.; Munhoz Essenfelder, G.; Kibar, Z.; Lanneluc, I.; Callouet, E.; Laoudj, D.; Lemaître, G.; Hand, C.; Hayflick, S.J.; Zonana, J.; et al. Mutations in GJB6 cause hidrotic ectodermal dysplasia. Nat. Genet. 2000, 26, 142–144. [Google Scholar] [CrossRef] [PubMed]
- Batissoco, A.C.; Abreu-Silva, R.S.; Braga, M.C.C.; Lezirovitz, K.; Della-Rosa, V.; Alfredo, T.; Otto, P.A.; Mingroni-Netto, R.C. Prevalence of GJB2 (connexin-26) and GJB6 (connexin-30) mutations in a cohort of 300 Brazilian hearing-impaired individuals: Implications for diagnosis and genetic counseling. Ear Hear. 2009, 30, 1–7. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, X.; Yan, H.; Zeng, J.; Ma, S.; Niu, Y.; Zhou, G.; Jiang, Y.; Chen, Y. Comparative transcriptome analysis of fetal skin reveals key genes related to hair follicle morphogenesis in cashmere goats. PLoS ONE 2016, 11, e0151118. [Google Scholar] [CrossRef]

| Run | Size Class (Mb) | Total Number/Total Length (Mb) | |
|---|---|---|---|
| Eghoria | Skopelos | ||
| ROH | 0–6 | 3363/2665.53 | 16,734/15,031.79 |
| 6–12 | 24/181.33 | 183/1532.23 | |
| 12–24 | 15/234.97 | 81/1303.51 | |
| 24–48 | 1/34.79 | 22/691.21 | |
| ROHet | 0.2–0.4 | 0/0 | 4/1.23 |
| 0.4–0.8 | 2309/1600.44 | 7770/5381.66 | |
| 0.8–1.6 | 1380/1291.94 | 4839/4502.14 | |
| 1.6–3.2 | 6/11.65 | 31/54.95 | |
| Breed | Chr | Start (bp) | End (bp) | Length (bp) | SNPs | ALD | Genes |
|---|---|---|---|---|---|---|---|
| ROH | |||||||
| EGH | 18 1 | 60,120,713 | 61,776,542 | 1,655,829 | 19 | 0.52 | 33 |
| SKO | 2 | 135,434,648 | 136,405,967 | 1,655,829 | 17 | 0.24 | 11 |
| 5 | 99,979,645 | 100,140,908 | 161,263 | 4 | 0.24 | 16 | |
| 12 | 43,598,144 | 44,611,581 | 1,013,437 | 19 | 0.28 | 1 | |
| 16 | 4,091,765 | 4,282,942 | 191,177 | 5 | 0.21 | 12 | |
| 18 1 | 60,120,713 | 61,813,628 | 1,692,915 | 21 | 0.39 | 33 | |
| ROHet | |||||||
| EGH | 1 2 | 131,931,194 | 132,736,269 | 805,075 | 17 | 0.28 | 9 |
| 3 | 24,908,534 | 25,076,519 | 167,985 | 5 | 0.26 | 3 | |
| 7 | 60,015,530 | 60,137,051 | 121,521 | 5 | 0.33 | 16 | |
| 18 2 | 36,283,603 | 37,295,931 | 1,012,328 | 18 | 0.27 | 60 | |
| 18 | 39,636,695 | 39,926,799 | 290,104 | 7 | 0.35 | 4 | |
| 20 | 13,847,556 | 14,250,543 | 402,987 | 9 | 0.29 | 11 | |
| SKO | 1 2 | 131,970,293 | 132,501,737 | 531,444 | 11 | 0.39 | 9 |
| 12 | 49,884,737 | 50,631,677 | 746,940 | 15 | 0.30 | 25 | |
| 18 2 | 36,360,921 | 37,006,578 | 645,657 | 12 | 0.30 | 46 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsartsianidou, V.; Otapasidis, A.; Papakostas, S.; Karaiskou, N.; Vouraki, S.; Triantafyllidis, A. Genome-Wide Patterns of Homozygosity and Heterozygosity and Candidate Genes in Greek Insular and Mainland Native Goats. Genes 2025, 16, 27. https://doi.org/10.3390/genes16010027
Tsartsianidou V, Otapasidis A, Papakostas S, Karaiskou N, Vouraki S, Triantafyllidis A. Genome-Wide Patterns of Homozygosity and Heterozygosity and Candidate Genes in Greek Insular and Mainland Native Goats. Genes. 2025; 16(1):27. https://doi.org/10.3390/genes16010027
Chicago/Turabian StyleTsartsianidou, Valentina, Antonis Otapasidis, Spiros Papakostas, Nikoleta Karaiskou, Sotiria Vouraki, and Alexandros Triantafyllidis. 2025. "Genome-Wide Patterns of Homozygosity and Heterozygosity and Candidate Genes in Greek Insular and Mainland Native Goats" Genes 16, no. 1: 27. https://doi.org/10.3390/genes16010027
APA StyleTsartsianidou, V., Otapasidis, A., Papakostas, S., Karaiskou, N., Vouraki, S., & Triantafyllidis, A. (2025). Genome-Wide Patterns of Homozygosity and Heterozygosity and Candidate Genes in Greek Insular and Mainland Native Goats. Genes, 16(1), 27. https://doi.org/10.3390/genes16010027






