Antioxidant Enzyme, Transcriptomic, and Metabolomic Changes in Lily (Lilium spp.) Leaves Induced by Aphis gossypii Glover
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Growth and Insect Colony
2.2. Insect Bioassays
2.3. Antioxidant Enzyme Activity Measurements
2.4. Metabolomic Analyses
2.5. Transcriptome Sequencing and Data Analysis
2.6. Validation of Genes by Quantitative Real-Time PCR
2.7. Integrated Metabolome and Transcriptome Analyses
3. Results
3.1. Effects of A. gossypii Infection on the Antioxidant Enzyme Activities of Lily
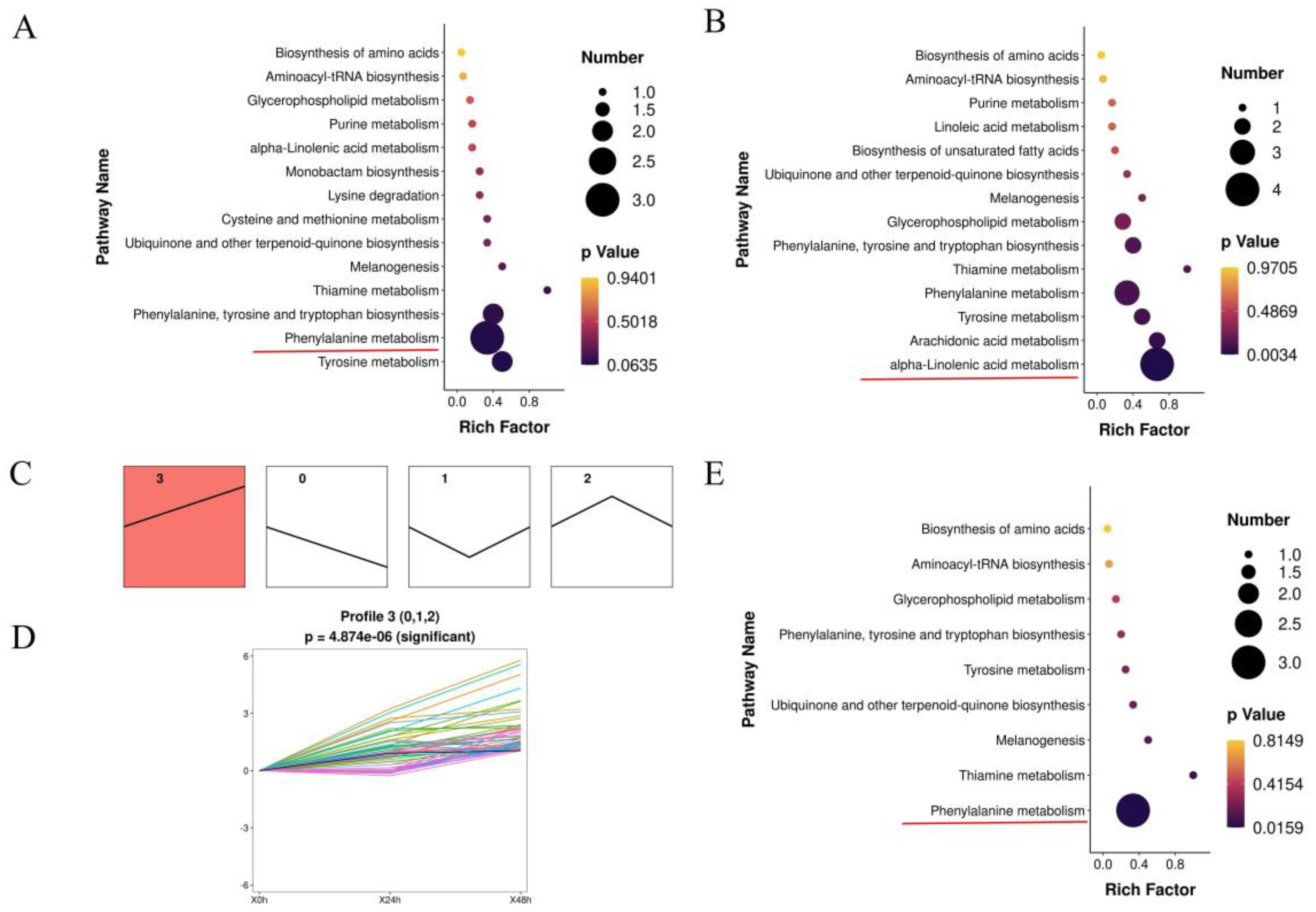
3.2. Summary of the Metabolomics of Lily under A. gossypii Infection
3.3. Transcriptomic Analysis of Lily Responses to A. gossypii Feeding
3.4. Integrated Analysis of Genes and Metabolites in Lily during A. gossypii Infection
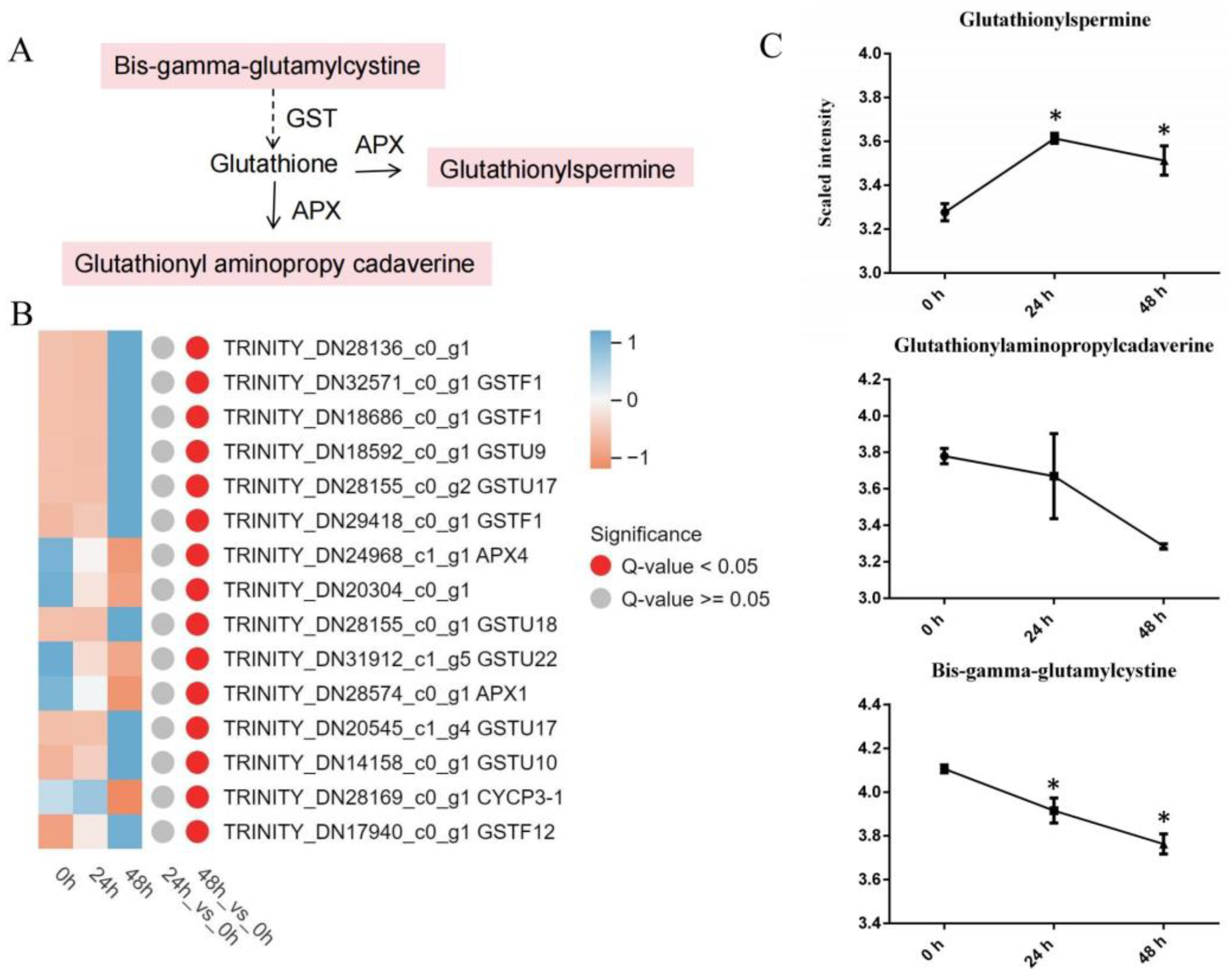
3.4.1. Glutathione Metabolism
3.4.2. Phenylalanine Metabolism
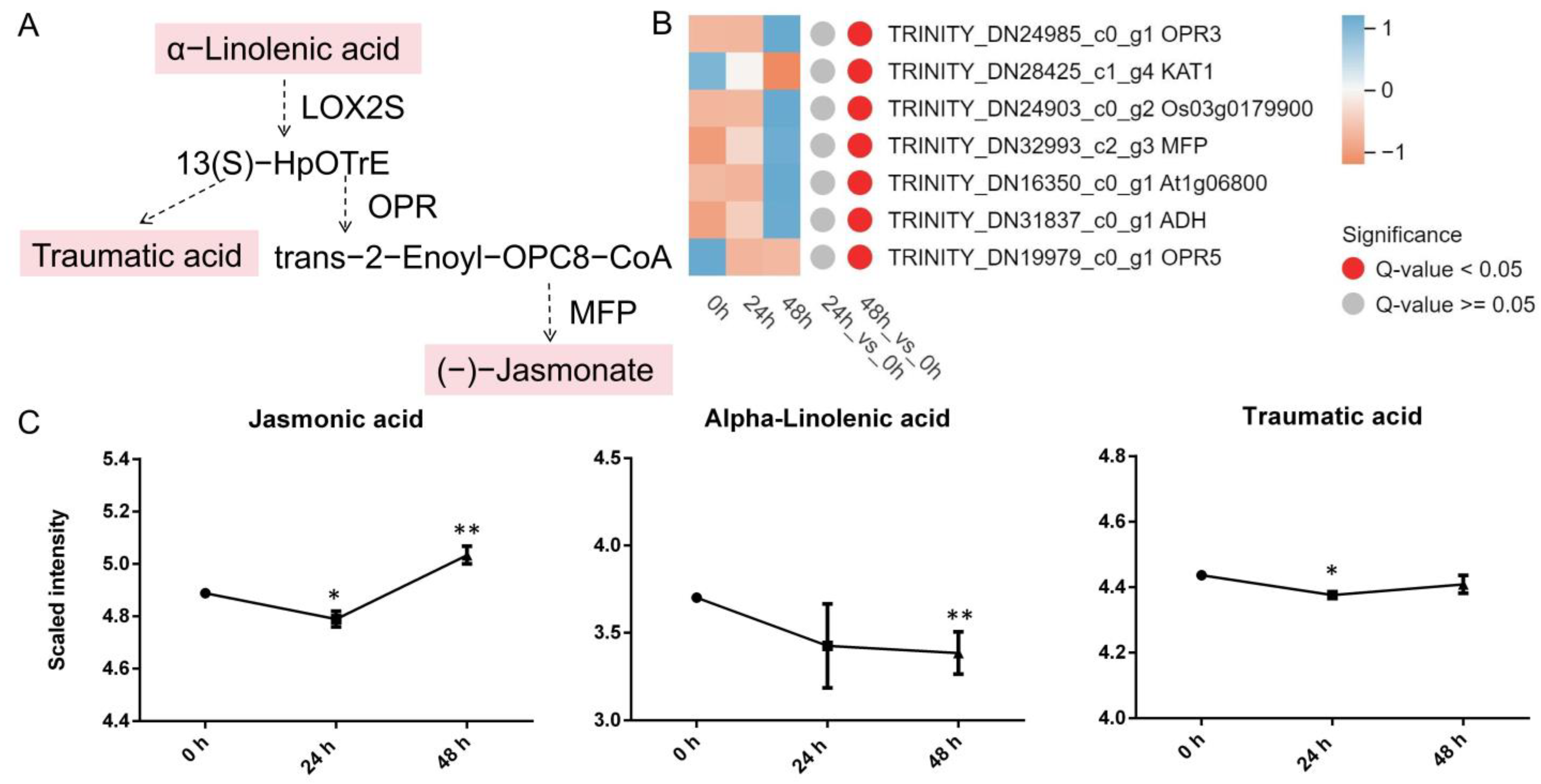
3.4.3. α-Linolenic Acid Metabolism
4. Discussion
4.1. Antioxidant Reactions in Lily after Cotton Aphid Infection
4.2. Phenylalanine Metabolism and Flavonoid Biosynthesis Pathways Involved in the Response to Aphid Stress
4.3. α-Linolenic Acid Metabolism Participates in the Stress Response
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carletto, J.; Martin, T.; Vanlerberghe-Msautti, F.; Brevaultc, T. Insecticide resistance traits differ among and within host races in Aphis gossypii. Pest Manag. Sci. 2010, 66, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Razmjou, J.; Vorburger, C.; Mohammadi, M.; Hassanpour, M. Influence of vermicompost and cucumber cultivar on population growth of Aphis gossypii glover. J. Appl. Entomol. 2012, 136, 568–575. [Google Scholar] [CrossRef]
- Furch, A.C.U.; van Bel, A.J.E.; Will, T. Aphid salivary proteases are capable of degrading sieve-tube proteins. J. Exp. Bot. 2015, 66, 533–539. [Google Scholar] [CrossRef]
- Sugio, A.; Dubreuil, G.; Giron, D.; Simon, J.-C. Plant-insect interactions under bacterial influence: Ecological implications and underlying mechanisms. J. Exp. Bot. 2015, 66, 467–478. [Google Scholar] [CrossRef]
- Whitfield, A.E.; Falk, B.W.; Rotenberg, D. Insect vector-mediated transmission of plant viruses. Virology 2015, 479, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Blackman, R.L.; Eastop, V.F. Aphids on the World’s Crops: An Identification and Information Guide, 2nd ed.; John Wiley & Sons: New Yorks, NY, USA, 2000. [Google Scholar]
- Shi, H.; Zhong, J.; Liang, Y.; Zhang, P.; Guo, L.; Wang, C.; Tang, Y.; Lu, Y.; Sun, M. Aphid resistance evaluation and constitutive resistance analysis of eighteen lilies. Insects 2023, 14, 936. [Google Scholar] [CrossRef]
- Shi, X.B.; Yan, S.; Zhang, C.; Zheng, L.M.; Zhang, Z.H.; Sun, S.E.; Gao, Y.; Tan, X.Q.; Zhang, D.Y.; Zhou, X.G. Aphid endosymbiont facilitates virus transmission by modulating the volatile profile of host plants. BMC Plant Biol. 2021, 21, 67. [Google Scholar] [CrossRef] [PubMed]
- Barakat, D.A.; Ibrahim, E.S.; Salama, M.R. Effectiveness and persistence of some synthetic insecticides and their nanoformulation against whitefly (Bemisia tabaci) and aphids (Aphis craccivora) on fennel plants and soil. Egypt. J. Chem. 2023, 66, 235–246. [Google Scholar]
- Sabater-Muñoz, B.; Legeai, F.; Rispe, C.; Bonhomme, J.; Dearden, P.; Dossat, C.; Duclert, A.; Gauthier, J.; Ducray, D.G.; Hunter, W.; et al. Large-scale gene discovery in the pea aphid Acyrthosiphon pisum (Hemiptera). Genome Biol. 2006, 7, R21. [Google Scholar] [CrossRef]
- Herron, G.A.; Wilson, L.J. Neonicotinoid resistance in Aphis gossypii Glover (Aphididae: Hemiptera) from Australian cotton. Aust. J. Entomol. 2011, 50, 93–98. [Google Scholar] [CrossRef]
- Kettles, G.J.; Drurey, C.; Schoonbeek, H.J.; Maule, A.J.; Hogenhout, S.A. Resistance of Arabidopsis thaliana to the green peach aphid, Myzus persicae, involves camalexin and is regulated by microRNAs. New Phytol. 2013, 198, 1178–1190. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Wang, G.; Huang, S.; Ma, Y.; Xia, L. Engineering plants for aphid resistance: Current status and future perspectives. Theor. Appl. Genet. 2014, 127, 2065–2083. [Google Scholar] [CrossRef] [PubMed]
- Erb, M.; Reymond, P. Molecular interactions between plants and insect herbivores. Annu. Rev. Plant Biol. 2019, 70, 527–557. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.M.; Zhang, Y.L. Plant immunity: Danger perception and signaling. Cell 2020, 181, 978–989. [Google Scholar] [CrossRef]
- Ngou, B.P.M.; Ahn, H.K.; Ding, P.T.; Jones, J.D.G. Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 2021, 592, 110–115. [Google Scholar] [CrossRef]
- Kessler, A.; Baldwin, I.T. Plant responses to insect herbivory: The emerging molecular analysis. Annu. Rev. Plant Biol. 2002, 53, 299–328. [Google Scholar] [CrossRef]
- Schuman, M.C.; Baldwin, I.T. The layers of plant responses to insect herbivores. Annu. Rev. Entomol. 2016, 61, 373–394. [Google Scholar] [CrossRef] [PubMed]
- Kerchev, P.I.; Fenton, B.; Foyer, C.H.; Hancock, R.D. Plant responses to insect herbivory: Interactions between photosynthsis, reactive oxygen species and hormonal signalling pathways. Plant Cell Environ. 2012, 35, 441–453. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Wang, W.; Karapetyan, S.; Mwimba, M.; Marques, J.; Buchler, N.E.; Dong, X. Redox rhythm reinforces the circadian clock to gate immune response. Nature 2015, 523, 472–476. [Google Scholar] [CrossRef]
- Penninckx, M.J. An overview on glutathione in Saccharomyces versus non-conventional yeasts. FEMS Yeast Res. 2002, 2, 295–305. [Google Scholar]
- Łukasik, I.; Wołoch, A.; Sytykiewicz, H.; Sprawka, I.; Goławska, S. Changes in the content of thiol compounds and the activity of glutathione s-transferase in maize seedlings in response to a rose-grass aphid infestation. PLoS ONE 2019, 14, e0221160. [Google Scholar] [CrossRef]
- Karban, R.; Baldwin, I.T. Induced Responses to Herbivory; The University of Chicago Press: Chicago, IL, USA, 1997. [Google Scholar]
- Åhman, I.; Kim, S.Y.; Zhu, L.H. Plant genes benefitting aphids-potential for exploitation in resistance breeding. Front. Plant Sci. 2019, 10, 1452. [Google Scholar] [CrossRef]
- Coppola, M.; Diretto, G.; Digilio, M.C.; Woo, S.L.; Giuliano, G.; Molisso, D.; Pennacchio, F.; Lorito, M.; Rao, R. Transcriptome and metabolome reprogramming in tomato plants by trichoderma harzianum stain T22 primes and enhances defense responses against aphids. Front. Physiol. 2019, 10, 745. [Google Scholar] [CrossRef]
- Thompson, G.A.; Goggin, F.L. Transcriptomics and functional genomics of plant defence induction by phloem-feeding insects. J. Exp. Bot. 2006, 57, 755–766. [Google Scholar] [CrossRef]
- Couldridge, C.; Newbury, H.J.; Ford-Lloyd, B.; Bale, J.; Pritchard, J. Exploring plant responses to aphid feeding using a full Arabidopsis microarray reveals a small number of genes with significantly altered expression. Bull. Ent. Res. 2007, 97, 523–532. [Google Scholar] [CrossRef]
- Coppola, V.; Coppola, M.; Rocco, M.; Digilio, M.C.; D’Ambrosio, C.; Renzone, G.; Martinelli, R.; Scaloni, A.; Pennacchio, F.; Rao, R.; et al. Transcriptomic and proteomic analysis of a compatible tomato-aphid interaction reveals a predominant salicylic acid-dependent plant response. BMC Genom. 2013, 14, 515. [Google Scholar] [CrossRef]
- Appel, H.M.; Fescemyer, H.; Ehlting, J.; Weston, D.; Rehrig, E.; Joshi, T.; Xu, D.; Bohlmann, J.; Schultz, J. Transcriptional responses of Arabidopsis thaliana to chewing and sucking insect herbivores. Front. Plant Sci. 2014, 5, 565. [Google Scholar] [CrossRef]
- Xia, X.; Shao, Y.; Jiang, J.; Ren, L.; Chen, F.; Fang, W.; Guan, Z.; Chen, S. Gene expression profiles responses to aphid feeding in chrysanthemum (Chrysanthemum morifolium). BMC Genom. 2014, 15, 1050. [Google Scholar] [CrossRef]
- Züst, T.; Agrawal, A.A. Mechanisms and evolution of plant resistance to aphids. Nat. Plants 2016, 2, 15206. [Google Scholar] [CrossRef]
- Batyrshina, Z.S.; Yaakov, B.; Shavit, R.; Singh, A.; Tzin, V. Comparative transcriptomic and metabolic analysis of wild and domesticated wheat genotypes reveals differences in chemical and physical defense responses against aphids. BMC Plant Biol. 2020, 20, 19. [Google Scholar] [CrossRef]
- Zhou, L.H.; Shi, H.; Yang, Y.D.; Cui, Y.H.; Yang, P.P.; Xu, L.F.; Ming, J. Identification of resistance to Aphis gossypii Glover and genetic diversity analysis of lily resources. J. Northeast. Agric. Univ. 2021, 52, 24–33. [Google Scholar]
- Liu, X.T.; Liu, H.F.; Tian, B.M.; Shi, G.Y.; Liu, C.; Guo, J.L.; Cao, G.Q.; Wei, F. Metabolome and transcriptome analyses of anthocyanin biosynthesis reveal key metabolites and candidate genes in purple wheat (Triticum aestivum L.). Physiol. Plant. 2023, 175, e13921. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using diamond. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 2008, 5, 621. [Google Scholar] [CrossRef]
- Robinson, M.D.; Mccarthy, D.J.; Smyth, G.K. EdgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Ernst, J.; Bar-Joseph, Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinform. 2006, 7, 191. [Google Scholar] [CrossRef]
- Tan, Y.; Bai, L.; Xiao, L.; Wei, S.; Zhao, H. Herbivore stress by Lygus lucorum inducing protective enzyme activity and MDA content on different cotton varieties. Cotton Sci. 2010, 22, 479–485. [Google Scholar]
- Rani, P.U.; Pratyusha, S. Defensive role of Gossypium hirsutum L. anti-oxidative enzymes and phenolic acids in response to Spodoptera litura F. feeding. J. Asia-Pac. Entomol. 2013, 16, 131–136. [Google Scholar] [CrossRef]
- Gao, Y.; Men, X.; Yu, Y.; Zhou, H. Changes of physiological indexes of jujube, peach, cherry and grape leaves damaged by Apolygus lucorum in Northern China. Sci. Agric. Sin. 2012, 45, 4627–4634. [Google Scholar]
- Rasool, B.; Karpinska, B.; Pascual, J.; Kangasjärvi, S.; Foyer, C.H. Catalase, glutathione, and protein phosphatase 2A-dependent organellar redox signalling regulate aphid fecundity under moderate and high irradiance. Plant Cell Environ. 2020, 43, 209–222. [Google Scholar] [CrossRef]
- Sytykiewicz, H. Transcriptional reprogramming of genes related to ascorbate and glutathione biosynthesis, turnover and translocation in aphid-challenged maize seedlings. Biochem. Syst. Ecol. 2016, 69, 236–251. [Google Scholar] [CrossRef]
- Moran, P.J.; Cheng, Y.; Cassell, J.L.; Thompson, G.A. Gene expression profiling of Arabidopsis thaliana in compatible plant-aphid interactions. Arch. Insect Biochem. Physiol. 2002, 51, 182–203. [Google Scholar] [CrossRef]
- Loschke, D.C.; Hadwiger, L.A.; Schroder, J.; Hahlbrock, K.; Loeshke, D.C. Effects of light and of Fusarium solani on synthesis and activity of phenylalanine ammonia-lyase in peas. Plant Physiol. 1981, 68, 680–685. [Google Scholar] [CrossRef]
- Orczyk, W.; Hipskind, J.; De Neergaard, E.; Goldsbrough, P.; Nicholson, R. Stimulation of phenylalanine ammonia-lyase in sorghum in response to inoculation with Bipolaris maydis. Physiol. Mol. Plant Pathol. 1996, 48, 55–64. [Google Scholar] [CrossRef]
- Giberti, S.; Bertea, C.M.; Narayana, R.; Maffei, M.E.; Forlani, G. Two phenylalanine ammonia lyase isoforms are involve in the elicitor-induced response of rice to the fugal pathogen Magnaporthe oryzae. J. Plant Physiol. 2012, 169, 249–254. [Google Scholar] [CrossRef]
- Cass, C.L.; Peraldi, A.; Dowd, P.F.; Mottiar, Y.; Santoro, N.; Karlen, S.D.; Bukhman, Y.V.; Foster, C.E.; Thrower, N.; Bruno, L.C.; et al. Effects of PHENYLALANINE AMMONIA LYASE (PAL) knockdown on cell wall composition, biomass digestibility, and biotic and abiotic stress responses in Brachypodium. J. Exp. Bot. 2015, 66, 4317–4335. [Google Scholar] [CrossRef]
- Pant, S.; Huang, Y. Genome-wide studies of PAL genes in sorghum and their responses to aphid infestation. Sci. Rep. 2022, 12, 22537. [Google Scholar] [CrossRef]
- Ferreyra, M.; Rius, S.P.; Casati, P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012, 3, 222. [Google Scholar]
- Walling, L.L. Avoiding effective defenses: Strategies employed by phloem-feeding insects. Plant Physiol. 2008, 146, 859–866. [Google Scholar] [CrossRef]
- Ateyyat, M.; Abu-Romman, S.; Abu-Darwish, M.; Ghabeinsh, I. Impact of flavonoids against woolly apple aphid, Eriosoma lanigerum (Hausmann) and its sole parasitoid, Aphelinus mali (Hald.). J. Agric Sci. 2012, 4, 227–236. [Google Scholar] [CrossRef]
- Kariyat, R.R.; Gaffoor, I.; Sattar, S.; Dixon, C.W.; Frock, N.; Moen, J.; De Moraes, C.M.; Mescher, M.C.; Thompson, G.A.; Chopra, S. Sorghum 3-Deoxyanthocyanidin flavonoids confer resistance against corn leaf aphid. J. Chem. Ecol. 2019, 45, 502–514. [Google Scholar] [CrossRef]
- War, A.R.; Sharma, S.P.; Sharma, H.C. Differential induction of flavonoids in groundnut in response to Helicoverpa armigera and Aphis craccivora infestation. Int. J. Insect Sci. 2016, 8, 55–64. [Google Scholar] [CrossRef]
- Stec, K.; Kordan, B.; Gabryś, B. Quercetin and rutin as modifiers of aphid probing behavior. Molecules 2021, 26, 3622. [Google Scholar] [CrossRef]
- Goławska, S.; Sprawka, I.; Lukasik, I.; Goławski, A. Are naringenin and quercetin useful chemicals in pest−management strategies? Pest Sci. 2014, 87, 173–180. [Google Scholar] [CrossRef]
- Morimoto, M.; Kumeda, S.; Komai, K. Insect antifeedant flavonoids from Gnaphalium affin. J. Agric. Food Chem. 2000, 48, 1888–1891. [Google Scholar] [CrossRef]
- Simmonds, M.S.J. Flavonoid-insect interactions: Recent advances in our knowledge. Phytochemistry 2003, 6, 21–30. [Google Scholar] [CrossRef]
- Czerniewicz, P.; Sytykiewicz, H.; Durak, R.; Borowiak-Sobkowiak, B.; Chrzanowski, G. Role of phenolic compounds during antioxidative responses of winter triticale to aphid and beetle attack. Plant Physilo. Biochem. 2017, 118, 529–540. [Google Scholar] [CrossRef]
- Farmer, E.E.; Ryan, C.A. Interplant communication: Airborne methyl jasmonate induces synthesis of proteinase inhibitors in plant leaves. Proc. Natl. Acad. Sci. USA 1990, 87, 7713–7716. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.G.; Zhang, X.D. Progress of jasmonates and its signal transduction pathway. Biotechnol. Bull. 2009, 2, 43–49. [Google Scholar]
- Lu, C.C. Mechanism Research on OsOPR7 Alternative Splicing Involved in the Interaction between Rice and Xanthomonas oryzae pv. oryzicola. Ph.D. Thesis, Shangdong Agricultural University, Taian, China, 2020. [Google Scholar]
- Mou, Y.F. Identification and Functional Analysis of Wheat OPR and LOX Gene Families in Response to Stresses. Ph.D. Thesis, Northwest A. and F. University, Yangling, China, 2019. [Google Scholar]
- Stintzi, A.; Browse, J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reducase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 2000, 97, 10625–10630. [Google Scholar] [CrossRef]
- Guo, H.M.; Li, H.C.; Zhou, S.R.; Xue, H.W.; Miao, X.X. Cis−12-oxo-phytodienoic acid stimulates rice defense response to a piercing-sucking insect. Mol. Plant 2014, 7, 1683–1692. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, L.; Wang, E.; Yang, Y.; Yang, P.; Xu, L.; Ming, J. Antioxidant Enzyme, Transcriptomic, and Metabolomic Changes in Lily (Lilium spp.) Leaves Induced by Aphis gossypii Glover. Genes 2024, 15, 1124. https://doi.org/10.3390/genes15091124
Zhou L, Wang E, Yang Y, Yang P, Xu L, Ming J. Antioxidant Enzyme, Transcriptomic, and Metabolomic Changes in Lily (Lilium spp.) Leaves Induced by Aphis gossypii Glover. Genes. 2024; 15(9):1124. https://doi.org/10.3390/genes15091124
Chicago/Turabian StyleZhou, Lihong, Erli Wang, Yingdong Yang, Panpan Yang, Leifeng Xu, and Jun Ming. 2024. "Antioxidant Enzyme, Transcriptomic, and Metabolomic Changes in Lily (Lilium spp.) Leaves Induced by Aphis gossypii Glover" Genes 15, no. 9: 1124. https://doi.org/10.3390/genes15091124
APA StyleZhou, L., Wang, E., Yang, Y., Yang, P., Xu, L., & Ming, J. (2024). Antioxidant Enzyme, Transcriptomic, and Metabolomic Changes in Lily (Lilium spp.) Leaves Induced by Aphis gossypii Glover. Genes, 15(9), 1124. https://doi.org/10.3390/genes15091124





