Genome-Wide Survey of the Potential Function of CrLBDs in Catharanthus roseus MIA Biosynthesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of LBD Gene Family in C. roseus
2.2. Multiple Sequence Alignment and Phylogenetic Analysis
2.3. Gene Structures, Conserved Motifs Analysis, and Cis-Element Analysis
2.4. Chromosomal Distribution and Gene Duplication of CrLBD Genes
2.5. Transcription Profiling Based on RNA-Seq Data and RT-qPCR Validation
2.6. Plant Materials, Growth Conditions, and MIA Accumulation
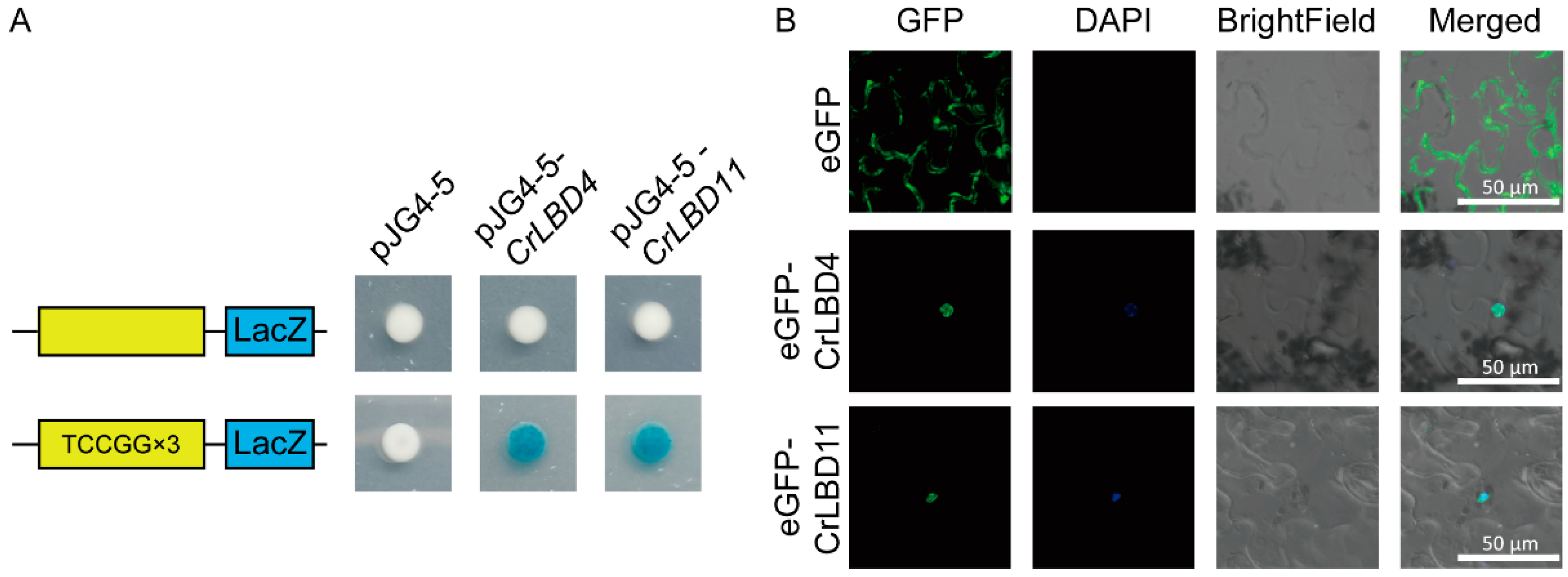
2.7. Y1H Assays and Subcellular Localization
2.8. Statistical Analysis
3. Results
3.1. Identification of the LBD Gene Family in C. roseus
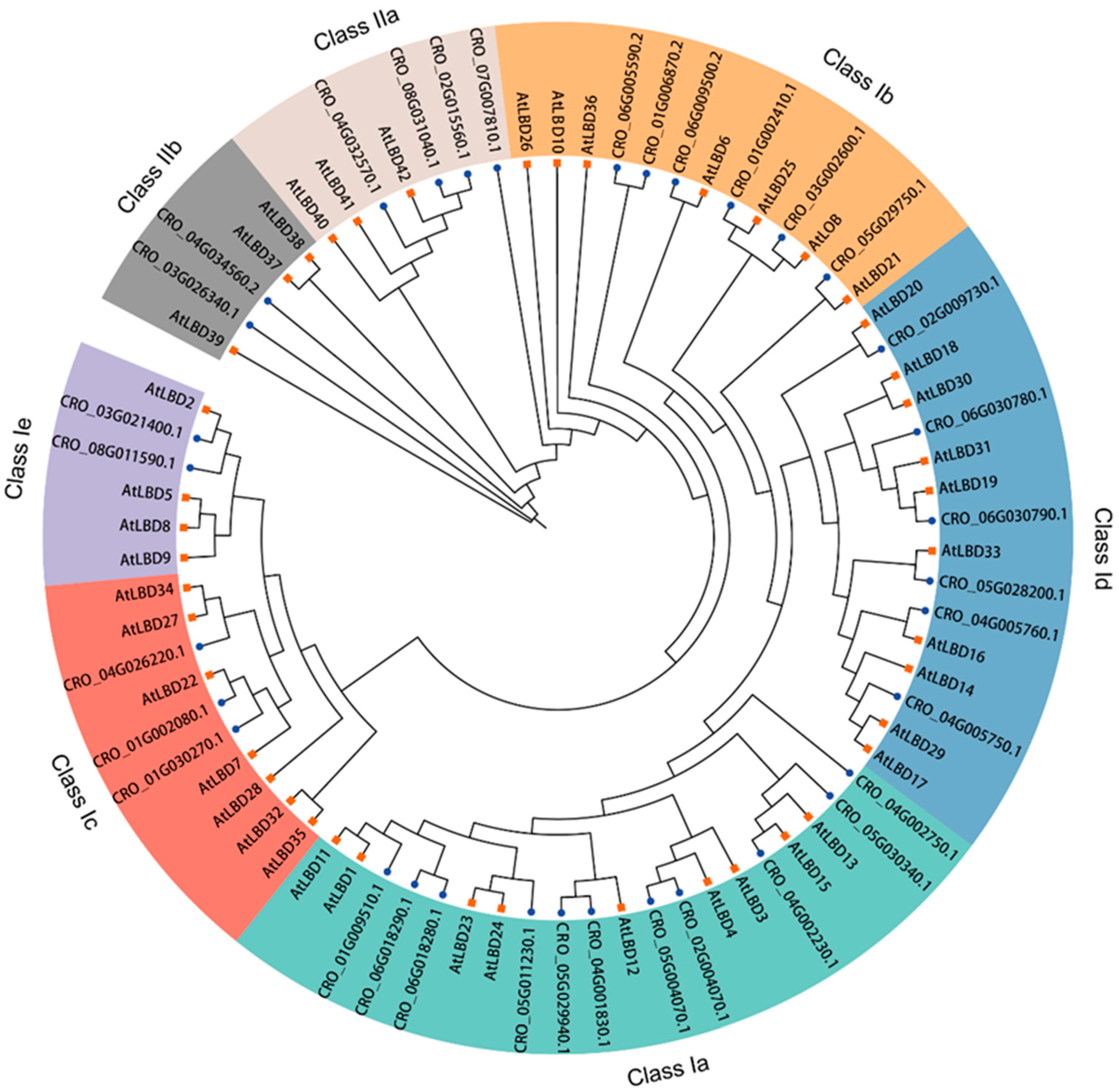
3.2. Phylogenetic and Conservative Analysis of the CrLBD Gene Family
3.3. Gene Structure and Motif Composition of the CrLBD Gene Family
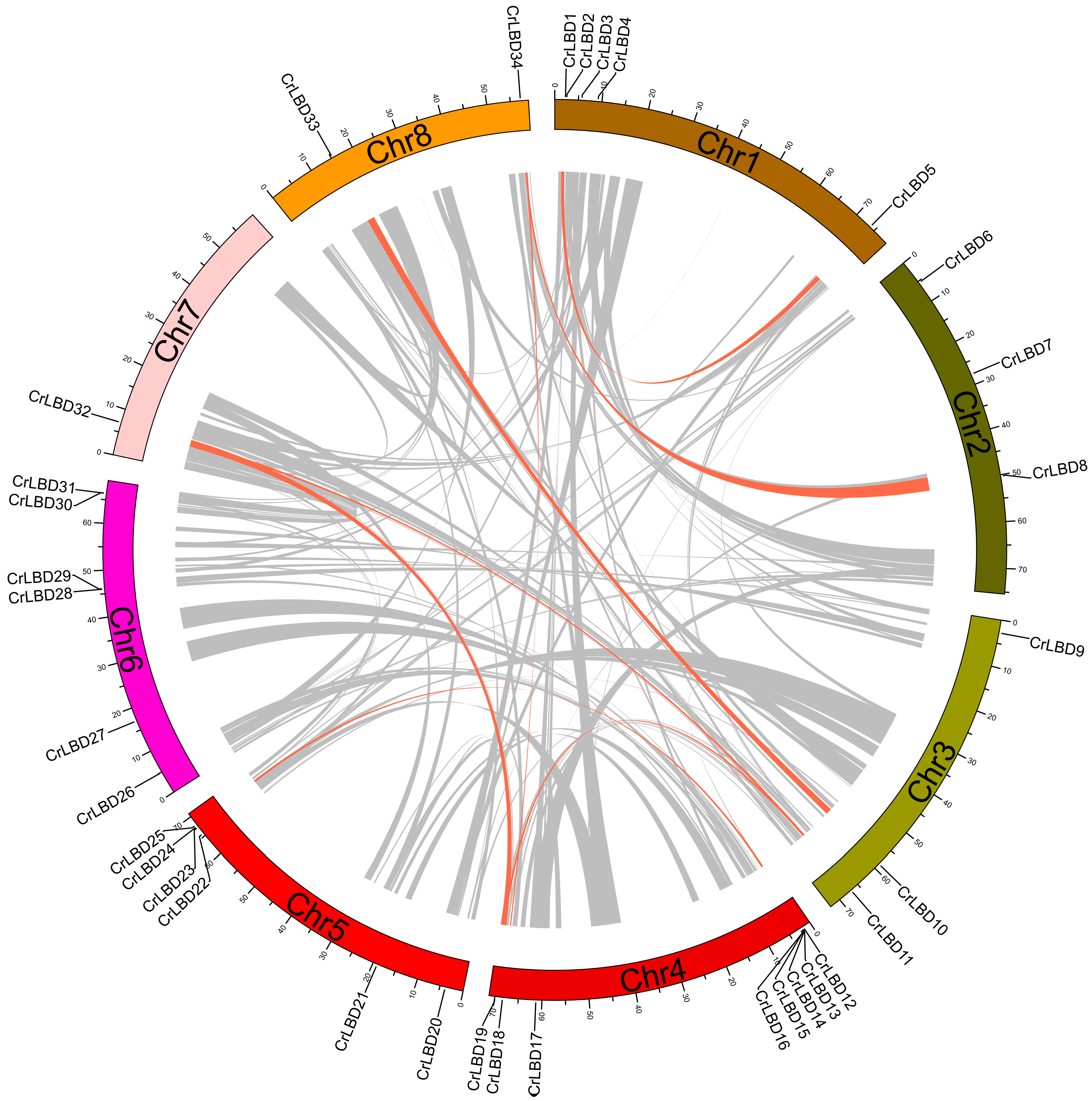
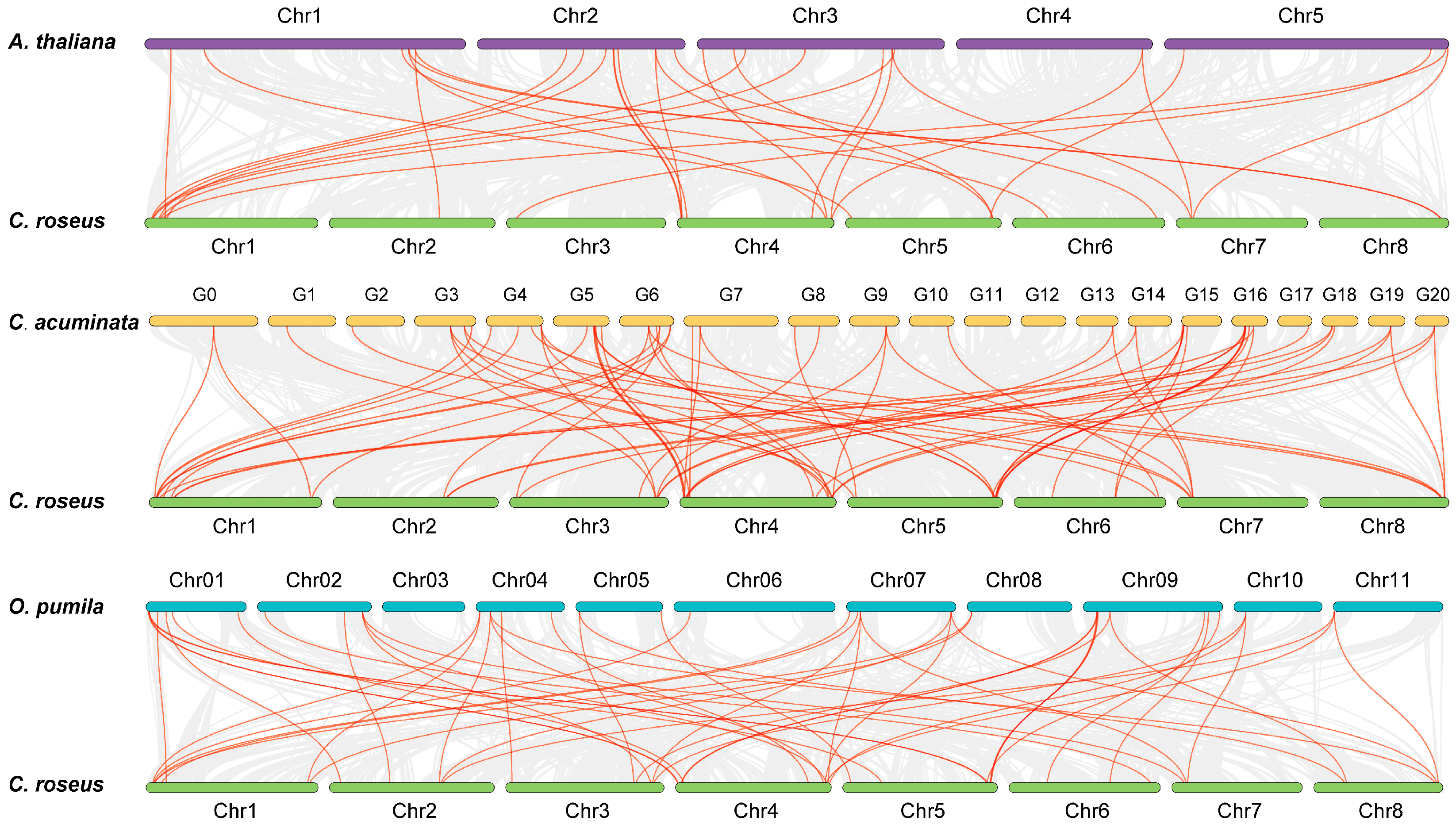
3.4. Chromosomal Distribution and Synteny Analysis of the CrLBD Gene Family
3.5. Cis-Element Analysis in the Putative Promoter of CrLBD Genes
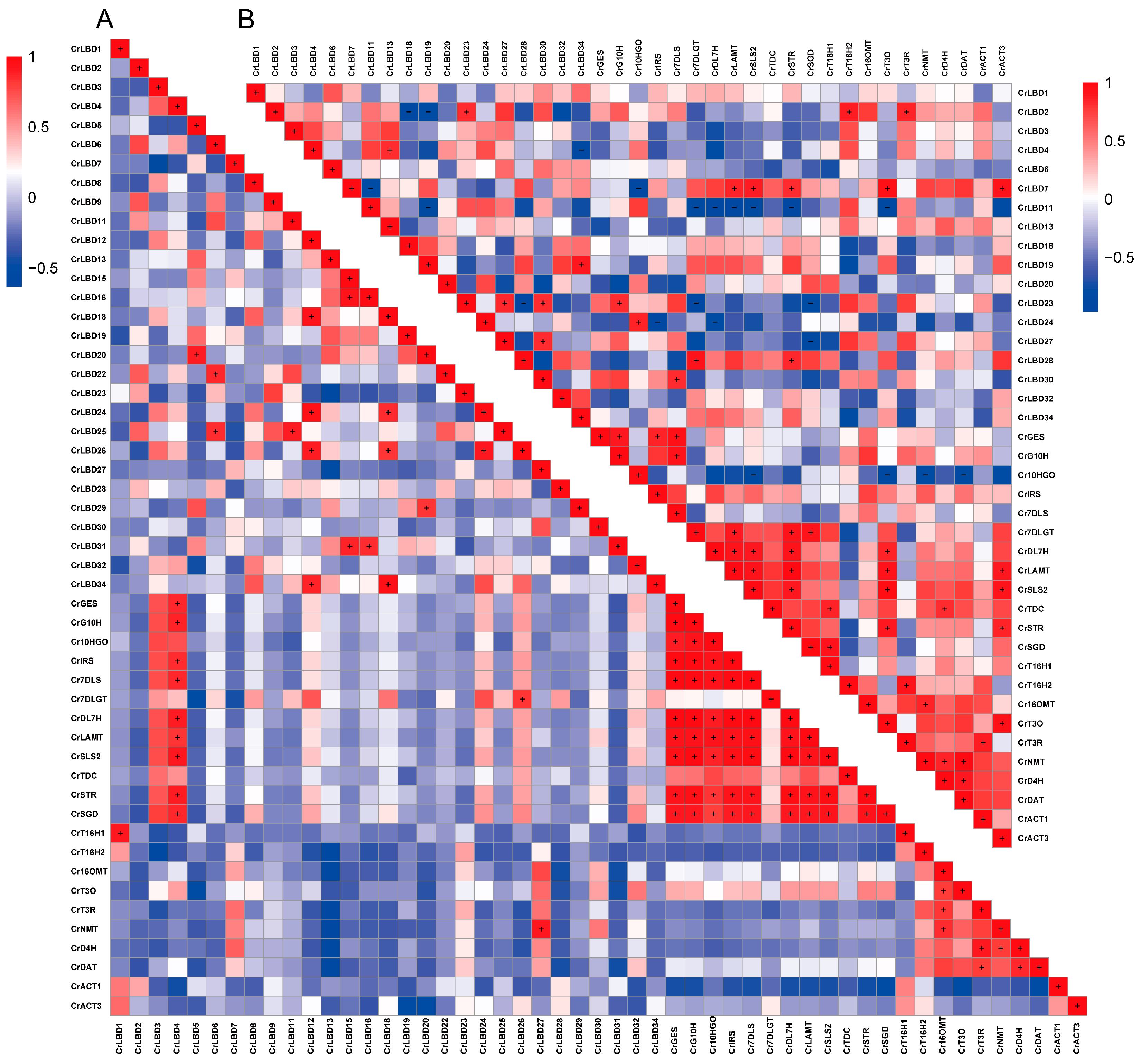
3.6. Potential Functions of CrLBDs in MIA Biosynthesis
3.7. CrLBD4/11 as Transcription Factors Might Regulate MIA Biosynthetic Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Kim, J.; Buell, C.R. A Revolution in Plant Metabolism: Genome-Enabled Pathway Discovery. Plant Physiol. 2015, 169, 1532–1539. [Google Scholar] [CrossRef] [PubMed]
- Nutzmann, H.W.; Huang, A.; Osbourn, A. Plant metabolic clusters—From genetics to genomics. New Phytol. 2016, 211, 771–789. [Google Scholar] [CrossRef] [PubMed]
- Zhan, C.; Shen, S.; Yang, C.; Liu, Z.; Fernie, A.R.; Graham, I.A.; Luo, J. Plant metabolic gene clusters in the multi-omics era. Trends Plant Sci. 2022, 27, 981–1001. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wood, J.C.; Vu, A.H.; Hamilton, J.P.; Rodriguez Lopez, C.E.; Payne, R.M.E.; Serna Guerrero, D.A.; Gase, K.; Yamamoto, K.; Vaillancourt, B.; et al. Single-cell multi-omics in the medicinal plant Catharanthus roseus. Nat. Chem. Biol. 2023, 19, 1031–1041. [Google Scholar] [CrossRef] [PubMed]
- Gladman, N.; Goodwin, S.; Chougule, K.; Richard McCombie, W.; Ware, D. Era of gapless plant genomes: Innovations in sequencing and mapping technologies revolutionize genomics and breeding. Curr. Opin. Biotechnol. 2023, 79, 102886. [Google Scholar] [CrossRef]
- Satam, H.; Joshi, K.; Mangrolia, U.; Waghoo, S.; Zaidi, G.; Rawool, S.; Thakare, R.P.; Banday, S.; Mishra, A.K.; Das, G.; et al. Next-Generation Sequencing Technology: Current Trends and Advancements. Biology 2023, 12, 997. [Google Scholar] [CrossRef]
- Wafula, E.K.; Zhang, H.; Von Kuster, G.; Leebens-Mack, J.H.; Honaas, L.A.; de Pamphilis, C.W. PlantTribes2: Tools for comparative gene family analysis in plant genomics. Front. Plant Sci. 2022, 13, 1011199. [Google Scholar] [CrossRef]
- Chen, C.; Chen, X.; Han, J.; Lu, W.; Ren, Z. Genome-wide analysis of the WRKY gene family in the cucumber genome and transcriptome-wide identification of WRKY transcription factors that respond to biotic and abiotic stresses. BMC Plant Biol. 2020, 20, 443. [Google Scholar] [CrossRef]
- Ayaz, A.; Saqib, S.; Huang, H.; Zaman, W.; Lü, S.; Zhao, H. Genome-wide comparative analysis of long-chain acyl-CoA synthetases (LACSs) gene family: A focus on identification, evolution and expression profiling related to lipid synthesis. Plant Physiol. Biochem. 2021, 161, 1–11. [Google Scholar] [CrossRef]
- Chakraborty, P.; Biswas, A.; Dey, S.; Bhattacharjee, T.; Chakrabarty, S. Cytochrome P450 Gene Families: Role in Plant Secondary Metabolites Production and Plant Defense. J. Xenobiotics 2023, 13, 402–423. [Google Scholar] [CrossRef]
- Feng, J.-X.; Liu, D.; Pan, Y.; Gong, W.; Ma, L.-G.; Luo, J.-C.; Deng, X.W.; Zhu, Y.-X. An annotation update via cDNA sequence analysis and comprehensive profiling of developmental, hormonal or environmental responsiveness of the Arabidopsis AP2/EREBP transcription factor gene family. Plant Mol. Biol. 2005, 59, 853–868. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Gu, M.; Lai, Z.; Fan, B.; Shi, K.; Zhou, Y.H.; Yu, J.Q.; Chen, Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Meng, Z.; Zhang, H.; Chu, Y.; Qiu, Y.; Jin, B.; Wang, L. Identification and characterization of thirteen gene families involved in flavonoid biosynthesis in Ginkgo biloba. Ind. Crops Prod. 2022, 188, 115576. [Google Scholar] [CrossRef]
- Li, C.; Zhu, S.; Zhang, H.; Chen, L.; Cai, M.; Wang, J.; Chai, J.; Wu, F.; Cheng, Z.; Guo, X.; et al. OsLBD37 and OsLBD38, two class II type LBD proteins, are involved in the regulation of heading date by controlling the expression of Ehd1 in rice. Biochem. Biophys. Res. Commun. 2017, 486, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Luo, F.; Hochholdinger, F. LOB domain proteins: Beyond lateral organ boundaries. Trends Plant Sci. 2016, 21, 159–167. [Google Scholar] [CrossRef]
- Yordanov, Y.S.; Regan, S.; Busov, V. Members of the LATERAL ORGAN BOUNDARIES DOMAIN transcription factor family are involved in the regulation of secondary growth in Populus. Plant Cell 2010, 22, 3662–3677. [Google Scholar] [CrossRef]
- Cho, C.; Jeon, E.; Pandey, S.K.; Ha, S.H.; Kim, J. LBD13 positively regulates lateral root formation in Arabidopsis. Planta 2019, 249, 1251–1258. [Google Scholar] [CrossRef]
- Lee, H.W.; Kim, N.Y.; Lee, D.J.; Kim, J. LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol. 2009, 151, 1377–1389. [Google Scholar] [CrossRef]
- Shuai, B.; Reynaga-Pena, C.G.; Springer, P.S. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 2002, 129, 747–761. [Google Scholar] [CrossRef]
- Lee, H.W.; Kim, M.J.; Kim, N.Y.; Lee, S.H.; Kim, J. LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis. Plant J. 2013, 73, 212–224. [Google Scholar] [CrossRef]
- Xiong, J.; Feng, X.; Zhang, W.; Wang, X.; Hu, Y.; Zhang, X.; Wu, F.; Guo, W.; Xie, W.; Wang, Q. Class II LBD genes ZmLBD5 and ZmLBD33 regulate gibberellin and abscisic acid biosynthesis. bioRxiv 2021. [Google Scholar] [CrossRef]
- Rubin, G.; Tohge, T.; Matsuda, F.; Saito, K.; Scheible, W.R. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 2009, 21, 3567–3584. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Yu, J.; Ge, Y.; Qin, P.; Xu, L. Pivotal role of LBD1 6 in root and root-like organ initiation. Cell. Mol. Life Sci. 2018, 75, 3329–3338. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.-Y.; Liang, X.-Y.; Li, X.; Shen, P.-X.; Cao, X.-Y.; Chen, C.; Song, S.-H.; Wang, D.-H.; Wang, Z.-Z.; Zhang, Y. Genome-wide characterisation and expression profiling of the LBD family in Salvia miltiorrhiza reveals the function of LBD50 in jasmonate signaling and phenolic biosynthesis. Ind. Crops Prod. 2020, 144, 112006. [Google Scholar] [CrossRef]
- Zeng, J.; Wang, J.; Liu, X.; Qin, J.; Lan, X.; Chen, M.; Liao, Z. An auxin-responsive transcription factor AbLBD1 promotes the development of lateral roots and reduces the biosynthesis of tropane alkaloids in Atropa belladonna. Plant Cell Tissue Organ Cult. (PCTOC) 2020, 142, 559–569. [Google Scholar] [CrossRef]
- Feng, Z.; Zhu, J.; Du, X.; Cui, X. Effects of three auxin-inducible LBD members on lateral root formation in Arabidopsis thaliana. Planta 2012, 236, 1227–1237. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, F.; Guo, J.; Zhang, X.S. Rice OsAS2 gene, a member of LOB domain family, functions in the regulation of shoot differentiation and leaf development. J. Plant Biol. 2009, 52, 374–381. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, S.; Su, L.; Liu, X.; Hao, Y. A genome-wide analysis of the LBD (LATERAL ORGAN BOUNDARIES domain) gene family in Malus domestica with a functional characterization of MdLBD11. PLoS ONE 2013, 8, e57044. [Google Scholar] [CrossRef]
- Yu, F.; De Luca, V. ATP-binding cassette transporter controls leaf surface secretion of anticancer drug components in Catharanthus roseus. Proc. Natl. Acad. Sci. USA 2013, 110, 15830–15835. [Google Scholar] [CrossRef]
- van der Fits, L.; Memelink, J. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 2000, 289, 295–297. [Google Scholar] [CrossRef]
- Suttipanta, N.; Pattanaik, S.; Kulshrestha, M.; Patra, B.; Singh, S.K.; Yuan, L. The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 2011, 157, 2081–2093. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Hedhili, S.; Montiel, G.; Zhang, Y.; Chatel, G.; Pre, M.; Gantet, P.; Memelink, J. The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. Plant J. 2011, 67, 61–71. [Google Scholar] [CrossRef]
- Garcia-Hernandez, M.; Berardini, T.; Chen, G.; Crist, D.; Doyle, A.; Huala, E.; Knee, E.; Lambrecht, M.; Miller, N.; Mueller, L.A. TAIR: A resource for integrated Arabidopsis data. Funct. Integr. Genom. 2002, 2, 239–253. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Voorrips, R. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Chou, K.-C.; Shen, H.-B. Cell-PLoc 2.0: An improved package of web-servers for predicting subcellular localization of proteins in various organisms. Nat. Sci. 2010, 2, 1090. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Xu, M.; Wu, C.; Zhao, L.; Wang, Y.; Wang, C.; Zhou, W.; Ming, Y.; Kai, G. WRKY transcription factor OpWRKY1 acts as a negative regulator of camptothecin biosynthesis in Ophiorrhiza pumila hairy roots. Plant Cell Tissue Organ Cult. (PCTOC) 2020, 142, 69–78. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Bai, H.; Han, Y.; Yu, F. An ABCG-Type Transporter Facilitates ABA Influx and Regulates Camptothecin Biosynthesis in Camptotheca acuminata. Int. J. Mol. Sci. 2022, 23, 16120. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.; Liu, Z.; Wang, Y.; Tang, Z.; Yu, F. A bZIP transcription factor, CaLMF, mediated light-regulated camptothecin biosynthesis in Camptotheca acuminata. Tree Physiol. 2019, 39, 372–380. [Google Scholar] [CrossRef]
- Jiang, S.-Y.; González, J.M.; Ramachandran, S. Comparative genomic and transcriptomic analysis of tandemly and segmentally duplicated genes in rice. PLoS ONE 2013, 8, e63551. [Google Scholar] [CrossRef]
- Larsen, B.; Fuller, V.L.; Pollier, J.; Van Moerkercke, A.; Schweizer, F.; Payne, R.; Colinas, M.; O’Connor, S.E.; Goossens, A.; Halkier, B.A. Identification of iridoid glucoside transporters in Catharanthus roseus. Plant Cell Physiol. 2017, 58, 1507–1518. [Google Scholar] [CrossRef]
- Chalfun-Junior, A.; Franken, J.; Mes, J.J.; Marsch-Martinez, N.; Pereira, A.; Angenent, G.C. ASYMMETRIC LEAVES2-LIKE1 gene, a member of the AS2/LOB family, controls proximal–distal patterning in Arabidopsis petals. Plant Mol. Biol. 2005, 57, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Ha, C.M.; Jun, J.H.; Nam, H.G.; Fletcher, J.C. BLADE-ON-PETIOLE1 and 2 control Arabidopsis lateral organ fate through regulation of LOB domain and adaxial-abaxial polarity genes. Plant Cell 2007, 19, 1809–1825. [Google Scholar] [CrossRef]
- Dang, T.V.T.; Lee, S.; Cho, H.; Choi, K.; Hwang, I. The LBD11-ROS feedback regulatory loop modulates vascular cambium proliferation and secondary growth in Arabidopsis. Mol. Plant 2023, 16, 1131–1145. [Google Scholar] [CrossRef]
- Ye, L.; Wang, X.; Lyu, M.; Siligato, R.; Eswaran, G.; Vainio, L.; Blomster, T.; Zhang, J.; Mähönen, A.P. Cytokinins initiate secondary growth in the Arabidopsis root through a set of LBD genes. Curr. Biol. 2021, 31, 3365–3373.e7. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, D.; Xie, L.; Zhou, T.; Zhao, J.; Zhang, Q.; Yang, M.; Wu, W.; Lian, X. Rice transcription factors OsLBD37/38/39 regulate nitrate uptake by repressing OsNRT2. 1/2.2/2.3 under high-nitrogen conditions. Crop J. 2022, 10, 1623–1632. [Google Scholar] [CrossRef]
- Salim, V.; Yu, F.; Altarejos, J.; De Luca, V. Virus-induced gene silencing identifies Catharanthus roseus 7-deoxyloganic acid-7-hydroxylase, a step in iridoid and monoterpene indole alkaloid biosynthesis. Plant J. 2013, 76, 754–765. [Google Scholar] [CrossRef] [PubMed]
- Menke, F.L.; Champion, A.; Kijne, J.W.; Memelink, J. A novel jasmonate- and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate- and elicitor-inducible AP2-domain transcription factor, ORCA2. EMBO J. 1999, 18, 4455–4463. [Google Scholar] [CrossRef]
- Van Der Fits, L.; Memelink, J. The jasmonate-inducible AP2/ERF-domain transcription factor ORCA3 activates gene expression via interaction with a jasmonate-responsive promoter element. Plant J. 2001, 25, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Wang, C.; Xiong, Z.; Wang, H.; Fu, X.; Shen, Q.; Peng, B.; Ma, Y.; Sun, X.; Tang, K. CrERF5, an AP2/ERF Transcription Factor, Positively Regulates the Biosynthesis of Bisindole Alkaloids and Their Precursors in Catharanthus roseus. Front. Plant Sci. 2019, 10, 931. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Gao, F.; Ren, J.; Lu, X.; Ren, G.; Wang, R. A Novel AP2/ERF Transcription Factor CR1 Regulates the Accumulation of Vindoline and Serpentine in Catharanthus roseus. Front. Plant Sci. 2017, 8, 2082. [Google Scholar] [CrossRef]
- Huang, B.; Huang, Z.; Ma, R.; Ramakrishnan, M.; Chen, J.; Zhang, Z.; Yrjälä, K. Genome-wide identification and expression analysis of LBD transcription factor genes in Moso bamboo (Phyllostachys edulis). BMC Plant Biol. 2021, 21, 296. [Google Scholar] [CrossRef]
- Kim, M.; Kim, M.-J.; Pandey, S.; Kim, J. Expression and protein interaction analyses reveal combinatorial interactions of LBD transcription factors during Arabidopsis pollen development. Plant Cell Physiol. 2016, 57, 2291–2299. [Google Scholar] [CrossRef]
- Xiong, J.; Zhang, W.; Zheng, D.; Xiong, H.; Feng, X.; Zhang, X.; Wang, Q.; Wu, F.; Xu, J.; Lu, Y. ZmLBD5 increases drought sensitivity by suppressing ROS accumulation in arabidopsis. Plants 2022, 11, 1382. [Google Scholar] [CrossRef]
- Burlat, V.; Oudin, A.; Courtois, M.; Rideau, M.; St-Pierre, B. Co-expression of three MEP pathway genes and geraniol 10-hydroxylase in internal phloem parenchyma of Catharanthus roseus implicates multicellular translocation of intermediates during the biosynthesis of monoterpene indole alkaloids and isoprenoid-derived primary metabolites. Plant J. 2004, 38, 131–141. [Google Scholar]
- Guirimand, G.; Guihur, A.; Poutrain, P.; Héricourt, F.; Mahroug, S.; St-Pierre, B.; Burlat, V.; Courdavault, V. Spatial organization of the vindoline biosynthetic pathway in Catharanthus roseus. J. Plant Physiol. 2011, 168, 549–557. [Google Scholar] [CrossRef]
- Miettinen, K.; Dong, L.; Navrot, N.; Schneider, T.; Burlat, V.; Pollier, J.; Woittiez, L.; Van Der Krol, S.; Lugan, R.; Ilc, T. The seco-iridoid pathway from Catharanthus roseus. Nat. Commun. 2014, 5, 3606. [Google Scholar] [CrossRef]
- Stavrinides, A.; Tatsis, E.C.; Foureau, E.; Caputi, L.; Kellner, F.; Courdavault, V.; O’Connor, S.E. Unlocking the diversity of alkaloids in Catharanthus roseus: Nuclear localization suggests metabolic channeling in secondary metabolism. Chem. Biol. 2015, 22, 336–341. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, C.; Yang, B.; Guo, X.; Gao, C.; Wang, B.; Zhao, X.; Tang, Z. Genome-Wide Survey of the Potential Function of CrLBDs in Catharanthus roseus MIA Biosynthesis. Genes 2024, 15, 1140. https://doi.org/10.3390/genes15091140
Chang C, Yang B, Guo X, Gao C, Wang B, Zhao X, Tang Z. Genome-Wide Survey of the Potential Function of CrLBDs in Catharanthus roseus MIA Biosynthesis. Genes. 2024; 15(9):1140. https://doi.org/10.3390/genes15091140
Chicago/Turabian StyleChang, Chunhao, Bingrun Yang, Xiaorui Guo, Chunyan Gao, Biying Wang, Xiaoju Zhao, and Zhonghua Tang. 2024. "Genome-Wide Survey of the Potential Function of CrLBDs in Catharanthus roseus MIA Biosynthesis" Genes 15, no. 9: 1140. https://doi.org/10.3390/genes15091140
APA StyleChang, C., Yang, B., Guo, X., Gao, C., Wang, B., Zhao, X., & Tang, Z. (2024). Genome-Wide Survey of the Potential Function of CrLBDs in Catharanthus roseus MIA Biosynthesis. Genes, 15(9), 1140. https://doi.org/10.3390/genes15091140






